Introduction
The value of data reuse is one of the founding postulates behind the Open Science movement yet remains an under-examined aspect of researchers’ experience of open data1. Global sharing of biological datasets became technically possible with the rise in access to the World Wide Web, and data reuse transitioned into an attractive option for researchers through benefits that came with an increasing number of available datasets and reuse applications2. Genomics data is particularly amenable to reuse, as many different types of structural and functional data are provided as DNA sequence, and many analytical tools have been developed to analyze and integrate genomics data types3. With constantly emerging sequence-based technologies, the language of nucleotides has become increasingly ubiquitous and useful. Alternatives for assays that traditionally have generated difficult-to-share data types, such as flow cytometry fluorescence, yield easy-to-share sequence-based data types to directly integrate RNA and protein modalities4. However, no dataset is perfect and data producers can only strive to satisfy the requirements for its initial use and reuse. Some researchers have identified the risks and challenges associated with data reuse in the life sciences5,6, which informs agricultural data management7, but a detailed assessment of the reuse issue in this area has not been conducted yet.
Recently, a report on the status of open data called attention to the importance of data availability in reuse1; however, barriers remain in making data amenable for reuse. Our objective in this perspective is to highlight concerns in data reuse across the agricultural genomics community to identify major challenges and viable solutions. We also provide our perspectives on best practices for sharing data to make it more accessible and reusable, as well as how to reuse publicly available data.
We define data reuse as the practice of utilizing existing data for a novel scientific purpose beyond their original scope. Although we recognize that this definition would include use of reference genome sequences, we find that their reuse comes with unique challenges beyond the scope of this paper. Furthermore, while the reuse of one’s own data fits under our definition, the recommendations and perspectives set out in this paper apply primarily to data reuse by researchers other than the data producer’s group.
While types of data in agricultural research are diverse and go beyond sequence-based datasets, the sequencing community harbors a long-standing tradition of data sharing. A major advantage of genomics data for agriculture is that most of such data has a common format and ontology (DNA sequence), allowing the reuse and tuning of tools developed in the well-funded biomedical sphere. Reuse in genomics research is largely facilitated by the International Nucleotide Sequence Database Collaboration (INSDC)8.The INSDC consists of the National Center for Biotechnology Information (NCBI), The European Bioinformatics Institute (EMBL-EBI) and DNA Data Bank of Japan (DDBJ), which collectively support the Sequence Read Archive (SRA) and the European Nucleotide Archive (ENA). Due to its predominance, we will focus on the reuse of sequencing data in this paper, while acknowledging the importance of other data types and emerging analysis technologies in the reuse research arena.
Reusing existing data brings significant benefits for scientific research, such as saving time and cost without generating new datasets, enabling meta-analyses and interdisciplinary research by combining data from multiple studies, or new discoveries by exploring novel hypotheses through integrating data from different sources or using innovative analytical techniques. More and more exciting publications
9,10 are being produced that highlight the value of data reuse, but still, many datasets are not reusable, or scientists may feel they do not trust or do not want to use the data
11. Several review articles have discussed the opportunities and challenges of data reuse
5,12–15, the latter highlighted in
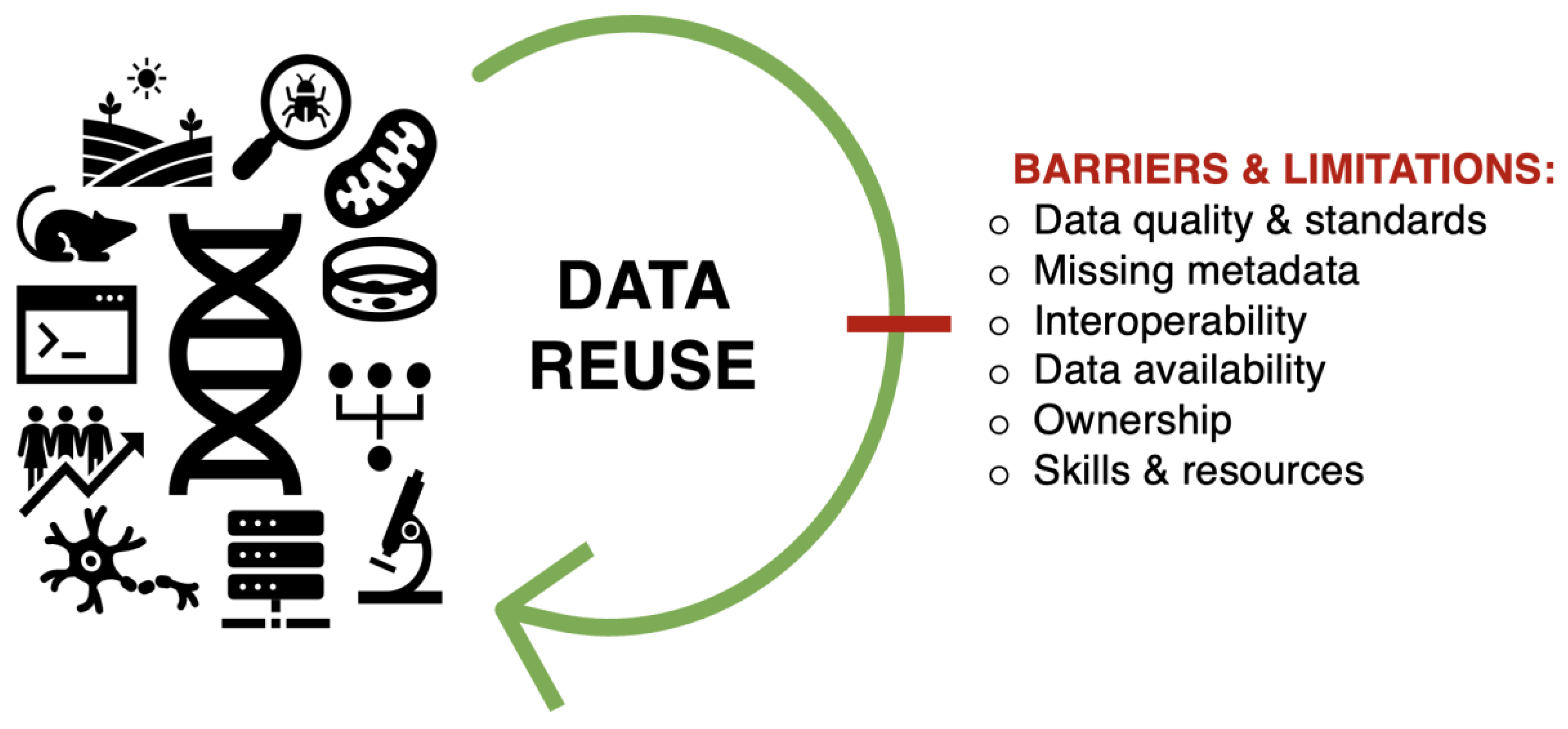
Figure 1.
Principles of Findability, Accessibility, Interoperability, and Reusability (FAIR) are essential to enable successful sharing and reuse of datasets in the ‘Big Data’ world16. The science community has also agreed to uphold data sharing practices that enable data reuse through accords and requirements that promote it17–21. Recognizing the value of reusable datasets and ubiquity of FAIR principles might lead one to believe they are universally accepted and applied. However, as any data stakeholder can testify, no dataset is without flaws6 and a multitude of problems can present themselves to a potential re-user.
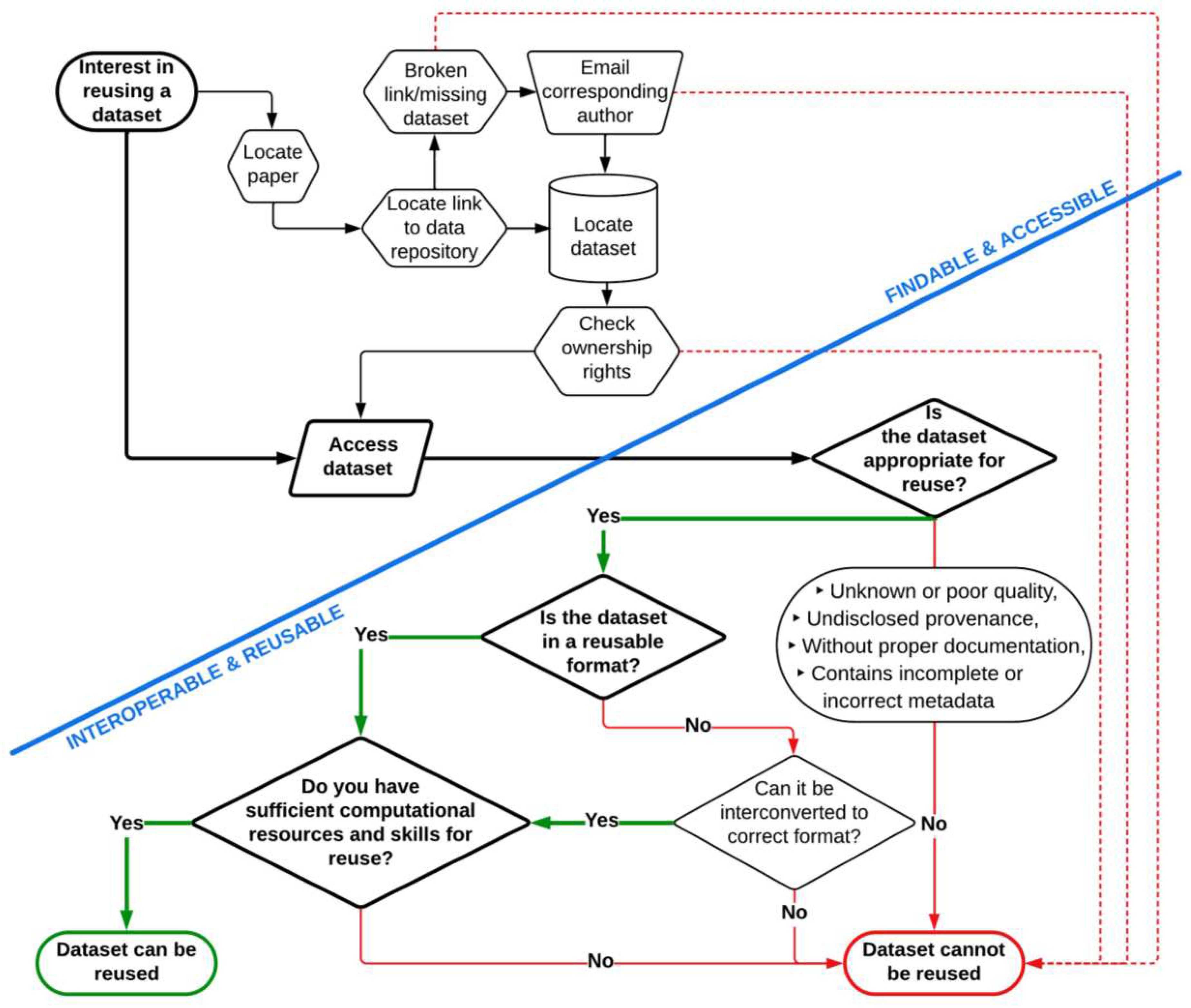
Consider a potential data re-user in agricultural research on their path to a dataset, as depicted in
Figure 2. They are seeking data from an experiment they learnt about at a conference are able to locate the paper the dataset was originally used in. Sometimes they need to email the corresponding author to overcome the broken link to the datasets, and they eventually find the dataset in an online repository. The dataset itself might be of unknown or poor quality, from undisclosed provenance, without proper documentation, or contain incomplete or even incorrect metadata. All these factors can generate confusion in the comprehension of the data and make their reuse challenging. Our re-user must assess whether their subjective requirements of “quality” are met before deciding to reuse the dataset. Data ownership rights must be checked and adhered to as well. The next problem the re-user might encounter is the format of the dataset and if it can be correctly and successfully interchanged into a configuration their downstream analysis supports, which might depend on their skill level. If they are attempting to retrieve large datasets from a study, they might not have access to sufficient computational resources to store the raw datasets or run the analysis. The intermediate results produced in the original study, which could partially remedy the storage problem, may not be available on the repository. It is also likely that intermediate results were produced based on an outdated version of the reference genome sequence or its annotation. Furthermore, the hopeful re-user could be a student, who seeks counsel from their advisor but is informed that the experiment (or public data in general) is untrustworthy, or unsuitable, because of ethics or proprietary constraints. For reuse of a dataset to be successful, these issues must be overcome. The prevalence of these problems can vary depending on the data type, prominence of the original study, the repository they are in, and user skills. However, most stakeholders acknowledge that these issues remain problematic
11, including in agricultural research.
Once initial challenges to sharing are overcome, reuse of existing datasets has numerous advantages5. Designing experiments, collecting samples, and generating data usually involve extensive time, effort, and funding. Retrieving datasets from a repository and reusing speeds up research as the analysis can be started immediately. Biologists can generate new hypotheses to inform their experiments or analyze existing data for preliminary results for emerging research proposals. Alternatively, they may analyze public datasets as additional evidence to test hypotheses in their studies. Through reuse of datasets from public domains, it is possible to investigate massive datasets for data-driven discovery that would not be viable to generate as part of an individual study or explore datasets of species that would not otherwise be accessible. Examples include datasets that were compiled over multiple years or represent a substantial number of species22 in a certain taxonomic group23. Finally, reused datasets enhance equity of science as they are available without substantial costs and allow anyone with sufficient computational resources to benefit from cost-effective data sharing, contributing to inclusion of early-career and underrepresented scientists5. Bioinformatic software developers can rely on publicly available datasets for their benchmarking studies, making it possible to evaluate the performance of novel bioinformatic tools based on real datasets. The power of data reuse is growing with emerging technologies and integration of enormous amounts of data24. This includes harnessing high quality datasets for analysis using machine learning and cloud computing25, as well as using real datasets as quality control for synthetic and artificial intelligence-generated datasets. Benefits of a shared infrastructure and avoidance of resource multiplicity26 enable productive and efficient investigations into new questions using ‘old’ data, a desirable future for agricultural research.
A unifying objective across biology is understanding the link from genome to phenotype (G2P) to move toward predictive biology; reuse of existing datasets will play an important role in this process. G2P initiatives both depend on, and act as a test of, existing data reuse standards and infrastructure. In this way, G2P will also identify where deficiencies exist in data reuse resources. Different funding organizations fund these long-term goals through requests for applications (RFAs). For example, the Genome to Phenome Blueprint
27 discusses the importance of data reuse for animal genetics as a 10 year research priority as identified by researchers at the United States Department of Agriculture (USDA), also reflected in their
Agricultural Genomes to Phenomes Initiative (AG2PI;
https://www.ag2pi.org)
28,29, while the National Science Foundation (NSF) runs the
Understanding the Rules of Life program (
https://www.nsf.gov/news/special_reports/big_ideas/life.jsp). These RFAs seek ways to improve data reuse as integration of data across diverse and expansive datatypes is needed to identify novel phenomena regarding genome function. Tuggle et al. describe the shared efforts of the animal and plant genomics communities to develop synergies and leverage strengths to advance genome to phenome research to make scientific advancements that will accelerate applications in agriculture to help feed a growing world under a variety of challenges
28,29. Comparative and evolutionary biology studies
22,23,30–32 are also important initiatives whose data will need to be amenable to integration and reuse to help in these efforts. While this perspective focuses on sequence-based data, it is important to acknowledge the issues facing phenotypic data reuse, particularly the prevalent
ad hoc formats, lack of archives for storing and accessing data, and inability to share phenotype and genotype data together (due to agreements with industry or lack of infrastructure). For G2P initiatives to be successful, sequence-based and phenotype datasets need to be combined, overcoming their respective barriers to reuse and challenges of integration.
To assess the data reuse needs and obstacles that this community faces, our working group explored the challenges associated with data reuse (and their potential solutions) through personal testimonies and discussions within the AgBioData consortium’s Data Reuse Working Group (DRWG), as well as a review of pertinent literature. The DRWG represents a diverse group of researchers with varied interests in species and scientific applications of data within the domain of agriculture. The AgBioData Consortium (
https://www.agbiodata.org/) is a group of genomics, genetics, and breeding databases and partners working to consolidate data standards and best practices
33–35. The issues and opportunities presented here were generated as part of regular meetings, conference presentations, and workshops held as part of a data reuse project funded by the USDA AG2PI initiative (
https://www.ag2pi.org).
Barriers to Data Reuse and Recommendations to Overcome Them
Data Quality Standards as a Solution
No dataset is perfect5,6, but that does not mean it is not suitable for reuse. As data are made publicly available regardless of the quality metrics, data quality assessment and standardization are important considerations6. Statisticians are well aware of this issue36, which is particularly problematic in the life sciences likely due to the complexity of biological systems, number of variables, and scale of experiments. The difficulty in obtaining and understanding the context surrounding the available data has been identified as a major obstacle to reuse in synthetic biology37 where interdisciplinarity is one of the defining features of the field. We can extrapolate similar issues to agricultural research, which often involves cross-disciplinary collaboration that combines diverse (meta)data types requiring integration and analysis.
To assess if and how a publicly available dataset can be used in analyses beyond its original purpose, a decision must be made of whether it is suitable for reuse. In a sequence-based context, data suitability can mean a variety of dataset properties, including coverage, depth, technical and biological replication, tissue type and sample collection method, extraction method and library preparation, and other criteria. Further, sequencing technology, platform, chemistry kits used, flowcell version and related information must be considered as is required by basecallers for conversion into sequence. All these technologies are also continuously under fast-paced development. With this in mind, whether a dataset is of sufficient quality and suitable to be reused is a difficult, and largely subjective decision
38 and varies between applications. While there are some data type-specific standards available (e.g.,
Genomic Data Commons; https://gdc.cancer.gov/), their scope is limited. Agricultural research is often multidisciplinary, has complex experimental designs, and spans many non-model species, which makes applying any universal standard very difficult.
Unified experimental protocols or bioinformatic pipelines for common data types and organisms are rare. This is not a problem in and of itself at the level of data production, although an off-the-shelf pipeline could streamline the process. The lack of standard protocols and pipelines becomes a hurdle when it comes to data reuse. Not only can obtaining the exact experimental protocol used be difficult (e.g. discussions of data reuse often result in anecdotes of lost protocols with unanswered emails and/or students who graduated), but meta-analyses are also hindered by a lack of standardization. Sharing experimental designs and protocols together with produced datasets is a challenge that the international data standards rarely address. Examples of minimum information standards being implemented by necessity include the Minimum Information About a Microarray Experiment (MIAME) and Minimum Information about a Sequencing Experiment (MINSEQE) (
https://www.ncbi.nlm.nih.gov/geo/info/MIAME.html).
Further, an important question that needs to be considered in the field is whether our experiments should be designed with future data reuse in mind. For example, while for the original data producer one biological replicate may have been sufficient for the purposes of gene prediction, a statistically robust meta-analysis of gene expression may require at least three39. Such meta-analyses must solve the important issue of handling batch effects, when merging data from multiple sources and attempting to use multi-source replication for statistical analysis. Not only can the complete datasets be harnessed in the future, but they can also limit the need for the same sample to be sequenced again, saving resources for dataset production and storage. However, considering the upfront cost to the data producer, it cannot be expected that the original experimental design considers future use, regardless of potential benefits to the field. A model for partially transferring the costs of the initial experiment from the individual to the community would be required as incentive for additional data generation. Additionally, future use objectives can be difficult to predict, and emerging technologies can make numerous datasets irrelevant. The most important consideration that can be made by the data producer to ensure future reuse is to submit complete metainformation, including recorded factors that were not relevant to the original study.
Looking to the example of the biomedical sphere in solving issues of data quality, the agricultural research community should adopt more standardization across the board. While file type standardization is common for sequence-based data (e.g., FASTA or FASTQ), there is a lack of experimental protocol, sample handling, computational pipeline, and statistical standards present in agricultural research. This makes assessing data quality one of the biggest barriers to dataset reuse. Unified recommendations (if not standards) for all aspects of data collection would enable more successful data reuse, increasing a dataset’s economic utility, with the added benefit of aiding the data producer in making their research more broadly comparable. Such standards need to be broadly applicable and not too severe, in a “legacy standard” format that does not hold back future stricter requirements and developments in the field.
On the Road to Complete Metadata through Incentives
The missing information about datasets available to a potential re-user exacerbates the problem of lacking metadata standards. Historically, the need for minimum metadata standards were recognized and implemented by many journal and funding agencies, but missing metadata is still one of the main barriers to data reuse cited by researchers5,38.
While most sequencing datasets are released through INSDCs databases33, there is a sparsity of metadata accompanying them. For example, the precise tissue type, cultivation conditions or developmental stage may not have been recorded. Complete metadata is especially important for RNA-seq datasets because the transcriptome responds quickly to the environmental conditions of the sampled individual. As DNA methylation can now be investigated based on Oxford Nanopore Technologies or Pacific Biosciences HiFi sequencing data, information about the conditions prior to DNA extraction gains importance. Re-users might want to study the methylation of DNA in response to certain environmental conditions or treatments. Further, methods used to minimize sample-to-sample variation due to sequencing methods, such as barcoding of pooled samples, must be clearly explained. If there is data from the same sample sequenced in different lanes to increase the sequencing coverage, this needs to be clearly annotated in the metadata table, as it can lead to confusion when distinguishing samples that were just sequenced in different lanes from replicates.
The paradigm of ontologies has enabled the interoperability and reuse of data in the genomics era
40. However, using available ontologies to describe data from agriculturally relevant species is often not appropriate, as such tools are model organism- and medical-based. Initiatives like the
Genomic Data Commons (
https://gdc.cancer.gov/) do provide scaffolds of metadata standards but are limited to a small number of data types and purposes. Furthermore, metadata submission templates tend to only work for some organisms or sample types, and do not enforce the use of controlled vocabularies. Smaller, community-based efforts are on the way to improve available ontologies (e.g. FAANG’s
Ontology Improver;
https://data.faang.org/ontology).
The biggest effort to integrate data and metadata with available controlled vocabulary standards is the INSDC (
https://www.ncbi.nlm.nih.gov/biosample/docs/attributes/). It enables extensive data sharing and interoperability, with the responsibility of quality and accuracy of the record naturally falling on the submitting author, not on the database
41. Interoperability standards in medicine for genotypic and phenotypic patient data
42 could be informative for agricultural research as well. These health information formats include metadata on the tests run, and sometimes even on the analyses not run, to enable healthcare providers to integrate results from diverse panels. Such complete metadata could generate large overhead in some circumstances and must be considered in the context of agricultural genomics. Various communities have proposed guidelines for standardizing metadata
43,44 and minimum information standards in experiments (MIAME and MINSEQE), but there is still a need for standardization of metadata across different databases, both in what is captured and how it is captured.
As the submission of metadata can require substantial work, there is a trade-off between collecting all datasets via a lenient submission system and mandating comprehensive metadata to boost the reuse potential of datasets
5. Without incentives or requirements, researchers often seek the lowest effort route to publication with minimal metadata. Ideally, submitting users would be supported by automatic completion of certain fields. Initiatives like
nfdi4plants (
https://www.nfdi4plants.de) in Germany are working to make data submission as convenient as possible. Data documentation takes extra effort, necessitating the need for a reward system to encourage production of datasets amenable to reuse. This could include dataset citations, credit for shared data in promotion, and other rewards for datasets that are reused often and successfully.
Towards Interoperability via Data Formatting
The genetics and genomics community converged rapidly on data format standards and is on the road to establishing standards for the metadata stored within data files
45 (Zhang, 2016). Widespread standardization of these file formats facilitates easy interconversion and use by analysis and visualization software, ensuring interoperability. The Sequence Alignment Map (SAM) format for high throughput sequence data, and its respective mapping results, requires the recording of a data dictionary with information on the reference genome sequence used for mapping, such that can ensure any subsequent analysis will be required to use the same reference
46. There are also provisions therein to record data processing information, such as the program and command line used to generate the mapped dataset and any post-processing, including sorting and PCR duplicate removal. Other standardized formats with enforced rules include the SAM compressed format Binary Alignment Map (BAM) (
https://samtools.github.io/hts-specs/SAMv1.pdf), the Variant Call Format (VCF)
47,48, the Gene Transfer Format (GTF) (
http://mblab.wustl.edu/GTF22.html), General Feature Format (GFF3) (
https://github.com/The-Sequence-Ontology/Specifications/blob/master/gff3.md) and Browser Extensible Data (BED) (
https://genome.ucsc.edu/FAQ/FAQformat.html#format1) files that allow for annotation of regions of a given genome sequence (
http://useast.ensembl.org/info/website/upload/gff.html). All these files can be coordinate indexed such that they may be searched, and subset easily by locus or loci.
As evidenced by the wide acceptance of universal data formats in genomics research, the limitation to the wider adoption of data reuse is not the lack of defined data formats, but the consistency of their use. Many datasets are deposited according to the parameters of the database chosen to hold the data. The database may allow for several types of files when it comes to, for example, transcriptomic studies. A researcher has the option of uploading the data in the form of a set of FASTQ files or maybe as a set of BAM files, with the choice made dictating how reusable the data can be for others. A possible solution is for the repositories to provide more re-user-friendly tools that facilitate interconversion between formats, for example, FASTQ and BAM, without accompanying loss of metadata.
Although the genomics datasets of types mentioned above have documented standards requiring information such as what reference genome sequence and what version were used for their analysis (standards enforced by assertions in analysis packages like the
Genome Analysis Toolkit;
https://gatk.broadinstitute.org/hc/en-us), mapping to reference genome sequences does create an impediment to interoperability with processed, or secondary datasets. Any solution to this problem would require reference-free analysis of data. This is an area of active research
49–51, and a future in which indices accompany raw datasets for rapid query and use in synchronous analyses that run at remote sites seems possible.
Interoperability with data from outdated wet lab and/or computational analysis methods can also present a challenge. A few tools have been built to bridge the data found in newer, standardized sequencing files with data encoded by older formats such as arrays and spa typing
52–54. To guard against data obsolescence, researchers need to incorporate thorough methodological metadata (for example, using tools like
https://www.protocols.io). Hence, interoperability is also supported by adherence to metadata and data quality standards described in previous sections.
To encourage interoperability, data warehouses and journals can raise their standards for data submission to require the inclusion of the outputs of primary analyses. This practice is often encouraged, but not required or enforced. Synthesis Centers (funded by the NSF) are examples of projects that highly promote data reuse and integration, demonstrating the economic efficiency of data exchange with incredible success26. Recent efforts have also been made to boost interoperability in the Bgee knowledge base by taking stock of file-based data exchange, programmatic interfaces, and automatic interoperability efforts55. The good news is that interoperability boosting seems to have a positive domino effect enabled by automation, which will hopefully lead to near total integration capabilities soon55.
Bridging the Data Availability Gap: A Role for All Stakeholders
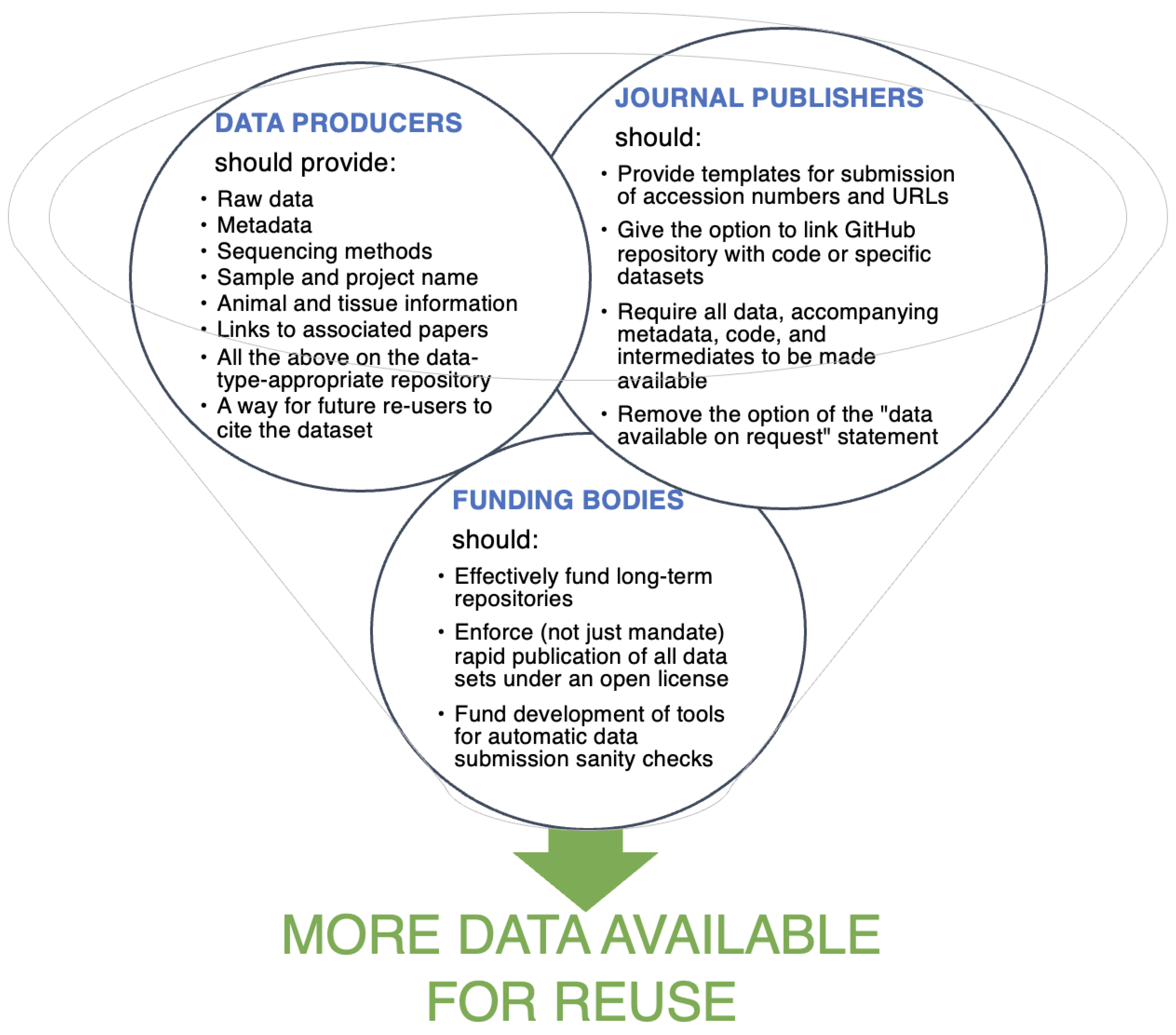
A major barrier to reuse is the availability of data with their accompanying metadata and sample information in repositories. It is crucial for data providers to include all samples and relevant information in a clear sequence, using the provided data format or metadata template when available. This includes raw data and metadata, including sequencing methods, sample name and tissue, organism, project, and associated papers. The information provided needs to be clear and comprehensive to facilitate the reproducibility of analyses. The commitment of all data stakeholders is crucial in narrowing the data availability gap as summarized in
Figure 3.
Many journals provide generic statements for authors to declare that all data are included in the supplementary files of the article or deposited in a public repository. However, such statements are not helpful without specific accessions or links that point readers to the respective datasets. A further contributor to this data availability gap is the “data available on request” statement present in many papers that do not provide a direct link to their data in a repository but ask the potential reuser to contact them to receive it. A study on data availability from papers published in Science and Nature in 2021 found that an alarming less than 50% of data stated to be “available upon request” could be effectively obtained from the original authors56. Further, about 20% of all metagenome assemblies are not easily accessible due to the lack of accession numbers in the publication or due to empty accession numbers57. Even if data are provided, it can take months to receive it56, with questions about storage and management arising. More encouragingly, after many attempts at contact, 83% of data was made available at least partially56.
Journals could improve the situation by providing more detailed templates that require researchers to fill in accessions or URLs. Options to link a GitHub repository with code or specific datasets to the submission would be another option. However, enforcing such data standards requires additional labor by editorial staff. While journals would be well placed to enforce a policy that would benefit reuse, funding bodies could be in an even stronger position to mandate rapid publication of all datasets under an open license. Automatic checks of the submitted datasets would be helpful to reduce the amount of work that reviewers need to invest on the technical aspects of a submission.
Datasets should be shared through the repository appropriate for the data type as summarized by Deng et al. (Table 1)33. For example, RNA-seq datasets should be submitted to Gene Expression Omnibus (GEO) to make precomputed count tables and the underlying raw sequence reads available. The reads are passed on to the SRA which also mirrors them through the ENA and the DDBJ. This ensures preservation of the data. A direct submission of RNA-seq datasets to the SRA/ENA/DDBJ is possible and common but does not allow the sharing of already computed count tables. This places a burden on researchers trying to reuse these datasets. Genomic sequencing data are best placed in this mirrored database to ensure availability to the community. Accession numbers for data submitted to repositories should also be included in publications.
All data published to sequence archives are data that have had some primary analyses, including quality control, performed on them. For next generation sequence data, nearly all will have been mapped to a reference genome sequence. Whole genome shotgun sequence data will likely have been variant called, and will have, at least a VCF file, in addition to the BAM file and the mapped FASTQ file. RNA-seq and epigenetic datasets will have been mapped, and likely have quantified transcripts and peak sizes respectively. For example, DNA methylation data will often supply only raw reads in FASTQ and differentially methylated regions, the latter representing the final output of highly variable and long pipelines. For the most part, the data that are being stored and are filling up public repositories are the raw FASTQ files. Due to the large sets of information and calculations needed to examine all manner of “omics” data, computational methods are employed for analyses. In some cases, the analyses require the authors to write code, yet they often do not share the code itself, diminishing the usefulness of the shared data.
For such datasets to be reused, scientists are required to not only download the raw data but also reprocess them. This re-analysis is likely to generate many identical pipeline intermediates and final datasets that were created by the original analysis. Being able to demonstrate reproducibility in an analysis is important, and too often proves impossible58, but it is equally important that the datasets achieve their full utility potential through reuse for novel purposes59. The processes that are performed to analyze the raw data are often beyond the computational resources and skills available to most researchers who could benefit from them. Therefore, it may be useful to make processed data, such as transcriptome and genome sequence assemblies, genomic variants, and peaks identified using technologies like chromatin immunoprecipitation sequencing (ChIP-seq), available along with the underlying reads whenever possible. However, storing intermediates and final products of pipelines comes at the cost of increasing the amount of necessary disk space, an important trade-off to consider. A possible partial solution to this bottleneck to reuse is to make all code used in the computational analysis available alongside raw and/or processed datasets.
Storing ever-growing datasets in a sustainable way is a current and growing challenge. Disk space and electric power consumption will continue to rise as database sizes increase and data reuse becomes more popular at research institutes and companies. There is a recent trend to move analyses to the data instead of moving the data, for example through cloud computing
60. Given the explosion in dataset sizes, this seems like a logical step to take, since many large datasets are already available within a cloud environment. However, this harbors the risk that datasets will be effectively locked behind paywalls, as users would be required to pay for the computational resources. Once fully established, such a system could lead to expensive charges beyond the costs of maintaining the cloud infrastructure. It would be important to have a publicly funded infrastructure or to ensure sufficient competition between several providers. Efforts for more sustainable and interconnected funding of biodata resources are already underway, for example, through the Global Biodata Coalition (
https://globalbiodata.org/).
As citations of scientific publications are considered the currency of science, citations of datasets could acquire a similar importance
61. Open Science Framework (
https://osf.io) provides scientists with options to easily share datasets that are citable and searchable through Digital Object Identifiers (DOIs). The benefits associated with publication of paper preprints extend to datasets mentioned in them, enabling instant dissemination and citation of DOIs. A cultural shift or requirement is needed to ensure that dataset identifiers are included in the main text of publications, enabling automatic readers to discover them. Additionally, automated literature tracking solutions could credit the impact of a dataset, by tracking whenever this dataset is mentioned in a subsequent publication. For meta-analyses that contain large numbers of datasets that cannot all be mentioned in-text, it would be necessary to develop an automatic screen that searches all supplementary files for mentioned DOIs. Such a screen could be extended to patents to analyze the commercial relevance of datasets.
Rewards for well documented data submissions could be a strategy to further improve the quality and quantity of publicly available datasets62. Among them could be an evaluation criterion for research proposals of data an investigator has shared in accordance with data sharing plans in previously funded research projects. Researchers spend substantial amounts of time and resources on generating and submitting datasets. This could be rewarded by tracking the number of studies re-using these datasets, as attempted by the Omics Discovery Index (OmicsDI)63. Funding agencies, universities, and companies would need to make hiring decisions based on this criterion, similarly to how they already do with publication citations. As this would be a rearward facing statistic, it would likely come with the same biases and issues of equity as citations of scientific publications, namely self-citation, gender, racial, and institutional bias64, but may still incentivize generation of more reusable datasets.
Data Ownership and Sharing Requirements
An important source of genetic material for research in plant and animal genomics is samples from genetic lines derived from breeding companies that have current commercial value or intellectual property. Often, arrangements to use such data for experiments are important for omics analyses to be relevant to species of agricultural importance. Breeding companies often have large populations with excellent metadata and can provide samples at little to no additional cost. However, these companies need to protect their investments in intellectual property and often prohibit researchers from making their sequence or omics data public (e.g., a recent dispute of intellectual property rights for improved seeds65). Unfortunately, this is a major barrier in reusing relevant agricultural data.
There is a challenge in having access to relevant, affordable study populations from breeding companies that can also be shared publicly as sequence or genotype data. The extent of sharing is also unknown as a reliable assessment of the economic importance of datasets would be difficult to achieve because most companies could not permit an analysis of internal data reuse to protect their intellectual property. Enabling a self-reporting system could be an approach to gain insights into data re-use within companies, in addition to the dataset citation reward system mentioned in the previous section. Finding common ground in precompetitive research spaces and ways to leverage industry data for scientific discovery, while protecting intellectual property, will help facilitate reuse of some industry data.
Maintaining the competitive value of industry data is important, thus, there is a need to develop novel data sharing solutions that protect intellectual property but facilitate more data sharing. Several methods have been proposed to overcome this problem, including homomorphic/monomorphic encryption and federated learning methods
66–69. The inability to share industry data inhibits publication in an increasing number of journals. Additionally, it also threatens to reduce public-private research partnerships funded by the US government as pending regulations will require all data funded by federal grants to be made public tentatively sometime in 2026 (
https://www.whitehouse.gov/ostp/news-updates/2022/08/25/ostp-issues-guidance-to-make-federally-funded-research-freely-available-without-delay/).
Agricultural industry datasets provide value to both the public and private sectors, and importantly facilitate innovative training of graduate students. The ability to reuse industry data impacts graduate student training since students are required to produce publications and demonstrate competency based on their expertise. Reduced access to industry data will diminish training sought by industry to work with industry relevant data. Thus, challenges related to data reuse of industry data have broad impact.
Another consideration is data generated from biological resources that are maintained by specific cultural groups (discussed below in The importance and benefits of equity and inclusion in agricultural data reuse). Landraces and traditional crops and crop wild relatives contain valuable genetic variation. There are weak systems in place to guarantee the engagement of these communities when their data is used and reused70,71. The human genomics community has experience in data privacy to maintain HIPAA compliance to ensure healthcare data remains both private and portable. The use of data management, sharing and processing tools developed for medical systems may be helpful in overcoming some of these challenges in agriculture.
There already exist numerous federal grant data sharing requirements. Genetic sequence data is an increasingly important consideration in policy regarding agricultural intellectual property rights and conservation (e.g., The Nagoya Protocol -
https://www.cbd.int/abs/, International Treaty on Plant Genetic Resources for Food and Agriculture -
https://www.fao.org/plant-treaty/en/), Africa BioGenome Project -
https://africanbiogenome.org/). The upcoming 2026 mandate to make research funded by the USA government publicly available will undoubtedly alter the landscape of data sharing and ownership further. When it comes to future publicly funded research, we believe that partnerships between public and private entities should prioritize collective benefits to ensure that the rewards of data reuse are reaped equitably.
Resource Availability and User Skill Level
With respect to high throughput sequence data, the data that are stored are typically unprocessed sequence datasets in FASTQ format. For most genetic or genomic studies, this format is the starting point for any analytical pipeline. The bioinformatics skills and computational resources required to store and transform FASTQ data into, for example, quantified expression levels, variants, or genotypes, exist in most larger research institutes. Therefore, we believe that many issues of data storage and computational resource availability are not the limiting factors in most US-based academic and government institutions any longer (which could be said a decade ago). However, world-wide many agricultural researchers and institutions do not have ready access to these resources. This constitutes a barrier to the reuse of these data, which for many, is insurmountable, constituting a major challenge to equity and inclusion in the future of data reuse.
Additionally, user skill level, awareness of resources and time investment into data management are likely inhibiting a lot of productive reuses and limiting how many resources are being made available for future reuse. A recent study
13 shows that, at least anecdotally, skill or perceived ability was identified by many participants as a major factor influencing reuse behavior. Concerning methods of data storage, sharing, and management was identified across all science sectors and types of research activities, with most respondents to a 2017-2018 global survey of scientists exhibiting “high and mediocre risk data practices'', for example storing data on USB drives
11. That same survey found that attitudes toward data reuse were mostly positive, but that practice does not always support data storage, sharing, and future reuse
11. Investment into data literacy early in science education will address these issues in the future generations of researchers
72. We agree with Tenopir et al.
11, namely that “
programs for both awareness and to help engender good data practices are clearly needed”. Further, data reuse can be incentivized using award systems for successful reuse cases, for example, the DataWorks! Prize (
https://www.herox.com/dataworks) or The Research Parasite Award (
https://researchparasite.com/).
The Importance and Benefits of Equity and Inclusion in Agricultural Data Reuse
The introduction of Big Data in agriculture has unlocked tremendous opportunities for advancements73. Equity considerations are essential to ensure that the benefits of agricultural data reuse are shared equitably among diverse stakeholders, including marginalized communities and vulnerable populations74.
Reuse of data can improve equity and inclusion by reducing costs and increasing dataset utility. Nonetheless, reuse of data requires computational capacity, internet access, digital literacy, and proficiency in dominant languages. Despite significant global disparities, nations are formulating policies and expanding infrastructure to reach remote, rural, and peri-urban communities. The percentage of people with internet access has been steadily increasing, although each locality has its own unique needs. The internet plays a pivotal role in bridging the gap to access a wealth of information.
The knowledge disparities can be narrowed by employing data visualization techniques and providing commentaries, detailed explanations, glossaries, and links to both basic and complex information. Data visualization, defined as “information which has been abstracted in some schematic form, including attributes or variables for the units of information” plays a pivotal role in assisting non-data scientists in comprehending and effectively reusing data75. In contemporary data science, professionals are increasingly incorporating advanced technologies into data visualization, including algorithms, human perception, animation, and the development of computer graphics and software. These innovations enable the discovery of valuable insights within vast datasets76.
Documentation of data is essential for facilitating reuse, and it is crucial to link the outcomes of data reuse with contextual information. Scientists require technical details regarding equipment and data procedures, maintenance of data formats, ontologies, and metadata within a specific field77. However, individuals with varying levels of knowledge disparity often need access to more information. To address this need, databases and repositories for reused data should be linked with institutional science communication websites, providing comprehensive explanations of fundamental concepts.
Equally, as numerous studies have shown, diversity breeds innovation
78 (
Figure 4). Thus, to harness the full power of a data-driven future in agriculture, the omics community needs to wrestle with the question of whether biases present in research citation patterns (prestige of the authors being cited, their gender, race, and nationality
64 are transferred to datasets which are selected for reuse.
It is also vital we adhere to and enforce the CARE (Collective Benefit, Authority to Control, Responsibility, and Ethics) principles for Indigenous data governance
79 of existing and future datasets. As Carroll et al.
79 note, we must acknowledge that many publicly available and reused datasets already use Indigenous resources and traditional knowledge. A great resource for data sovereignty enhancing research is the Local Contexts initiative (
https://localcontexts.org), providing “
a digital infrastructure for community governance of Indigenous data”. Our recommendation to the community is to engage with Indigenous communities, practice responsible data stewardship and use Indigenous ethics to determine data access
80. This includes the use of appropriate digital identifiers and inquiry into and respect of ownership rights. Traditional Knowledge Labels “
improve the quality of provenance, encourage communities to enrich records with their own traditional knowledge, and increase capacity for better understanding of equity and decision-making regarding re-use and circulation” 80. Provenance of any biocultural samples, collections, datasets, and traditional knowledge should be noted in full in metadata.
Although limited research has been conducted on access to agricultural omics benefits74, we can learn from ethics frameworks for health and biomedical data, which can be adapted to the agricultural domain81. For example, Tiffin et al.82 emphasize the need for data governance that protects vulnerable populations, especially in low-income and middle-income countries, when utilizing digital health data. Further, Mott et al.68 discuss the use of homomorphic encryption for secure data sharing, which can facilitate the inclusion of private or sensitive data without compromising data confidentiality. This technology could be a key enabler in making data sharing more inclusive, especially when dealing with sensitive information from indigenous communities, as highlighted by Carroll et al.80
On the heels of many studies quantifying discrimination in academia, which was largely ignored by institutions for decades, the big data community has a unique opportunity to build a field of research with fewer such biases. Efforts should be directed towards creating centralized repositories that host diverse agricultural datasets, making it easier for researchers to locate and access relevant information. Addressing issues related to data ownership and equitable access is vital if we are to reap all the benefits of data reuse as a global genomics community.
The Future of Data Reuse Is Bright
Here, we have assessed challenges to reuse sequence-based agricultural datasets and presented possible future solutions regarding (meta)data availability, ownership, user resources, and equity. There is a growing demand for the reuse of published datasets and reinforcing the importance of well-structured databases to increase these numbers in the future. A change in global research culture that emphasizes the ‘R’ for reuse in FAIR would cause significant increases in data submissions, accompanied by more frequent reuse.
One of the biggest challenges of data reuse is to establish standards across the board. Defined data standards and recommendations would address the issues of data quality, availability, sparsity of metadata, and formatting in the agricultural genomics field. The number of omics datasets is increasing every year and to keep the data well organized, following some standards can be helpful to enable reproducibility, with the added benefit of being good scientific practice. Other traditional knowledge management domains such as libraries, specifically data librarians may ultimately guide the creation of organizational standards. Maintaining these standards, as well as detailing important information that was cited throughout this article, may facilitate the reuse of omics data for future analysis. It may also aid in bringing all areas of agricultural research on equal footing when it comes to the benefits of open science83. This will benefit future scientists and developers of applications and databases, contributing to science.
The focus of this (over)view of the status of data reuse in agricultural research have been sequence-based datasets. However, we acknowledge that many challenges and opportunities associated with these types of biological data are shared with non-sequence-based datasets. Indeed, these diverse data types come with their own unique set of challenges and rewards of reuse. Examples of these datasets include, and are not limited to, phenomes, metabolomes, proteomes, interactomes, enviromes, microbiomes, lipidomes, and glycomes. Additionally, many analyses include geographic, climate, and ecological data, which must also be considered for reuse purposes. Advances in artificial intelligence promise to allow for more knowledge to be gleaned from large, shared, interdisciplinary datasets. The omics revolution is clearly still ongoing, and we must keep emerging data types in mind when considering reuse standards and platforms. It will be important to consider how such data types can be integrated with sequence-based data for future applications, further emphasizing the importance of complete metadata and biosample information currently deposited in databases. We, in the AgBioData DRWG, believe the future of data reuse is bright as more datasets are reused successfully, contributing to sustainability of agricultural research in the omics era.
Conclusions
Data reuse is beginning to yield exciting science across disciplines. Harnessing the power of large agricultural omics projects, like FarmGTEx24 and Rice3K25, has demonstrated the detailed knowledge that can be obtained from reuse. As many barriers to reuse keep falling, the biggest obstacle may continue to be the labor investment needed from the data producer (e.g., submitting data to repositories) and re-user (e.g., often convoluted process of obtaining data). Establishing more standards across data production, management, and sharing would pave the way to lowering the barrier of entry to benefits of reuse. Many data producers are sharing their data, but there is a need for more incentives to encourage complete metadata sharing to facilitate reuse. Researcher skill level, one of the major barriers to reuse, needs to be bolstered with guidance and training programs, ensuring equity across all stakeholders in the global agricultural community. In addition, to ensure the maintenance of data availability, it is imperative that the scientific community continues to invest in data management infrastructure and resources. The future of data reuse will also benefit from the development of user-friendly tools and platforms that facilitate data discovery, access, and analysis.
The benefits are clear; data reuse facilitates the ability to ask big questions and provide community resources about genomes and phenomes that one group alone cannot achieve. As more funding agencies and RFAs are promoting data reuse, more scientists will see the exciting opportunities to solve grand challenges in biology. The next big breakthrough in predictive biology will likely require the integration of many diverse datasets. The future of data reuse in agriculture hinges on a collective commitment to data management, standards, infrastructure development and collaboration between researchers. The open science principles are necessary to improve the innovative research and sustainable agricultural practices. The data is out there to reuse; it is time to develop your innovative idea and run with the exciting datasets that are already available. The sky's the limit!
Contributions
JEK initiated the collaboration, contributed to writing the manuscript, obtained funding for data reuse workshops, and chaired a working group to discuss data reuse needs and challenges. AH contributed to writing the manuscript, made the figures, and co-chaired the working group. CGE contributed to writing the manuscript. VLD contributed to writing the text for the interoperability section and revising the manuscript. PWH contributed to writing text for the metadata and ontologies section, and the future of data reuse section. BPu contributed to writing text for the sections benefits of data reuse, data availability, and future of data reuse. contributed to writing the text. TK contributed to writing text for “Towards interoperability via data formatting” and “Resource availability and user skill level”. BPe contributed to the editing and revising the manuscript and future of data reuse section. EQR contributed to writing the equity and inclusion section. CD, DM, and CT contributed to editing and revising the manuscript.
Acknowledgements
The authors wish to thank the AgBioData group for support and assistance in the logistics of the data reuse subgroup meetings. We acknowledge funding from the USDA NIFA-AG2PI seed grant entitled “Harnessing Ag Genomics Data to link genotype to phenotype” as part of the USDA-NIFA awards 2020-70412-32615 and 2021-70412-35233, and to the AgBioData Consortium through the NSF for the Research Coordination Network project award abstract #2126334.
References
- Science Digital et al. The State of Open Data 2023. https://digitalscience.figshare.com/articles/report/The_State_of_Open_Data_2023/24428194/1 (2023). [CrossRef]
- McKiernan, E. C. et al. How open science helps researchers succeed. eLife 5, e16800 (2016). [CrossRef]
- Satam, H. et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 12, 997 (2023). [CrossRef]
- Wu, S. Z. et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet 53, 1334–1347 (2021). [CrossRef]
- Sielemann, K., Hafner, A. & Pucker, B. The reuse of public datasets in the life sciences: potential risks and rewards. PeerJ 8, e9954 (2020). [CrossRef]
- Fernández-Ardèvol, M. & Rosales, A. Quality Assessment and Biases in Reused Data. American Behavioral Scientist 000276422211448 (2022). [CrossRef]
- Devare, M., Arnaud, E., Antezana, E. & King, B. Governing Agricultural Data: Challenges and Recommendations. in Towards Responsible Plant Data Linkage: Data Challenges for Agricultural Research and Development (eds. Williamson, H. F. & Leonelli, S.) 201–222 (Springer International Publishing, 2023). [CrossRef]
- Arita, M., Karsch-Mizrachi, I. & Cochrane, G. The international nucleotide sequence database collaboration. Nucleic Acids Research 49, D121–D124 (2021). [CrossRef]
- Liu, S. et al. A multi-tissue atlas of regulatory variants in cattle. Nature Genetics 54, 1438–1447 (2022). [CrossRef]
- Papoutsoglou, E. A. et al. Enabling reusability of plant phenomic datasets with MIAPPE 1.1. New Phytol 227, 260–273 (2020). [CrossRef]
- Tenopir, C. et al. Data sharing, management, use, and reuse: Practices and perceptions of scientists worldwide. PLoS ONE 15, e0229003 (2020). [CrossRef]
- Gomes, D. G. E. et al. Why don’t we share data and code? Perceived barriers and benefits to public archiving practices. (2022).
- LaFlamme, M., Poetz, M. & Spichtinger, D. Seeing oneself as a data reuser: How subjectification activates the drivers of data reuse in science. PLoS ONE 17, e0272153 (2022). [CrossRef]
- Senft, M., Stahl, U. & Svoboda, N. Research data management in agricultural sciences in Germany: We are not yet where we want to be. PLoS ONE 17, e0274677 (2022). [CrossRef]
- Verhulst, S. & Young, A. Identifying and addressing data asymmetries so as to enable (better) science. Front. Big Data 5, 888384 (2022). [CrossRef]
- Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018 (2016). [CrossRef]
- Announcement: Where are the data? Nature 537, 138–138 (2016). [CrossRef]
- Open Data in a Big Data World. Chemistry International 38, 17–17 (2016). [CrossRef]
- CODATA, Hodson, Simon, Mons, Barend, Uhlir, Paul, & Zhang, Lili. The Beijing Declaration on Research Data. in (2019). [CrossRef]
- Nosek, B. A. et al. Promoting an open research culture. Science 348, 1422–1425 (2015). [CrossRef]
- OECD. Enhanced Access to Publicly Funded Data for Science, Technology and Innovation. (OECD, 2020). [CrossRef]
- Lewin, H. A. et al. The Earth BioGenome Project 2020: Starting the clock. Proceedings of the National Academy of Sciences 119, e2115635118 (2022). [CrossRef]
- Vertebrate Genomes Project. Nature https://www.nature.com/collections/cabiagjdfj (2021).
- The CattleGTEx atlas reveals regulatory mechanisms underlying complex traits. Nature Genetics 54, 1273–1274 (2022). [CrossRef]
- Day, A. & Poplin, R. Analyzing 3024 rice genomes characterized by DeepVariant. Google Cloud Blog https://cloud.google.com/blog/products/data-analytics/analyzing-3024-rice-genomes-characterized-by-deepvariant (2019).
- Rodrigo, A. et al. Science Incubators: Synthesis Centers and Their Role in the Research Ecosystem. PLOS Biology 11, e1001468 (2013). [CrossRef]
- Rexroad, C. et al. Genome to Phenome: Improving Animal Health, Production, and Well-Being – A New USDA Blueprint for Animal Genome Research 2018–2027. Frontiers in Genetics 10, (2019).
- Tuggle, C. K. et al. Current challenges and future of agricultural genomes to phenomes in the USA. Genome Biol 25, 8 (2024). [CrossRef]
- Tuggle, C. K. et al. The Agricultural Genome to Phenome Initiative (AG2PI): creating a shared vision across crop and livestock research communities. Genome Biology 23, 3 (2022). [CrossRef]
- Chen, L. et al. Large-scale ruminant genome sequencing provides insights into their evolution and distinct traits. Science 364, eaav6202 (2019). [CrossRef]
- Leebens-Mack, J. H. et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019). [CrossRef]
- Zhang, G. Bird sequencing project takes off. Nature 522, 34–34 (2015). [CrossRef]
- Deng, C. H. et al. Genotype and phenotype data standardization, utilization and integration in the big data era for agricultural sciences. Database 2023, baad088 (2023). [CrossRef]
- Harper, L. et al. AgBioData consortium recommendations for sustainable genomics and genetics databases for agriculture. Database (Oxford) 2018, (2018). [CrossRef]
- Saha, S. et al. Recommendations for extending the GFF3 specification for improved interoperability of genomic data. arXiv (2022).
- Moorhead, J. E., Rao, P. V. & Anusavice, K. J. Guidelines for experimental studies. Dental Materials 10, 45–51 (1994). [CrossRef]
- Delgado, A. An economy of details: standards and data reusability. Synthetic Biology 8, ysac030 (2023). [CrossRef]
- Curty, R. G., Crowston, K., Specht, A., Grant, B. W. & Dalton, E. D. Attitudes and norms affecting scientists’ data reuse. PLOS ONE 12, e0189288 (2017). [CrossRef]
- Schurch, N. J. et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 22, 839–851 (2016). [CrossRef]
- Schuurman, N. & Leszczynski, A. Ontologies for Bioinformatics. Bioinform Biol Insights 2, 187–200 (2008).
- Brunak, S. et al. Nucleotide Sequence Database Policies. Science 298, 1333–1333 (2002). [CrossRef]
- Deckard, J., McDonald, C. J. & Vreeman, D. J. Supporting interoperability of genetic data with LOINC. Journal of the American Medical Informatics Association 22, 621–627 (2015). [CrossRef]
- Ćwiek-Kupczyńska, H. et al. Measures for interoperability of phenotypic data: minimum information requirements and formatting. Plant Methods 12, 44 (2016). [CrossRef]
- Jenkins, G. B. et al. Reproducibility in ecology and evolution: Minimum standards for data and code. Ecology and Evolution 13, e9961 (2023). [CrossRef]
- Zhang, H. Overview of Sequence Data Formats. in Statistical Genomics (eds. Mathé, E. & Davis, S.) vol. 1418 3–17 (Springer New York, 2016).
- Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [CrossRef]
- Beier, S. et al. Recommendations for the formatting of Variant Call Format (VCF) files to make plant genotyping data FAIR [version 2; peer review: 2 approved]. F1000Research 11, (2022). [CrossRef]
- Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [CrossRef]
- Lee, S.-G., Na, D. & Park, C. Comparability of reference-based and reference-free transcriptome analysis approaches at the gene expression level. BMC Bioinformatics 22, 310 (2021). [CrossRef]
- Parra-Salazar, A., Gomez, J., Lozano-Arce, D., Reyes-Herrera, P. H. & Duitama, J. Robust and efficient software for reference-free genomic diversity analysis of genotyping-by-sequencing data on diploid and polyploid species. Molecular Ecology Resources 22, 439–454 (2022). [CrossRef]
- Petri, A. J. & Sahlin, K. isONform: reference-free transcriptome reconstruction from Oxford Nanopore data. Bioinformatics 39, i222–i231 (2023). [CrossRef]
- Ambroise, J. et al. Backward compatibility of whole genome sequencing data with MLVA typing using a new MLVAtype shiny application for Vibrio cholerae. PLOS ONE 14, e0225848 (2019). [CrossRef]
- Bletz, S., Mellmann, A., Rothgänger, J. & Harmsen, D. Ensuring backwards compatibility: traditional genotyping efforts in the era of whole genome sequencing. Clinical Microbiology and Infection 21, 347.e1-347.e4 (2015). [CrossRef]
- Gordon, M., Yakunin, E., Valinsky, L., Chalifa-Caspi, V. & Moran-Gilad, J. A bioinformatics tool for ensuring the backwards compatibility of Legionella pneumophila typing in the genomic era. Clinical Microbiology and Infection 23, 306–310 (2017). [CrossRef]
- de Farias, T. M., Wollbrett, J., Robinson-Rechavi, M. & Bastian, F. Lessons learned to boost a bioinformatics knowledge base reusability, the Bgee experience. arXiv (2023).
- Tedersoo, L. et al. Data sharing practices and data availability upon request differ across scientific disciplines. Sci Data 8, 192 (2021). [CrossRef]
- Eckert, E. M. et al. Every fifth published metagenome is not available to science. PLoS Biol 18, e3000698 (2020). [CrossRef]
- Stodden, V., Seiler, J. & Ma, Z. An empirical analysis of journal policy effectiveness for computational reproducibility. Proc Natl Acad Sci U S A 115, 2584–2589 (2018). [CrossRef]
- Ahmed, M., Kim, H. J. & Kim, D. R. Maximizing the utility of public data. Front. Genet. 14, 1106631 (2023). [CrossRef]
- Koppad, S., B, A., Gkoutos, G. V. & Acharjee, A. Cloud Computing Enabled Big Multi-Omics Data Analytics. Bioinform Biol Insights 15, 11779322211035921 (2021). [CrossRef]
- Groth, P., Cousijn, H., Clark, T. & Goble, C. FAIR Data Reuse – the Path through Data Citation. Data Intellegence 2, 78–86 (2020). [CrossRef]
- Wood-Charlson, E. M., Crockett, Z., Erdmann, C., Arkin, A. P. & Robinson, C. B. Ten simple rules for getting and giving credit for data. PLoS Comput Biol 18, e1010476 (2022). [CrossRef]
- Perez-Riverol, Y. et al. Quantifying the impact of public omics data. Nature Communications 10, 3512 (2019). [CrossRef]
- Ray, K. S., Zurn, P., Dworkin, J. D., Bassett, D. S. & Resnik, D. B. Citation bias, diversity, and ethics. Accountability in Research 0, 1–15 (2022). [CrossRef]
- Zimmerman, S. Corteva lawsuit accuses gene-editing startup of stealing seeds. Agriculture Dive https://www.agriculturedive.com/news/corteva-lawsuit-inari-steal-seeds-gene-editing/695605/ (2023).
- Blatt, M., Gusev, A., Polyakov, Y. & Goldwasser, S. Secure large-scale genome-wide association studies using homomorphic encryption. Proc Natl Acad Sci U S A 117, 11608–11613 (2020). [CrossRef]
- Konečný, J., McMahan, B. & Ramage, D. Federated Optimization:Distributed Optimization Beyond the Datacenter. arXiv (2015).
- Mott, R., Fischer, C., Prins, P. & Davies, R. W. Private Genomes and Public SNPs: Homomorphic Encryption of Genotypes and Phenotypes for Shared Quantitative Genetics. Genetics 215, 359–372 (2020). [CrossRef]
- Zhao, T., Wang, F., Mott, R., Dekkers, J. & Cheng, H. Using encrypted genotypes and phenotypes for collaborative genomic analyses to maintain data confidentiality. Genetics iyad210 (2023). [CrossRef]
- Smyth, S. J., Macall, D. M., Phillips, P. W. B. & de Beer, J. Implications of biological information digitization: Access and benefit sharing of plant genetic resources. The Journal of World Intellectual Property 23, 267–287 (2020). [CrossRef]
- Wynberg, R. et al. Farmers’ Rights and Digital Sequence Information: Crisis or Opportunity to Reclaim Stewardship Over Agrobiodiversity? Frontiers in Plant Science 12, (2021).
- Wolff, K., Friedhoff, R., Schwarzer, F. & Pucker, B. Data literacy in genome research. Journal of Integrative Bioinformatics 0, 20230033 (2023). [CrossRef]
- Weersink, A., Fraser, E., Pannell, D., Duncan, E. & Rotz, S. Opportunities and Challenges for Big Data in Agricultural and Environmental Analysis. Annual Review of Resource Economics 10, 19–37 (2018). [CrossRef]
- Harris, J., Tan, W., Mitchell, B. & Zayed, D. Equity in agriculture-nutrition-health research: a scoping review. Nutrition Reviews 80, 78–90 (2022). [CrossRef]
- Friendly, M. & Denis, D. J. Milestones in the history of thematic cartography, statistical graphics, and data visualization. http://www.datavis.ca/milestones/ (2001).
- Li, Q. Embodying Data: Chinese Aesthetics, Interactive Visualization and Gaming Technologies. (Springer, 2020). [CrossRef]
- Pasquetto, I. V., Borgman, C. L. & Wofford, M. F. Uses and Reuses of Scientific Data: The Data Creators’ Advantage. Harvard Data Science Review 1, (2019). [CrossRef]
- Hofstra, B. et al. The Diversity–Innovation Paradox in Science. Proceedings of the National Academy of Sciences 117, 9284–9291 (2020). [CrossRef]
- Carroll, S. R. et al. The CARE Principles for Indigenous Data Governance. Data Science Journal (2020). [CrossRef]
- Carroll, S. R., Herczog, E., Hudson, M., Russell, K. & Stall, S. Operationalizing the CARE and FAIR Principles for Indigenous data futures. Scientific Data 8, 108 (2021). [CrossRef]
- Xafis, V. et al. An Ethics Framework for Big Data in Health and Research. Asian Bioeth Rev 11, 227–254 (2019). [CrossRef]
- Tiffin, N., George, A. & LeFevre, A. E. How to use relevant data for maximal benefit with minimal risk: digital health data governance to protect vulnerable populations in low-income and middle-income countries. BMJ Global Health 4, e001395 (2019). [CrossRef]
- Muñoz-Tamayo, R. et al. Seven steps to enhance Open Science practices in animal science. PNAS Nexus 1, pgac106 (2022). [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).