Submitted:
08 January 2024
Posted:
09 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
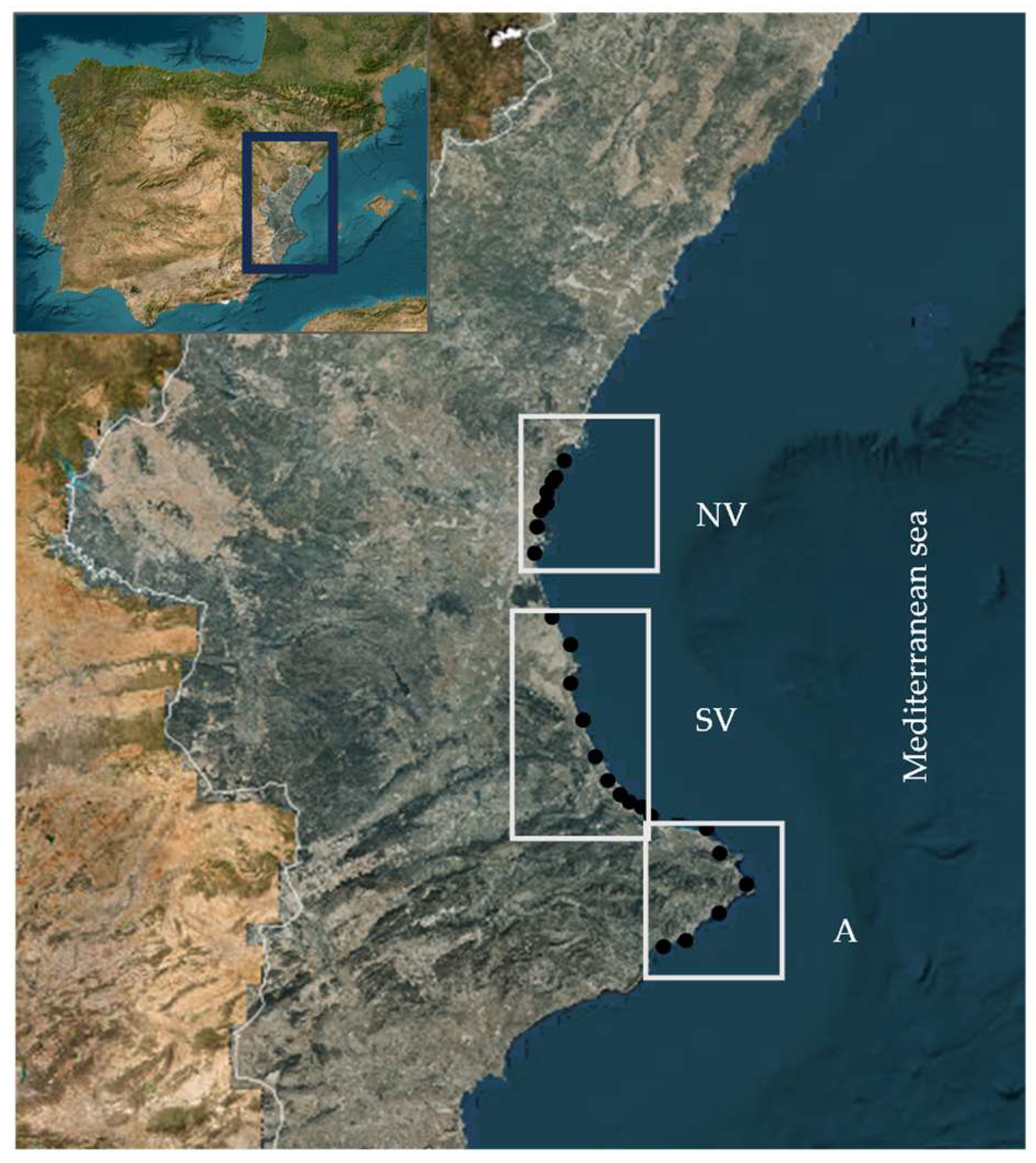
2.1. Samples
2.2. Bacterial analysis
2.3. Determination of antibiotic resistance
2.4. Detection of ARG
3. Results
3.1. Microbiological determination of E. coli and Enterococcus sp. in marine water samples
3.2. Antibiotic resistance determination of bacterial isolates
3.3. Antibiotic resistant gene determination in water samples and bacterial isolates
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef]
- Waseem, H.; Williams, M.R.; Jameel, S.; Hashsham, S.A. Antimicrobial Resistance in the Environment. Water Environ. Res. 2018, 90, 865–884. [Google Scholar] [CrossRef]
- Bondarczuk, K.; Piotrowska-Seget, Z. Microbial diversity and antibiotic resistance in a final effluent-receiving lake. Sci.Total Environ. 2019, 650, 2951–2961. [Google Scholar] [CrossRef]
- Gibson, MK: Forsberg, K.J.; Dantas, G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015, 9, 207–216. [CrossRef]
- Martínez, J.L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321, 365–367. [Google Scholar] [CrossRef]
- Alduina, R. Antibiotics and environment. Antibiotics 2020, 9, 202. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E. and Kaur, P. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.; Gaze, W.; Pruden, A.; Samalla, K.; Tiedje, J.M.; Zhu, Y-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Cycón, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the soil environment—Degradation and their impact on microbial activity and diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef]
- Ahmad, N.; Joji, R.M.; Shahid, M. Evolution and implementation of One Health to control the dissemination of antibiotic-resistant bacteria and resistance genes: A review. Front. Cell Infect. Microbiol. 2023, 12, 1065796. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, J.; Li, Z.; Guo, S.; Li, K.; Xu, P.; Ok, Y.S.; Jones, D.L.; Zou, J. Antibiotics and antibiotic resistance genes in agricultural soils: A systematic analysis. Crit. Rev. Environ. Sci. Technol. 2023, 53, 847–864. [Google Scholar] [CrossRef]
- Jiménez-Belenguer, A.I.; Ferrús, M.A.; Hernández, M.; García-Hernández, J.; Moreno, Y.; Castillo, M.A. Prevalence and characterization of beta-lactam and carbapenem-resistant bacteria isolated from organic fresh produce retailed in Eastern Spain. Antibiotics 2023, 12, 387. [Google Scholar] [CrossRef]
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: a need for quantitative risk assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai, China. J. Hazard. Mater. 2012, 235, 178–185. [Google Scholar] [CrossRef]
- Gwenzi,W.; Musiyiwa, K.; Mangori, L. Sources, behaviour and health risks of antimicrobial resistance genes in wastewaters: a hotspot reservoir. J. Environ. Chem. Eng. 2020, 8, 102220. [CrossRef]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.; Powell, J.; O’Connell, N.H.; Murphy, L.; Dunne, C.P. Antimicrobial resistance is prevalent in E. coli and other Enterobacterales isolated from public and private drinking water supplies in the Republic of Ireland. Microorganisms 2023, 11, 1224. [Google Scholar] [CrossRef] [PubMed]
- Odonkor, S.T.; Addo, K.K. Prevalence of multidrug-resistant Escherichia coli isolated from drinking water sources. Int. J. Microbiol. 2018, 2018, 7204013. [Google Scholar] [CrossRef]
- Fernando, D.M.; Tun, H.M.; Poole, J.; Patidar, R.; Li, R.; Mi, R.; Amarawansha, G.E.A.; Fernando, W.G.D.; Khafipour, E.; Farenhorst, A.; Kumar, A. Detection of antibiotic resistance genes in source and drinking water samples from a First Nations community in Canada. Appl. Environ. Microbiol. 2016, 82, 4767–4775. [Google Scholar] [CrossRef]
- Coleman, B.L.; Salvadori, M.I.; McGeer, A.J.; Sibley, K.A.; Neumann, N.F.; Bondy, S. J. , Gutmanis, I.A.; McEwen, S.A.; Lavoie, M.; Strong, D.; Johnson, I.; Jamieson, F.B.; Louie, M. The role of drinking water in the transmission of antimicrobial-resistant E. coli. Epidemiol. Infect. 2012, 140, 633–642. [Google Scholar] [CrossRef]
- Wang, R-N.; Zhang, Y.; Cao, Z-H.; Wang, X-Y.; Ma, B.; Wu, W-B.; Hu, N.; Huo, Z-Y.; Yuan, Q-B. Occurrence of super antibiotic resistance genes in the downstream of the Yangtze River in China: prevalence and antibiotic resistance profiles. Sci. Total Environ. 2019, 651, 1946–1957. [CrossRef]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sanchez-Melsio, A.; Borrego, C.M.; Barcelo, D.; Balcazar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef]
- Stoll, C.; Sidhu, J.P.S.; Tiehm, A.; Toze, S. Prevalence of clinically relevant antibiotic resistance genes in surface water samples collected from Germany and Australia. Environ. Sci. Technol. 2012, 46, 9716–9726. [Google Scholar] [CrossRef] [PubMed]
- Amato, M.; Dasí, D.; González, A.; Ferrús, M.A.; Castillo, M.A. Occurrence of antibiotic resistant bacteria and resistance genes in agricultural irrigation waters from Valencia city (Spain). Agric. Water Manag. 2021, 256, 107097. [Google Scholar] [CrossRef]
- Araújo, S.; Silva, I.A.T.; Tacao, M.; Patinha, C.; Alves, A.; Henriques, I. Characterization of antibiotic resistant and pathogenic Escherichia coli in irrigation water and vegetables in household farms. Int. J. Food Microbiol. 2017, 257, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Amos, G. C. A.; Hawkey, P. M.; Gaze, W. H.; Wellington, E. M. Wastewater effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob. Chemother. 2014, 69, 1785–1791. [Google Scholar] [CrossRef]
- Yuan, X.; Lv, Z.; Zhang, Z.; Han, Y.; Liu, Z.; Zhang, H. A review of antibiotics, antibiotic resistant bacteria, and resistance genes in aquaculture: occurrence, contamination, and transmission. Toxics 2023, 11, 420. [Google Scholar] [CrossRef] [PubMed]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- Su, Z.; Li, A.; Chen, J.; Huang, B.; Mu, Q.; Chen, L.; Wen, D. Wastewater discharge drives ARGs spread in the coastal area: a case study in Hangzhou Bay, China. Mar. Pollut. Bull. 2020, 151, 110856. [Google Scholar] [CrossRef] [PubMed]
- Carney, R.L.; Labbate, M.; Siboni, N.; Tagg, K.A.; Mitrovic, S.M.; Seymour, J.R. Urban beaches are environmental hotspots for antibiotic resistance following rainfall. Water Res. 2019, 167, 11508. [Google Scholar] [CrossRef]
- Leonard, A.F.; Zhang, L.; Balfour, A.J.; Garside, R.; Gaze, W.H. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ. Int. 2015, 82, 92–100. [Google Scholar] [CrossRef]
- Makkaew, P.; Kongprajug, A.; Chyerochana, N.; Sresung, M.; Precha, N.; Mongkolsuk, S.; Sirikanchana, K. Persisting antibiotic resistance gene pollution and its association with human sewage sources in tropical marine beach waters. Int. J. Hyg. Environ. Health 2021, 238, 113859. [Google Scholar] [CrossRef]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ Sci Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef]
- World Health Organization. Global Action Plan on Antimicrobial Resistance. 2015 http://apps.who.int/iris/bitstream/10665/193736/1/9789241509763eng.pdf?ua=1.
- Singer, A.C.; Shaw, H.; Rhodes, V.; Hart, A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 2016, 7, 1–22. [Google Scholar] [CrossRef]
- Zheng, D.; Yin, G.; Liu, M.; Chen, C.; Jiang, Y.; Hou, L.; Zheng, Y. A systematic review of antibiotics and antibiotic resistance genes in estuarine and coastal environments. Sci. Total Environ. 2021, 777, 146009. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. In Twenty-Fifth Informational Supplement; CLSI DocumentM100-S25; Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2015. [Google Scholar]
- Xi, C.; Zhang, Y.; Marrs, C.F.; Ye, W.; Simon, C.; Foxman, B.; Nriagu, J. Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl. Environ. Microbiol. 2009, 75, 5714–5718. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Z.T.; Michel, F.C., Jr.; Wittum, T.; Morrison, M. Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl. Environ. Microbiol. 2007, 73, 4407–4416. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Mozaz, S.; Chamorro, S.; Martí, E.; Huerta, B.; Gros, M.; Sánchez-Melsió, A.; Borrego, C.M.; Barceló, D.; Balcázar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Kim, S.C.; Carlson, K.H.; Pruden, A. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef]
- Aminov, R.I.; Garrigues-Jeanjean, N.; Mackie, R.I. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 2001, 67, 22–32. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; Paterson, D.L.; Rice, L.B.; Stelling, J.; Struelens, M.J.; Vatopoulos, A.; Weber, J.T.; Monnet, D.L. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Belding, C.; Boopathy, R. Presence of antibiotic-resistant bacteria and antibiotic resistance genes in coastal recreational waters of southeast Louisiana, USA. J. Water Supply Res. Technol. AQUA 2018, 67, 800–809. [Google Scholar] [CrossRef]
- Korajkic, A.; McMinn, B.R.; Harwood, V.J. Relationships between microbial indicators and pathogens in recreational water settings. Int. J. Environ. Res. Public Health 2018, 15, 2842. [Google Scholar] [CrossRef]
- Aragonés, L.; López, I.; Palazón, A.; López-Úbeda, R.; García, C. Evaluation of the quality of coastal bathing waters in Spain through fecal bacteria Escherichia coli and Enterococcus. Sci. Total Environ. 2016, 566–567, 288–297. [Google Scholar] [CrossRef]
- Ariza, E.; Jimenez, J.A.; Sarda, R.; Villares, M.; Pinto, J.; Fraguell, R.; Roca, E.; Marti, C.; Valdemoro, H.; Ballester, R. Proposal for an integral quality index for urban and urbanized beaches. Environ. Manag. 2010, 45, 998–1013. [Google Scholar] [CrossRef]
- McLellan, S.L. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 2004, 70, 4658–4665. [Google Scholar] [CrossRef]
- May, C.W.; Horner, R.R.; Karr, J.R.; Mar, B.W.; Welch, E.B. Effects of urbanization on small streams in the Puget Sound ecoregion. Watershed Prot. Tech. 1999, 2, 79–84. [Google Scholar]
- Winter, J.G.; Duthie, H.C. Effects of urbanization on water quality, periphyton and invertebrate communities in a southern Ontario stream. Can. Water Resour. J. 1998, 23, 245–257. [Google Scholar] [CrossRef]
- Abdelzaher, A.M.; Wright, M.E.; Ortega, C.; Solo-Gabriele, H.M.; Miller, G.; Elmir, S.; Newman, S.; Shih, P.; Bonilla, J.A.; Bonilla, T.D.; Palmer, C.J.; Scott, T.; Lukasik, J.; Harwood, V.J.; McQuaig, S.; Sinigalliano, C.; Gidley, M.; Plano, L.R.W.; Zhu, X.; Wang, J.D.; Fleming, L.E. Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Appl. Environ. Microbiol. 2010, 76, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Haugland, R.A.; Siefring, S.C.; Wymer, L.J.; Brenner, K.P.; Dufour, A.P. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 2005, 39, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Whitman, R.L.; Nevers, M.B. , Korinek, G.C.; Byappanahalli, M.N. Solar and temporal effects on Escherichia coli concentration at a Lake Michigan swimming beach. Appl. Environ. Microbiol. 2004, 70, 4276–4285. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.S.; Pereira, A.; Araújo, S.M.; Castro, B.B.; Correia, A.C.M.; Henriques, I. Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes. Front. Microbiol. 2014, 5, 426. [Google Scholar] [CrossRef]
- Bennani, M.; Amarouch, H.; Oubrim, N.; Cohen, N. Identification and antimicrobial resistance of fecal Enterococci isolated in coastal Mediterranean environments of Morocco. Eur J Sci Res. 2012, 70, 266–275. [Google Scholar]
- da Costa Andrade, V.; Zampieri Bdel, B.; Ballesteros, E.R.; Pinto, A.B.; de Oliveira, A.J. Densities and antimicrobial resistance of Escherichia coli isolated from marine waters and beach sands. Environ Monit Assess. 2015, 187, 342. [Google Scholar] [CrossRef]
- Na,G. ; Zhang, W.; Zhou, S.; Gao, H.; Lu,Z.; Wu, X.; Li, R.; Qiu, L.; Cai, Y.; Yao, Z. Sulfonamide antibiotics in the Northern Yellow Sea are related to resistant bacteria: Implications for antibiotic resistance genes. Mar. Poll. Bull. 2014, 84, 70–75. [Google Scholar] [CrossRef]
- Letchumanan, V.; Yin, W.F.; Lee, L.H.; Chan, K.G. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front Microbiol. 2015, 6, 33. [Google Scholar] [CrossRef]
- Şanlıtürk, G.; Güran, M. Monitoring of microbiological dynamics in beach sand and seawater samples from recreational and non-recreational beaches over a two-year period. Int. J. Environ. Health Res. 2022, 32, 1973–1985. [Google Scholar] [CrossRef]
- Hernández, F.; Calısto-Ulloa, N.; Gómez-Fuentes, C.; Gómez, M.; Ferrer, J.; González-Rocha, G.; Bello-Toledo, H.; Botero-Coy, A.M.; Boıx, C.; Ibáñez, M.; Montory, M. Occurrence of antibiotics and bacterial resistance in wastewater and sea water from the Antarctic. J. Hazard. Mater. 2019, 363, 447–456. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Antimicrobial resistance in the EU/EEA (EARS-Net) -Annual Epidemiological Report 2021. Stockholm: ECDC; 2022.
- Vignaroli, C.; Pasquaroli, S.; Citterio, B.; Di Cesare, A.; Mangiaterra, G.; Fattorini, D.; Biavasco, F. Antibiotic and heavy metal resistance in enterococci from coastal marine sediment. Environ Pollut. 2018, 237, 406–413. [Google Scholar] [CrossRef]
- Alibi, S.; Crespo, D.; Navas, J. Plant-derivatives small molecules with antibacterial activity. Antibiotics 2021, 10, 231. [Google Scholar] [CrossRef]
- Li, G.; Walker, M.J.; De Oliveira, D.M.P. Vancomycin resistance in Enterococcus and Staphylococcus aureus. Microorganisms 2022, 11, 24. [Google Scholar] [CrossRef]
- Vignaroli, C.; Luna, G.M.; Pasquaroli, S.; Di Cesare, A.; Petruzzella, R.; Paroncini, P.; Biavasco, F. Epidemic Escherichia coli ST131 and Enterococcus faecium ST17 in coastal marine sediments from an Italian beach. Environ. Sci. Technol. 2013, 47, 13772–13780. [Google Scholar] [CrossRef]
- Di Cesare, A.; Luna, G.M.; Vignaroli, C.; Pasquaroli, S.; Tota, S.; Paroncini, P.; Biavasco, F. Aquaculture can promote the presence and spread of antibiotic-resistant Enterococci in marine sediments. PLoS ONE 2013, 8, e62838. [Google Scholar] [CrossRef]
- Gambino, D.; Savoca, D.; Sucato, A.; Gargano, V.; Gentile, A.; Pantano, L.; Vicari, D.; Alduina, R. Occurrence of antibiotic resistance in the Mediterranean Sea. Antibiotics 2022, 11, 332. [Google Scholar] [CrossRef]
- Pepi, M.; Focardi, S. Antibiotic-resistant bacteria in aquaculture and climate change: a challenge for health in the Mediterranean area. Int. J. Environ. Res. Public Health 2021, 18, 5723. [Google Scholar] [CrossRef]
- Makkaew, P.; Kongprajug, A.; Chyerochana, N.; Sresung, M.; Precha, N.; Mongkolsuk, S.; Sirikanchana,K. Persisting antibiotic resistance gene pollution and its association with human sewage sources in tropical marine beach waters. Int. J. Hyg. Environ. Health 2021, 238, 113859. [Google Scholar] [CrossRef]
- Zheng, D.; Yin, G.; Liu, M.; Chen, C.; Jiang, Y.; Hou, L.; Zheng, Y. A systematic review of antibiotics and antibiotic resistance genes in estuarine and coastal environments. Sci. Total Environ. 2021, 777, 146009. [Google Scholar] [CrossRef]
- Li, L.G.; Huang, Q.; Yin, X.; Zhang, T. Source tracking of antibiotic resistance genes in the environment — challenges, progress, and prospects. Water Res. 2020, 185, 116127. [Google Scholar] [CrossRef]
- Divya, S.P.; Hatha, A.A.M. Screening of tropical estuarine water in south-west coast of India reveals emergence of ARGs-harboring hypervirulent Escherichia coli of global significance. Int. J. Hyg Environ. Health 2019, 222, 235–248. [Google Scholar] [CrossRef]
- Amato, M.; Dasí, D.; González, A.; Ferrús, M.A.; Castillo, M.A. Occurrence of antibiotic resistant bacteria and resistance genes in agricultural irrigation waters from Valencia city (Spain). Agric. Water Manag. 2021, 256, 107097. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef]

| Target gene | Sequence | Conditions | Reference |
|---|---|---|---|
| blaTEM | 5’ -GCKGCCAACTTACTTCTGACAACG- 3’ 5’ -CTTTATCCGCCTCCATCCAGTCTA- 3’ |
95ºC 3 min (1 cycle); 95ºC 15 s and 60ºC 20 s (40 cycles); 72 ºC 1 min | [38] |
| ermB | 5’ -GATACCGTTTACGAAATTGG- 3’ 5’ -GAATCGAGACTTGAGTGTGC- 3’ |
95ºC 3 min (1 cycle); 95ºC 15 s and 58ºC 20 s (40 cycles); 72 ºC 1 min | [39] |
| qnrS | 5’ -GACGTGCTAACTTGCGTGAT- 3’ 5′ -TGGCATTGTTGGAAACTTG- 3’ |
95ºC 3 min (1 cycle); 95ºC 15 s and 62ºC 20 s (40 cycles); 72 ºC 1 min |
[40] |
| sulI | 5’ -CGCACCGGAAACATCGCTGCAC- 3’ 5’ -TGAAGTTCCGCCGCAAGGCTCG- 3’ |
95ºC 3 min (1 cycle); 95ºC 15 s and 65ºC 20 s (40 cycles); 72 ºC 1 min | [41] |
| tetW | 5’ -GAGAGCCTGCTATATGCCAGC- 3′ 5’ -CTTTATCCGCCTCCATCCAGTCTA- 3’ |
95ºC 3 min (1 cycle); 95ºC 15 s and 60ºC 20 s (40 cycles); 72 ºC 1 min | [42] |
| Zone | No. tested isolates | Number of E. coli resistant isolates | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SMX | TMP | CIP | TET | MER | AZI | NAL | FOT | CHL | TGC | TAZ | COL | AMP | GEN | MDRa | ||
| NV | 14 | 2 | 3 | 2 | 2 | 0 | 1 | 3 | 0 | 0 | 0 | 1 | 0 | 4 | 1 | 2 |
| SV | 23 | 4 | 4 | 2 | 2 | 1 | 3 | 4 | 2 | 0 | 0 | 4 | 1 | 12 | 0 | 2 |
| A | 20 | 3 | 2 | 4 | 4 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 1 |
| Total | 57 | 9 | 9 | 10 | 8 | 1 | 5 | 8 | 2 | 0 | 0 | 5 | 1 | 20 | 1 | 5 |
| % of resistant isolates | 15.8 | 15.8 | 17.5 | 10.4 | 1.8 | 8.8 | 14 | 3.5 | 0 | 0 | 8.8 | 1.8 | 35.1 | 1.8 | 14.7 | |
| Zone | No. samples | Number of positive samples | ||||
|---|---|---|---|---|---|---|
| blaTEM | qnrS | ermB | sulI | tetW | ||
| NV | 21 | 6 | 5 | 0 | 3 | 5 |
| SV | 30 | 15 | 9 | 0 | 5 | 0 |
| A | 26 | 15 | 11 | 1 | 10 | 8 |
| TOTAL (%) | 77 | 36 (46.7) | 25 (32.5) | 1 (1.2) | 18 (23.4) | 13 (16.9) |
| Zone | No. samples | Number of positive samples | ||||
|---|---|---|---|---|---|---|
| blaTEM | qnrS | ermB | sulI | tetW | ||
| NV | 14 | 13 | 4 | 0 | 13 | 2 |
| SV | 23 | 21 | 11 | 1 | 21 | 11 |
| A | 20 | 19 | 6 | 2 | 19 | 10 |
| TOTAL (%) | 57 | 53 (92.9) | 21 (36.8) | 3 (5.2) | 53 (92.9) | 23 (45.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





