Submitted:
08 January 2024
Posted:
09 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Birds
2.2. Identification of Staphylococcus strains
2.3. Determination of bacterial sensitivity to antibiotics and chemotherapeutics based on minimum inhibitory concentration (MIC)
2.3.1. Phenotypic determination of S. aureus sensitivity to cefoxitin
2.4. Molecular analysis
2.4.1. Genomic DNA Extraction
2.4.2. PCR for identification of Staphylococcus isolates and genotypic analysis of virulence
2.4.3. Pulsed-field gel electrophoresis (PFGE)
3. Results
3.1. Staphylococcus strains
3.2. Phenotypic susceptibility of isolated bacteria to selected antimicrobial agents
- Resistance of S. aureus strains to oxacillin
3.3. Molecular analysis
- Molecular identification of Staphylococcus isolates and genotypic analysis of virulence
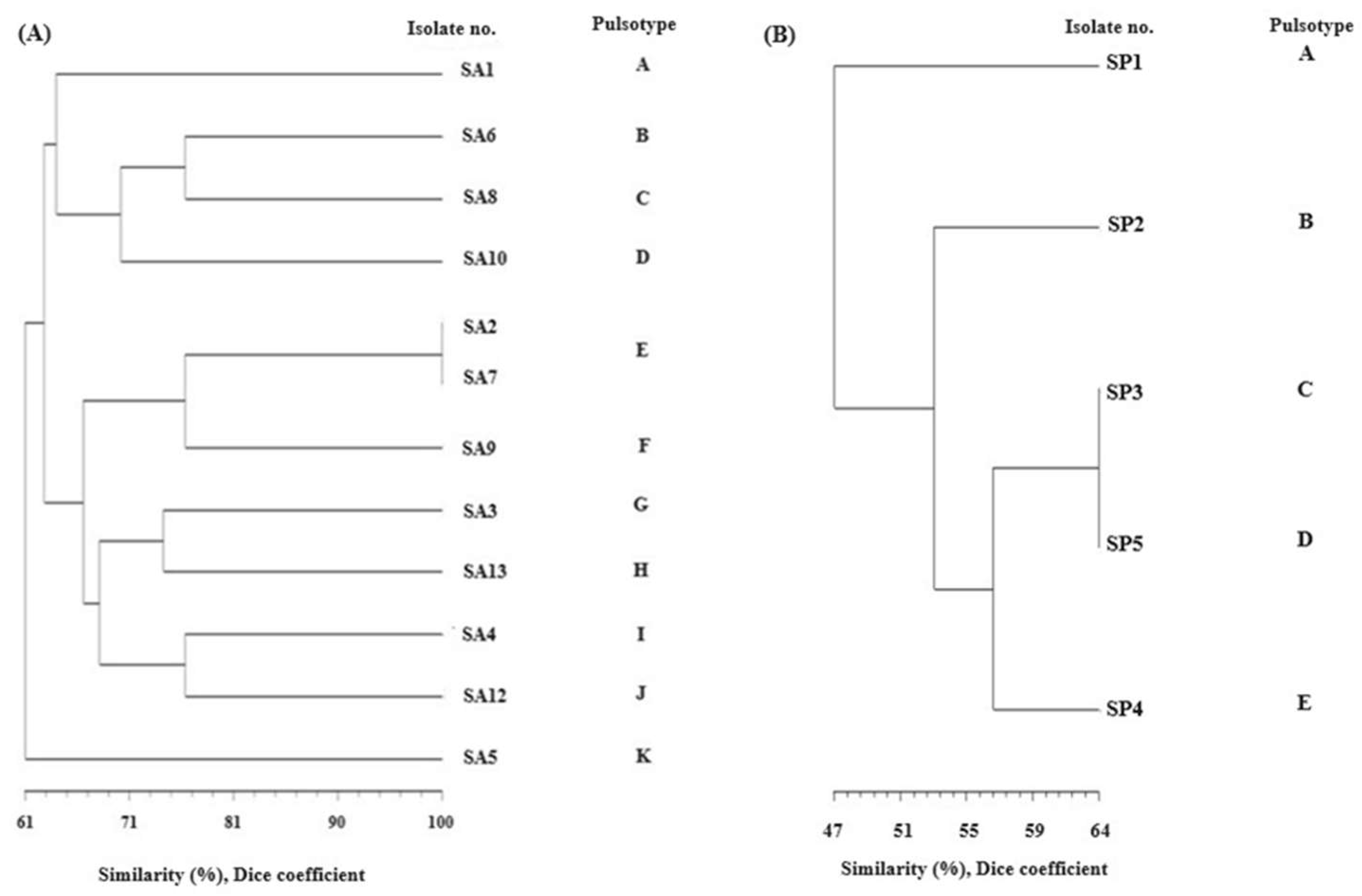
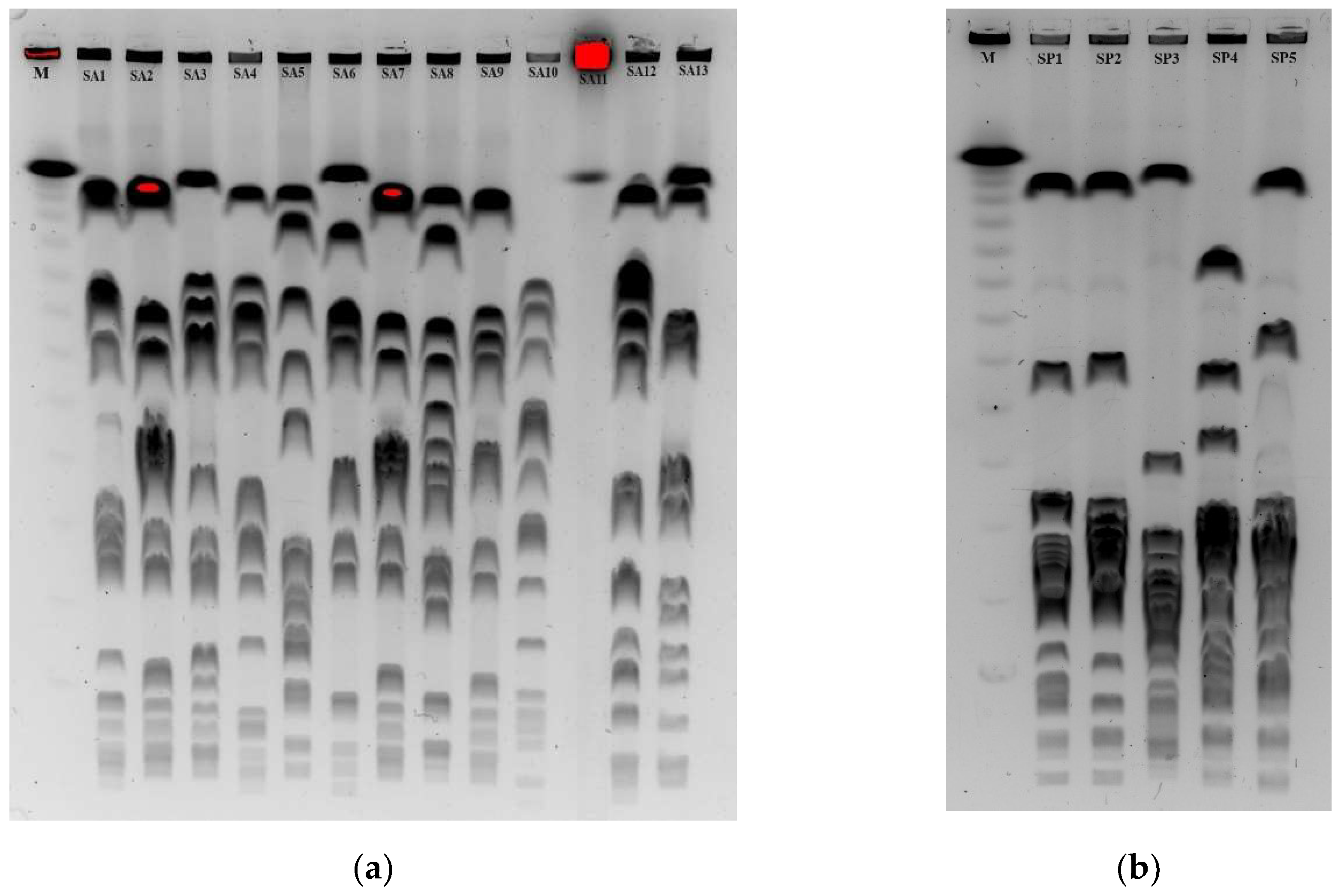
- Pulsed field gel electrophoresis (PFGE)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parte, A.C. LPSN – list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 2014, 42, D613–D616. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Karasawa, T.; Piao, C.; Itoda, I.; Hidai, H.; Yamaura, H.; Totsuka, K.; Morikawa, T.; Takayama, M. Molecular confirmation of transmission route of Staphylococcus intermedius in mastoid cavity infection from dog saliva. J. Infect. Chemother. 2004, 10, 46–48. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.K.; Lee, J.; Bendall, R.; Zhang, L.; Sunde, M.; Slettemeås, J.S.; Gaze, W.; Page, A.J.; Vos, M. Staphylococcus cornubiensis sp. nov., a member of the Staphylococcus intermedius Group (SIG). Int J Syst Evol Microbiol 2018, 68, 3404–3408. [Google Scholar] [CrossRef] [PubMed]
- Perreten, V.; Kania, S.A; Bemis, D. Staphylococcus ursi sp. nov., a new member of the ‘ Staphylococcus intermedius group’ isolated from healthy black bears. Int J Syst Evol Microbiol. 2020, 70, 4637–4645. [Google Scholar] [CrossRef]
- Gerstadt, K.; Daly, J.S.; Mitchell, M.; Wessolossky, M.; Cheeseman, S.H. Methicillin-resistant Staphylococcus intermedius pneumonia following coronary artery bypass grafting. Clin. Infect. Dis. 1999, 29, 218–219. [Google Scholar] [CrossRef] [PubMed]
- Mahoudeau, I.; Delabranche, X.; Prevost, G.; Monteil, H.; Piemont, Y. Frequency of isolation of Staphylococcus intermedius from humans. J. Clin. Microbiol. 1997, 35, 2153–2155. [Google Scholar] [CrossRef]
- Decristophoris, P.; Fasola, A.; Benagli, C.; Tonolla, M.; Petrini, O. Identification of Staphylococcus intermedius Group by MALDI--TOF MS. Syst. Appl. Microbiol. 2011, 34, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.; Hale, J.H.; Smith, M.M. The role of coagulase in staphylococcal infections. Brit J Exp Pathol 1947, 28, 57–67. [Google Scholar]
- Fehrer, S.L.; Boyle, M.D.; Halliwell, R.E. Identification of protein A from Staphylococcus intermedius isolated from canine skin. Am J Vet Res. 1988, 49, 697–701. [Google Scholar]
- Vandenesch, F.; Lina, G.; Henry, T. Staphylococcus aureus Hemolysins, bi-component Leukocidins, and Cytolytic Peptides: A Redundant Arsenal of Membrane-Damaging Virulence Factors? Front Cell Infect Microbiol. 2012, 2, 12. [Google Scholar] [CrossRef]
- Ruscher, C.; Lübke-Becker, A.; Semmler, T.; Wleklinski, C.G.; Paasch, A.; Soba, A.; Stamm, I.; Kopp, P.; Wieler, L.H.; Walther, B. Widespread rapid emergence of a distinct methicillin- and Multidrug-resistant Staphylococcus pseudintermedius (MRSP) genetic lineage in Europe. Vet. Microbiol. 2010, 144, 340–346. [Google Scholar] [CrossRef]
- Ünal, N.; Çinar, O.D. Detection of staphylococcal enterotoxin, methicillin-resistant and Panton–Valentine leukocidin genes in coagulase-negative staphylococci isolated from cows and ewes with subclinical mastitis. Trop Anim Health Prod 2012, 44, 369–375. [Google Scholar] [CrossRef]
- Weese, J.S.; van Duijkeren, E. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet. Microbiol. 2010, 140, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Bean, D.C.; Wigmore, S.M. Carriage rate and antibiotic susceptibility of coagulase-positive staphylococci isolated from healthy dogs in Victoria, Australia. Australia. Aust Vet J. 2016, 94, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Marek, A.; Stępień-Pyśniak, D.; Pyzik, E.; Adaszek, Ł.; Wilczyński, J.; Winiarczyk, S. Occurrence and characterization of Staphylococcus bacteria isolated from poultry in Western Poland. Berl Munch Tierarztl Wochenschr 2016, 129, 147–152. [Google Scholar] [CrossRef]
- Morgan, M. Methicillin-resistant Staphylococcus aureus and animals: zoonosis or humanosis? J Antimicrob Chemother. 2008, 62, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- Robb, A.; Pennycott, T.; Duncan, G.; Foster, G. Staphylococcus aureus carrying divergent mecA homologue (mecA LGA251) isolated from a free-ranging wild bird. Vet. Microbiol. 2013, 162, 300–301. [Google Scholar] [CrossRef]
- Van Hoovels, L.; Vankeerberghen, A.; Boel, A.; Van Vaerenbergh, K.; De Beenhouwer, H. First case of Staphylococcus pseudintermedius infection in a human. J Clin Microbiol. 2006, 44, 4609–12. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, D.B. The Staphylococcus intermedius group of bacterial pathogens: species re-classification, pathogenesis and the emergence of methicillin resistance. Vet. Dermatol. 2009, 20, 490–495. [Google Scholar] [CrossRef]
- Gharsa, H.; Slama, K.B.; Gómez-Sanz, E.; Gómez, P.; Klibi, N.; Zarazaga, M.; Boudabous, A.; Torres, C. Characterisation of nasal Staphylococcus delphini and Staphylococcus pesudintermedius isolates from healthy donkeys in Tunisia. Equine Vet. J. 2014, 47, 463–466. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Gomez, P.; Alonso, C.A.; Camacho, M.C.; de la Puente, J.; Fernandez-Fernandez, R.; Ramiro, Y.; Quivedo, M.A.; Blanco, J.M.; Zarazaga, M.; Höfle, U.; Torres, C. Detection of MRSA of Lineages CC130-mecC and CC398-mecA and Staphylococcus delphini-lnu(A) in Magpies and Cinereous Vultures in Spain. Microb. Ecol. 2019, 78, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Gary, J.M.; Langohr, I.M.; Lim, A.; Bolin, S.; Bolin, C.; Moore, I.; Kiupel, M. Enteric colonization by Staphylococcus delphini in four ferret kits with diarrhoea. J. Comp. Pathol. 2014, 151, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Magleby, R.; Bemis; D.A.; Kim, D.; Carroll, K.C.; Castanheira, M.; Kania, S.A.; Jenkins, S.G.; Westblade, L.F. First reported human isolation of Staphylococcus delphini. Diagn Microbiol Infect Dis. 2019, 94, 274–276. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Othman, E.H.; Merza, S.N.; Jubrael, S.M. Nucleotide sequence analysis of methicillin-resistant Staphylococcus aureus in Kurtdistan Region-Iraq. J. Univ. Zakho. 2014, 2, 1–13. [Google Scholar]
- Murakami, K.; Minamide, W.; Wada, K.; Nakamura, E.; Teraoka, H.; Watanabe, S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol 1991, 29, 2240–2244. [Google Scholar] [CrossRef] [PubMed]
- Stegger, M.; Andersen, P.S.; Kearns, A.; Pichon, B.; Holmes, M.A.; Edwards, G.; Laurent, F.; Teale, C.; Skov, R.; Larsen, A.R. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin Microbiol Infect 2012, 18, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, M.; Wang, G.; Johnson, W.M. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol 2000, 38, 1032–1035. [Google Scholar] [CrossRef] [PubMed]
- Cremonesi, P.; Perez, G.; Pisoni, G.; Moroni, P.; Morandi, S.; Luzzana, M.; Brasca, M.; Castiglioni, B. Detection of enterotoxigenic Staphylococcus aureus isolates in raw milk cheese. Lett Appl Microbiol 2007, 45, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Futagawa-Saito, K.; Sugiyama, T.; Karube, S.; Sakurai, N.; Ba-Thein, W.; Fukuyasu, T. Prevalence and characterization of leukotoxin-producing Staphylococcus intermedius in isolates from dogs and pigeons. J. Clin. Microbiol. 2004, 42, 5324–5326. [Google Scholar] [CrossRef]
- Sung, J. M.; Lloyd, D.H.; Lindsay, J.A. Staphylococcus aureus host specificity: Comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology, 2008; 154, 1949–1959. [Google Scholar]
- Jarraud, S.; Mougel, C.; Thioulouse, J.; Lina, G.; Meugnier, H.; Forey, F.; Nesme, X.; Etienne, J.; Vandenesch, F. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 2002, 70, 631–641. [Google Scholar] [CrossRef]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995, 33, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Gomez, P.; Lozano, C.; Camacho, M.C.; Lima-Barbero, J.F.; Hernandez, J.M.; Zarazaga, M.; Hofle, U.; Torres, C. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J. Antimicrob. Chemother. 2016, 71, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.; Silva, N.; Igrejas, G.; Silva, F.; Sargo, R.; Alegria, N.; Benito, D.; Gomez, P.; Lozano, C.; Gomez-Sanz, E.; Torres, C.; Caniça, M.; Poeta, P. Antimicrobial resistance determinants in Staphylococcus spp. recovered from birds of prey in Portugal. Vet. Microbiol. 2014, 171, 436–440. [Google Scholar] [PubMed]
- Guardabassi, L.; Schmidt, K.R.; Petersen, T.S.; Espinosa-Gongora, C.; Moodley, A.; Agerso, Y.; Olsen, J.E. Mustelidae are natural hosts of Staphylococcus delphini group A. Vet Microbiol 2012, 159, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Devriese, L.; Vancanneyt, M.; Baele, M.; Vaneechoutte, M.; De Graef, E.; Snauwaert, C.; Cleenwerck, I.; Dawyndt, P.; Swings, J.; Decostere, A.; Haesebrouck, F. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int. J. Syst. Evol. Microbiol. 2005, 55, 1569–1573. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Kikuchi, K.; Tanaka, Y.; Takahashi, N.; Kamata, S.; Hiramatsu, K. Reclassification of phenotypically identified Staphylococcus intermedius strains. J. Clin. Microbiol. 2007, 45, 2770–2778. [Google Scholar] [CrossRef] [PubMed]
- Sawhney, S.S.; Vargas, R.C.; Wallace, M.A.; Muenks, C.E.; Lubbers, B.V.; Stephanie, A.; Fritz, S.A.; Burnham, C.D.; Dantas, G. Diagnostic and commensal Staphylococcus pseudintermedius genomes reveal niche adaptation through parallel selection of defense mechanisms. Nature Communications 2023, 14, 7065. [Google Scholar] [CrossRef] [PubMed]
- Doškař, J.; Pantůček, R.; Růžičková, V.; Sedláček, I. Molecular diagnostics of Staphylococcus aureus. W: Detection of bacteria, viruses, parasites and fungi. Bioterrorism prevention, red. M.V. Magni, Springer, Dordrecht 2010, 139-184.
- Bens, C.P.M.; Voss, A.; Klaassen, C.H.W. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterprettable results in standard pulsed-field gel electrophoresis analysis. J Clin Microbiol 2006, 44, 1875–1876. [Google Scholar] [CrossRef] [PubMed]
- Awad-Alla, M.E.; Abdien, H.M.; Dessouki, A.A. Prevalence of bacteria and parasites in White Ibis in Egypt. Vet. Ital. 2010, 46, 277–286. [Google Scholar]
- Monecke, S.; Gavier-Widen, D.; Hotzel, H.; Peters, M.; Guenther, S.; Lazaris, A.; Loncaric, I.; Muller, E.; Reissig, A.; Ruppelt-Lorz, A.; Walter, B.; Coleman, D.C.; Ehricht, R. Diversity of Staphylococcus aureus Isolates in European Wildlife. PLoS ONE 2016, 11, e0168433. [Google Scholar] [CrossRef]
- Porrero, M.C.; Mentaberre, G.; Sanchez, S.; Fernandez-Llario, P.; Casas-Diaz, E.; Mateos, A.; Vidal, D.; Lavin, S.; Fernández-Garayzábal, J.-F.; Dominguez, L. Carriage of Staphylococcus aureus by free-living wild animals in Spain. Appl. Environ. Microbiol. 2014, 80, 4865–4870. [Google Scholar] [CrossRef] [PubMed]
- Porrero, M.C.; Mentaberre, G.; Sanchez, S.; Fernandez-Llario, P.; Gomez-Barrero, S.; Navarro-Gonzalez, N.; Serrano, E.; Casas-Diaz, E.; Marco, I.; Fernández-Garayzabal, J.F.; Mateos, A.; Vidal, D.; Lavín, S.; Domínguez, L. Methicillin resistant Staphylococcus aureus (MRSA) carriage in different free-living wild animal species in Spain. Vet. J. 2013, 198, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Silvanose, C.D.; Bailey, T.A.; Naldo, J.L.; Howlett, J.C. Bacterial flora of the conjunctiva and nasal cavity in normal and diseased captive bustards. Avian Dis. 2001, 45, 447–451. [Google Scholar] [CrossRef]
- Thapaliya, D.; Dalman, M.; Kadariya, J.; Little, K.; Mansell, V.; Taha, M.Y.; Grenier, D.; Smith, T.C. Characterization of Staphylococcus aureus in Goose Feces from State Parks in Northeast Ohio. Ecohealth 2017, 14, 303–309. [Google Scholar] [CrossRef]
- Vidal, A.; Baldoma, L.; Molina-Lopez, R.A.; Martin, M.; Darwich, L. Microbiological diagnosis and antimicrobial sensitivity profiles in diseased free-living raptors. Avian Pathol. 2017, 46, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Wardyn, S.E.; Kauffman, L.K.; Smith, T.C. Methicillin-resistant Staphylococcus aureus in central Iowa wildlife. J. Wildl. Dis. 2012, 48, 1069–1073. [Google Scholar] [CrossRef]
- Silva, V.; Lopes, A.F.; Soeiro, V.; Caniça, M.; Manageiro, V.; Pereira, J.E.; Maltez, L.; Capelo, J.L.; Igrejas, G.; Poeta, P. Nocturnal Birds of Prey as Carriers of Staphylococcus aureus and Other Staphylococci: Diversity, Antimicrobial Resistance and Clonal Lineages. Antibiotics 2022, 11, 240. [Google Scholar] [CrossRef] [PubMed]
- Chrobak, D.; Kizerwetter-Świda, M.; Rzewuska, M.; Moodley, A.; Guardabassi, L.; Binek, M. Molecular characterization of Staphylococcus pseudintermedius strains isolated from clinical samples of animal origin. Folia Microbiol. 2011, 56, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Chae, M.; Yoon, J.; Kim, S.; Lee, S.; Yoo, J.; Park, H.M. Antibiotic resistance and molecular characterization of ophthalmic Staphylococcus pseudintermedius isolates from dogs. J. Vet. Sci. 2014, 15, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.; Vijay, A.K.; Stapleton, F.; Willcox, M. Virulence genes of Staphylococcus aureus associated with keratitis, conjunctivitis and contact lens-assocaied inflammation. Transl. Vis. Sci. Technol. 2022, 11, 5. [Google Scholar] [CrossRef]
- Novick, R. P. Pathogenicity factors and their regulation. In V. A. Fuschetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, & J. I. Rood (Eds.), Gram-positive pathogens, 2000, 392–407. Washington, DC: AMS Press.
- Prévost, G.; Cribier, B.; Couppié, P.; Petiau, P.; Supersac, G.; Finck-Barbançon, V.; Monteil, H.; & Piémont, Y. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infection and Immunity 1995, 63, 4121–4129. [Google Scholar] [CrossRef] [PubMed]
- Goerke, C.; Wirtz, C.; Flückiger, U.; Wolz, C. Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol Microbiol 2006, 61, 1673–1685. [Google Scholar] [CrossRef]
- Matthews, A.M.; Novick, R.P. Staphylococcal phages in Phages. Their role in bacterial pathogenesis and biotechnology. Waldor MK, Friedman DI, Adhya SL (eds). Washington, DC: American Society for Microbiology Press,2005,297–318.
- Humphreys, H.; Coleman, D.C. Contribution of whole-genome sequencing to understanding of the epidemiology and control of meticillin-resistant Staphylococcus aureus. J. Hosp. Infect. 2019, 102, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, M.; Ohta, T.; Uchiyama, I.; Baba, T.; Yuzawa, H.; Kobayashi, I.; Cui, L.; Oguchi, A.; Aoki, K.; Nagai, Y.; Lian, J.; Ito, T.; Kanamori, M.; Matsumaru, H.; Maruyama, A.; Murakami, H.; Hosoyama, A.; Mizutani-Ui, Y.; Takahashi, N.K.; Sawano, T.; Hiramatsu, K. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 2001, 357, 1225–1240. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Keller, B.; von Eiff, C.; Brück, M.; Lubritz, G.; Etienne, J.; Peters, G. Enterotoxigenic potential of Staphylococcus intermedius. Appl Environ Microbiol. 2001, 67, 5551–7. [Google Scholar] [CrossRef] [PubMed]
- Pyzik, E.; Marek, A.; Stępień-Pyśniak, D.; Urban-Chmiel, R.; Jarosz, Ł.S.; Jagiełło-Podębska, I. Detection of Antibiotic Resistance and Classical Enterotoxin Genes in Coagulase -negative Staphylococci Isolated from Poultry in Poland. J Vet Res. 2019, 12;63, 183-190. [CrossRef]
- Kang, M.; Chae, M.; Yoon, J.; Lee, S.; Yoo, J.; Park, H.M. Resistance to fluoroquinolones and methicillin in ophthalmic isolates of Staphylococcus pseudintermedius from companion animals. Vet. Rec. 2014, 55, 678–682. [Google Scholar]
- Tareen, A.R.; Zahra, R. Community acquired methicillin resistant Staphylococci (CA-MRS) in fecal matter of wild birds – A ‘one health’ point of concern. J Infect Public Health. 2023, 16, 877–883. [Google Scholar] [CrossRef]
| Primer name | Oligonucleotide sequence (5′-3′)* | Amplicon size (bp) | Target gene | PCR conditions | Reference |
|---|---|---|---|---|---|
| nuc | GCGATTGATGGTGATACGGTTAGCCAAGCCTTGACGAACTAAAGC | 270 | nuc | 94°C, 5 min, 37 cycles of 94°C for 1 min, 55°C for 30 s, 72°C for 1 min, final extension 72°C for 7 min | [25] |
| mecA | AAAATCGATGGTAAAGGTTGGCAGTTCTGGCACTACCGGATTTGC | 533 | mecA | 94℃, 5 min, 40 cycles of 94℃ for 1 min, 58℃ for 1 min, 72℃ for 2 min, final extension 72℃ for 5 min | [26] |
| mecC | GAA AAA AAG GCT TAG AAC GCC TCGAA GAT CTT TTC CGT TTT CAG C | 138 | mecC | 94℃, 15 min, 30 cycles of 94℃ for 30 s, 59℃ for 1 min, 72℃ for 1 min, final extension for 10 min | [27] |
| sea | ACGATCAATTTTTACAGCTGCATGTTTTCAGAGTTAATC | 544 | enterotoxins | 94℃, 5 min, 35 cycles of 94℃ for 2 min, 57℃ for 2 min, 72℃ for 1 min, final extension 72℃ for 7 min | [28] |
| seb | GAATGATATTAATTCGCATCTCTTTGTCGTAAGATAAACTTC | 416 | |||
| sec | GACATAAAAGCTAGGAATTTAAATCGGATTAACATTATCCA | 257 | |||
| sed | TTACTAGTTTGGTAATATCTCCTTCCACCATAACAATTAATGC | 334 | |||
| see | ATAGATAAAGTTAAAACAAGCAATAACTTACCGTGGACCC | 170 | |||
| seg | CCACCTGTTGAAGGAAGAGGTGCAGAACCATCAAACTCGT | 432 | 94°C for 2 min, 30 cycles of 94°C for 25 s, 50°C for 20 s, 72°C for 40 s, final extension 72°C for 6 min | [29] | |
| seh | TCACATCATATGCGAAAGCAGTCGGACAATATTTTTCTGATCTTT | 463 | |||
| sei | CTCAAGGTGATATTGGTGTAGGCAGGCAGTCCATCTCCTGTA | 529 | |||
| sej | GGTTTTCAATGTTCTGGTGGTAACCAACGGTTCTTTTGAGG | 306 | |||
| sel | CACCAGAATCACACCGCTTACTGTTTGATGCTTGCCATTG | 240 | |||
| tst | ACCCCTGTTCCCTTATCATCTTTTCAGTATTTGTAACGCC | 326 | tsst-1 | 95℃, 5 min, 30 cycles of 94℃ for 1 min, 55℃ for 30 s, 72℃ for 1 min, final extension at 72℃ for 5 min | [12] |
| pvl | ATCATTAGGTAAAATGTCTGGACATGATCCAGCATCAASTGTATTGGATAGCAAAAGC | 433 | pvl | ||
| eta | GCAGGTGTTGATTTAGCATTAGATGTCCCTATTTTTGCTG | 93 | etaetb | ||
| etb | ACAAGCAAAAGAATACAGCGGTTTTTGGCTGCTTCTCTTG | 226 | |||
| lukS | TGTAAGCAGCAGAAAATGGGGGCCCGATAGGACTTCTTACAA | 503 | lukS | 94°C, 3 min, 35 cycles of 94°C for 1 min, 57°C for1 min, 72°C for1Min, final extension 72°C for7 min | [30] |
| lukF | CCTGTCTATGCCGCTAATCAAAGGTCATGGAAGCTATCTCGA | 572 | lukF | ||
| sak | TGAGGTAAGTGCATCAAGTTCACCTTTGTAATTAAGTTGAATCCAGG | 403 | sak | 94°C, 15 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 2 min | [31] |
| hla | CTGATTACTATCCAAGAAATTCGATTGCTTTCCAGCCTACTTTTTTATCAGT | 209 | haemolysins | 95 °C for 5 min, 30 cycles of 95 °C for 1 min, 58 °C for 1 min and 72 °C for 2 min, final extension of 72 °C for 10 min | [32] |
| hlb | GTGCACTTACTGACAATAGTGCGTTGATGAGTAGCTACCTTCAG | 309 | |||
| hld | AAGAATTTTTATCTTAATTAAGGAAGGAGTGTTAGTGAATTTGTTCACTGTGTCGA | 111 | |||
| hlg | GTCAYAGAGTCCATAATGCATTTAACACCAAATGTATAGCCTAAAGTG | 535 |
| Staphylococcus species | bird species | organ | MIC breakpointsμg/ml | Presence of virulence genes | ||||||||||
| MALDI-TOF MSBiotyperLog (Score) | Penicillin | Amoxicillin | Gentamicin | Chloramphenicol | Tetracycline | Enrofloxacin | Erythromycin | Trimethoprim | Sulfamethoxazole | |||||
| S. pseudintermedius | SP1 | 2.100 | Capercaillie | conjunctiva | ≤ 0.062 | ≤ 0.062 | ≤ 0.5 | ≥32 | ≤ 0.125 | ≤ 0.125 | ≤ 0.125 | ≥ 16 | ≤ 19 | - |
| SP2 | 2.096 | Capercaillie | joint | ≤ 0.062 | ≤ 0.062 | ≤ 0.5 | ≤ 4 | ≤ 0.125 | ≤ 0.125 | ≤ 0.5 | ≤ 0.5 | ≤ 38 | sec, hla, hld | |
| SP3 | 2.367 | Capercaillie | liver | ≥ 64 | ≥ 1 | ≤ 0.250 | ≥ 64 | ≥ 32 | ≤ 0.5 | ≥ 256 | ≤ 1 | ≤ 38 | - | |
| SP4 | 2.168 | Meadow pipit | joint | ≤ 0.125 | ≤ 0.125 | ≤ 0.250 | ≥ 32 | ≤ 0.250 | ≤ 0.125 | ≥ 16 | ≤ 0.5 | ≤ 38 | - | |
| SP5 | 2.008 | White stork | conjunctiva | ≥16 | ≥ 0.250 | ≤ 0.250 | ≤ 4 | ≤ 0.125 | ≤ 0.5 | ≤ 0.250 | ≤ 1 | ≤ 19 | - | |
| S. aureus | SA1 | 2.205 | Capercaillie | embryo | ≥ 32 | ≥ 64 | ≤ 1 | ≤ 4 | ≤ 0.125 | ≤ 0.125 | ≤ 0.5 | ≤ 0.5 | ≤ 19 | nuc, hla, hld |
| SA2 | 2.260 | Common kestrel | wound | ≤ 0.062 | ≤ 0.062 | ≤ 1 | ≤ 4 | ≤ 0.125 | ≤ 1 | ≤ 0.5 | ≤ 2 | ≤ 38 | nuc, seg, hla, hlb | |
| SA3 | 2.249 | Song thrush | palatal fissure | ≥ 8 | ≥ 4 | ≤ 1 | ≤ 8 | ≤ 0.250 | ≤ 0.250 | ≤ 0.5 | ≤ 0.250 | ≤ 19 | nuc, see, sej, sak | |
| SA4 | 2.314 | Capercaillie | conjunctiva | ≤ 0.062 | ≤ 0.062 | ≤ 1 | ≤ 4 | ≤ 0.250 | ≤ 0.125 | ≤ 0.5 | ≤ 0.5 | ≤ 38 | nuc, hla, hlb, hld | |
| SA5 | 2.419 | Capercaillie | embryo | ≤ 0.125 | ≤ 0.125 | ≤ 0.5 | ≤ 4 | ≤ 0.125 | ≤ 0.125 | ≤ 0.250 | ≤ 2 | ≤ 19 | nuc, hla, hlb | |
| SA6 | 2.363 | Common kestrel | palatal fissure | ≤ 0.062 | ≤ 0.062 | ≤ 0.5 | ≤ 8 | ≤ 0.250 | ≤ 0.125 | ≤ 0.5 | ≤ 1 | ≤ 19 | nuc, hla, hlb, hld | |
| SA7 | 2.054 | Common buzzard | palatal fissure | ≤ 0.062 | ≤ 0.062 | ≤ 1 | ≤ 4 | ≤ 0.125 | ≤ 0.5 | ≤ 0.5 | ≤ 2 | ≤ 9,5 | nuc, seg, hla, hlb | |
| SA8 | 2.146 | Capercaillie | liver | ≤ 0.062 | ≤ 0.062 | ≤ 0.250 | ≤ 4 | ≤ 0.5 | ≤ 0.125 | ≤ 0.5 | ≤ 0.5 | ≤ 19 | nuc, hla, hlb,hld | |
| SA9 | 2.326 | Capercaillie | embryo | ≤0.062 | ≤ 0.062 | ≤ 1 | 16 | ≥ 16 | ≤ 0.250 | ≤ 0.5 | ≥ 16 | ≤ 38 | nuc, hla, hlb | |
| SA10 | 2.359 | Common kestrel | liver | ≥ 0.5 | ≥ 0.5 | ≤ 1 | ≤ 4 | ≤ 0.5 | ≤ 0.125 | ≤ 0.5 | ≤ 0.5 | ≤ 19 | nuc, hla, hlb, hld | |
| SA11 | 2.230 | Capercaillie | palatal fissure | ≥ 4 | ≥ 2 | ≤ 1 | ≥ 256 | ≥ 256 | ≤ 0.125 | ≥ 256 | ≥ 8 | ≤ 38 | nuc, hlg | |
| SA12 | 2.030 | Common buzzard | spleen | ≤ 0.125 | ≤ 0.125 | ≤ 1 | ≤ 8 | ≤ 1 | ≤ 0.125 | ≤ 0.5 | ≤ 2 | ≤ 38 | nuc, seb, hld | |
| SA13 | 2.342 | Common buzzard | joint | ≥ 0.250 | ≥ 0.250 | ≤ 1 | ≤ 8 | ≤ 0.250 | ≤ 0.125 | ≤ 0.5 | ≤ 0.5 | ≤ 19 | nuc, hla, hld | |
| S. delphini | SD1 | 2.028 | Capercaillie | yolk sac | ≤ 0.062 | ≤ 0.062 | ≤ 0.250 | ≤ 4 | ≤ 0.125 | ≤ 0.125 | ≤ 0.5 | ≤ 2 | ≤ 38 | see, hlb, hld |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





