1. Introduction
The growing demand for food and agricultural products, fueled by the expanding human population, has adverse effects on soil, atmosphere, and water sources [
1]. Addressing these challenges requires solutions that not only increase agricultural productivity in terms of land area but also optimize resource usage.
One strategy to boost productivity is the cultivation of high-vigor seeds. The quality of these seeds, determined by genetic, physical, sanitary, and physiological factors, enhances crop development, directly impacting plant species yield. Traditional germination tests, while reliable for seed production standards, have limitations in predicting seedling emergence under less favorable environmental conditions.
To address this, evaluating the internal morphology of seeds has become crucial for identifying issues related to their physiological potential. Image analysis techniques offer non-destructive methods to understand various aspects of seed development, establishing a link between internal morphology and structural integrity. This approach allows the determination of the physiological potential of seed lots [19].
A well-known image acquisition method is based on X-ray differential absorption by seed tissues, which varies with tissue thickness, density, composition, and radiation wavelength [
2,
3]. The process involves exposing seeds to X-rays, creating a latent image on a photosensitive film [5].
Manual measurement of parameters contributing to seed vigor remains a challenge in seed image analysis, despite the support of computational tools. Seeking automation solutions, recent literature has explored various computational methods correlating image-derived parameters with seed quality standards [
2,
4,
5].
In this context, Computer Vision techniques have been widely applied in digital agriculture. Recent applications range from seed vigor testing [15,16] to precision beekeeping [17,18]. Moreover, recent progress in Deep Learning, especially convolutional neural networks such as YOLO [20] and R-CNN, has garnered considerable attention for their capability to identify occurrences in various regions of images [
6,
7,
8].
Here we propose a method to automate the analysis of X-ray images of Brachiaria seeds (
Urochloa brizantha cv. Xaraés) using YOLOv8 [
6] with a post-segmentation module. This approach aims to segment images and derive quality descriptors based on internal morphology, providing a quick and non-destructive estimate of seed germination potential.
2. Materials and Methods
In this section, we outline the entire process we followed to create the seeds image database and train the computer vision method used for segmentation and classification.
2.1. Seeds
The brachiaria (
Urochloa brizantha cv. Xaraés) is a highly cultivated forage grass globally [
5], making the automated assessment of its seed quality of significant interest to the agricultural industry. In this paper, random samples of brachiaria seeds were used to capture radiographic images.
Figure 1 illustrates the small size of these seeds under examination, highlighting the need for an automated evaluation approach.
2.2. Dataset
The dataset images were obtained using Faxitron digital equipment, model MX 20 DC-12, connected to a Core 2 Duo computer (3.16 GHz, 2 GB RAM, 160 GB hard drive), and a MultiSync monitor (17-inch LCD1190SX). The seeds underwent 20 seconds of radiation at 20 kV during the acquisition process.
For each batch, ten repetitions of 20 seeds were used, arranged on transparent plastic sheets and secured in position with double-sided adhesive tape. This setup ensured clear visualization of seed parts, including the embryo, endosperm, seed coat, and empty spaces. After the process, 30 images (each containing 20 seeds, as shown in
Figure 2) were saved on an external storage device for subsequent batch processing.
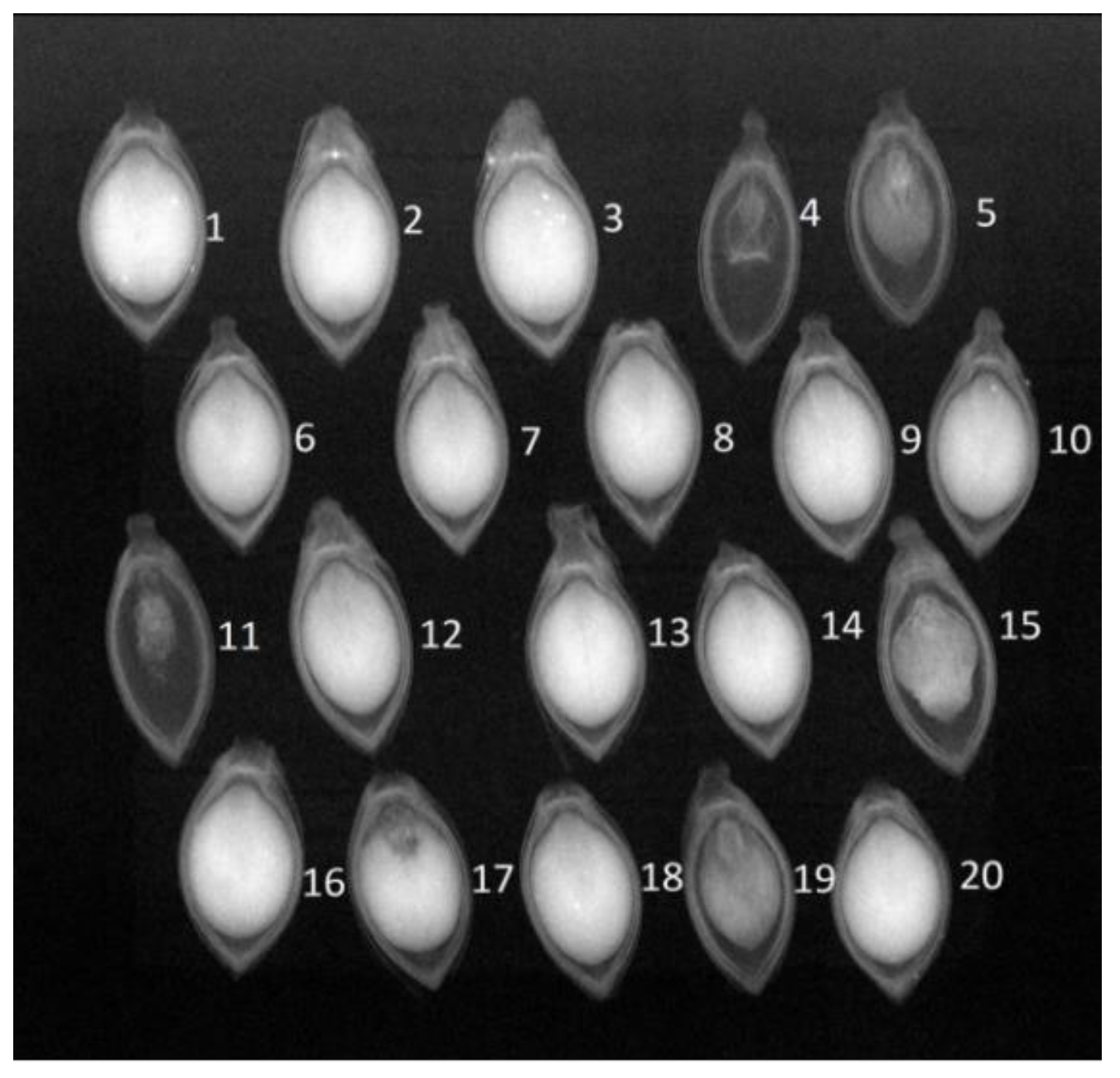
In
Figure 2, radiographic images of Urochloa brizantha seeds, cv. Xaraes, reveal crucial details of internal morphology. This allows the assessment of characteristics such as low quality (#5 #15 #19), poor formation (#4 and #11), and the presence of embryo damage (#17). These factors significantly impact seed lot quality, directly influencing germination.
To perform seed segmentation, we chose a supervised training approach. Using the Labelme tool, we labeled the 20 seeds in each image into two categories: seed and endosperm. This labeling was validated by a professional in Agronomy.
2.3. YOLOv8
To segment regions of interest, we chose to develop a model based on YOLOv8 for its practical training method and high accuracy. YOLOv8, a member of the YOLO (You Only Look Once) series of object detection algorithms, was introduced by Ultralytics in late 2022, improving upon their previous work with YOLOv5. YOLOv8 brings architectural changes that enhance speed and accuracy compared to earlier versions. It incorporates a new C2F module, replacing YOLOv5's C3 module, generating more precise feature maps for object detection. Also, YOLOv8 predicts object center points directly instead of using predefined anchor boxes like older YOLO versions, resulting in more accurate detections. Tests confirm YOLOv8's cutting-edge performance in various object detection benchmarks, making it suitable for real-time applications such as video surveillance and autonomous driving due to its speed and compact size [
9].
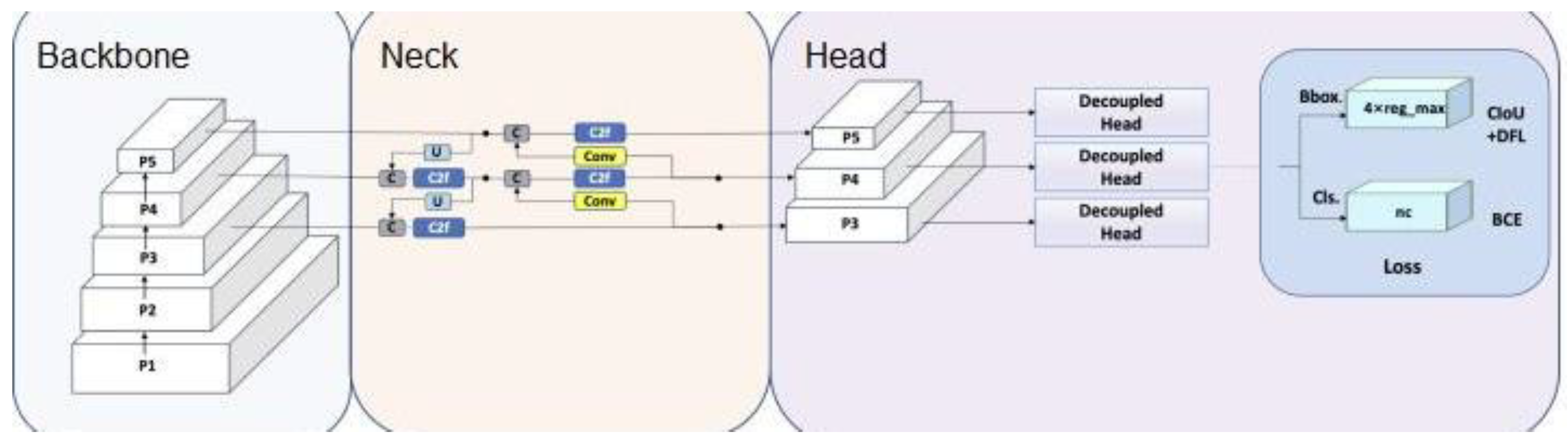
The YOLOv8 architecture is part of the "You Only Look Once" (YOLO) object detection family [
9]. It consists of three main parts: the backbone network, neck network, and detection head. The backbone extracts feature maps from the input image, while the neck network and main network infer bounding boxes and object labels on these feature maps.
Figure 3 provides an overview of the YOLOv8 architecture.
During this study, the model underwent training using an augmented dataset of initial images, split randomly into 70% for training and 30% for testing. Additionally, we employed k-fold cross-validation, dividing the image dataset into five sets for more reliable evaluation. The Ultralytics' version X implementation of the model included measures to prevent overfitting, such as dropout to balance network weights, early stopping at 50 training epochs, and weight transfer from pre-training on the COCO image dataset. The model underwent training at different epochs (500, 1500, and 3000) using the SGD optimizer, a learning rate of 0.01, and dynamic weight decay.
2.4. Human Analysis of Seed Vigor
To validate the data obtained from our proposed method, images from the dataset underwent human visual analysis. This analysis focused on studying the internal morphology of the seeds, identifying areas occupied by the embryo and endosperm. Additionally, it included the detection of deteriorated tissues and malformations, characteristics that can lead to reduced germination.
Following the visual analysis, the germination test was conducted using the same seeds utilized in the X-ray test, maintaining the seed distribution order. Each batch underwent ten repetitions of 20 seeds. The germination test employed germitest® paper sheets premoistened with distilled water, equivalent to 2.5 times the mass of the dry substrate. Seed-containing paper rolls were placed in a BOD-type germinator with alternating temperatures of 15-35°C and a photoperiod of 8 hours of light and 16 hours of darkness. Evaluations were conducted seven days after sowing, which is the established date for the first germination count [13].
Figure 4.
Measurement of Brachiaria seed structures using the ImageJ software.
Figure 4.
Measurement of Brachiaria seed structures using the ImageJ software.
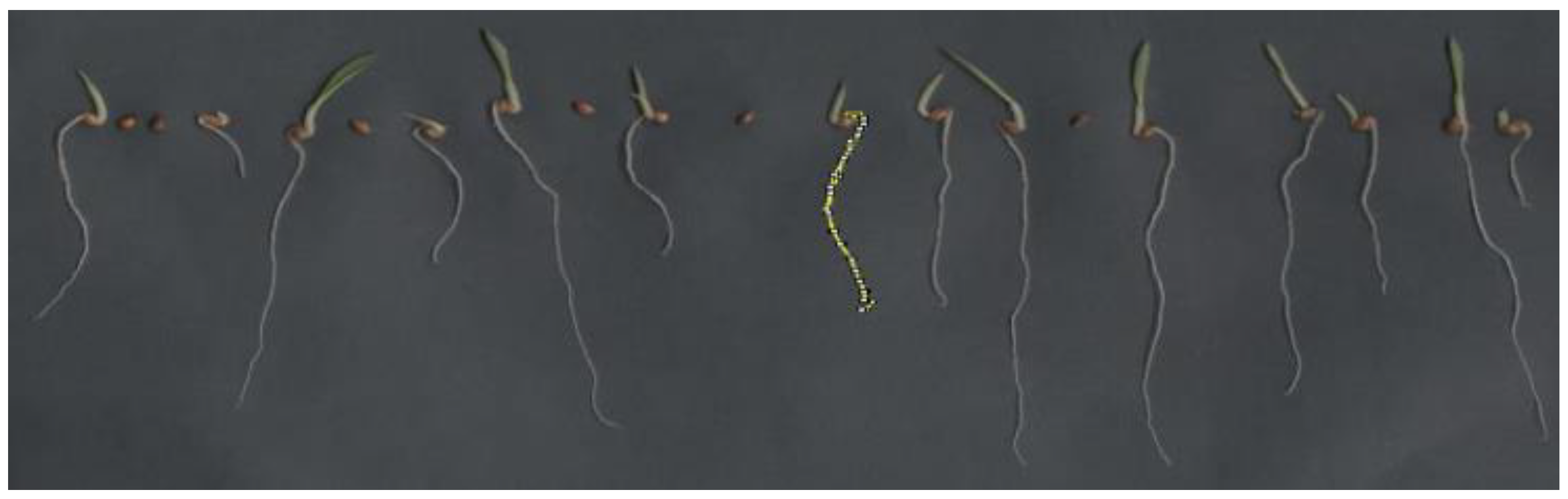
After seven days post-sowing, we moved normal and abnormal seedlings, along with dead seeds, onto black A3 paper. We captured images using an HP Scanjet 2004 scanner, which was adapted in an inverted manner within an aluminum box (see
Figure 5). The images were scanned at 300 dpi and stored for later analysis.
Figure 4.
Image capture system for seedlings.
Figure 4.
Image capture system for seedlings.
We individually measured the length of each seedling using the ImageJ® software tool. This tool enabled us to outline specific parts of the analyzed material, as indicated in yellow in
Figure 5. The length was expressed in millimeters per seedling.
According to the seeds frequency distribution, we have created four classes based on the internal area occupied by the seed endosperm as follows:
Class I: Seeds with internal area ranging from 0.391 to 1.667 mm².
Class II: Seeds with internal area ranging from 1.668 to 2.944 mm².
Class III: Seeds with internal area ranging from 2.945 to 4.221 mm².
Class IV: Seeds with internal area ranging from 4.222 to 5.497 mm².
We used the dataset from an experiment design with Unbalanced Completely Randomized Design (UCRD). For a seedling length variable y, the data underwent transformation by applying , a necessary step to perform parametric analysis of variance if the homoscedasticity assumption was not met [20]. The variables were then analyzed using analysis of variance (ANOVA) at a 5% significance level. Variables showing significant differences underwent Tukey's test for mean comparison.
Table 1 displays the morphological characteristics of Brachiaria seeds, in which means followed by the same letter (a, b, c or d) in the same column are statistically similar to each other. In addition to seedling traits observed seven days after the vigor test, it was noted that a larger area occupied by the embryo is associated with higher seed germination capacity (
Table 2). Means followed by the same letter in the same column are statistically similar to each other. It is important to mention that various factors, such as mechanical damage during harvest, damage from insects, pathogens, and others, can also impact seed germination.
Total seed area showed that Class IV differed significantly from the other three classes in the Tukey test at a 5% probability, with a value of 8.50 mm². Larger seeds can accommodate a greater endosperm area due to their size. A larger endosperm area provides more reserves for the germination process, increasing the likelihood of germination.
The ratio of internal area to total area (IA/TA) exhibited an increasing trend based on seed class. Class I had the smallest ratio, while Class IV had the largest. Seed length reached its peak in Class I (
Table 1), while the other classes were statistically similar.
Examining seedling length revealed that Class I had smaller seedlings, as the endosperm is the primary supplier of reserves needed for germination in Brachiaria seeds. The other classes did not differ from each other.
In summary, vigor increased with endosperm area classes. Larger endosperm areas indicate greater vigor, as reflected in seedling length. Analyzing seed images through X-ray morphological studies and classifying seeds into different filling levels allows the correlation of morphological characteristics with physiological potential. This highlights the importance of methods that automate this process, providing significant contributions to the seed industry.
2.5. Proposed Method
To assess the strength of Brachiaria brizantha cv Xaraés seeds using X-ray images, we introduced a post-segmentation module to the YOLO model. This module, known as the Morphological Analysis Module (MAM), applies a digital image processing algorithm to calculate various variables: total area (mm²), embryo and endosperm area (mm²), internal area/total area ratio (mm²), length (mm), and width of the seeds (mm).
The area is determined using Green's theorem, implemented numerically in the OpenCV library. Length and width are derived from the seed's angle and the extreme limits of the rotated bounding box, obtained through OpenCV methods based on O'Rourke's et al [
10] and Klee and Laskowski's [
11] algorithms.
Since the measurements are initially in pixels or pixels², the MAM converts these values to mm and mm², respectively, by calibrating the measurement system with a reference measurement. In
Figure 4, an X-ray image on a razor blade with a known measurement of 3.5 mm is marked. By counting the pixels along this measurement, we establish the mm/pixels ratio for the images.
2.5. Experiment Analysis
For the experiment analysis, we used a randomized block design based on the divisions within the X-ray database. This approach aims to reduce bias or confounding factors that might affect our findings. Initially, we evaluated the overall model performance by blocking based on the fold factor. However, for a more detailed assessment at the class level, we maintained the randomized block design, still considering the fold, but this time stratifying by class. To check the normality of the residuals, we conducted the Shapiro-Wilk test [
12], and for assessing homogeneity of variances, we used the O’Neill-Mathews test [13]. While the majority of the analyses in
Section 3 met the assumptions of normality and homogeneity of variances at the 5% significance level, a few did not comply with one or both assumptions. Therefore, for these cases, a non-parametric approach, specifically the Wilcoxon test [14], was employed.
3. Results
We chose the YOLOv8 architecture due to its balanced performance in segmentation accuracy and efficient training times. The model underwent training on the seed X-ray dataset for 500, 1500, and 3000 epochs, covering dataset sizes of 5, 15, and 30 combinations. This training utilized an NVIDIA Tesla T4 GPU with the PyTorch framework.
3.1. Segmentation Performance
The YOLOv8 model achieved a maximum overall Average Precision (AP) of 97.3% in segmenting seeds and endosperm in the X-ray images (see
Table 2). This implies correct delineation and classification of seed and endosperm regions 97.3% of the time.
Table 2 and
Table 3 show the complete segmentation performance results across training epochs and dataset sizes.
Table 2.
YOLOv8 overall segmentation performance (AP, AP50, AP75) by training epochs (500, 1500, 3000) and dataset size.
Table 2.
YOLOv8 overall segmentation performance (AP, AP50, AP75) by training epochs (500, 1500, 3000) and dataset size.
| Dataset Size |
|
AP |
|
|
AP50 |
|
|
AP75 |
|
| 500 |
1500 |
3000 |
500 |
1500 |
3000 |
500 |
15001
|
30001
|
| 5 |
92.8% |
96.3% |
97.2% |
99.1% |
99.2% |
99.3% |
98.5% |
98.6% |
99.2% |
| 15 |
93.7% |
96.9% |
97.3% |
98.8% |
99.5% |
99.4% |
98.0% |
99.1% |
99.0% |
| 30 |
93.2% |
96.7% |
96.6% |
98.9% |
99.1% |
99.2% |
98.4% |
98.7% |
98.8% |
On a per-class level, seeds were identified with a higher AP than endosperm, 98.9% vs. 95.5%, respectively, at 3,000 epochs (refer to
Table 3). Despite this slight difference, endosperm inference performance remained high, with a minimum 91.7% AP at just 500 training epochs. This showcases flexibility in handling variability and visual ambiguities in seed X-rays, particularly in the endosperm area, which varies more than the seed area (see
Figure 6).
The Tukey multiple comparison test found no statistically significant differences in AP metrics across varying epoch levels (p > 0.05), maintaining dataset size fixed. This indicates stability in performance and suggests suitable convergence without overfitting, even for smaller training set sizes.
3.2. Seed Classification and Vigor Prediction
The YOLOv8 model's segmented seed and endosperm areas provide the basis for an automated classification system for seed vigor levels, relying on internal morphology. The size of the endosperm, being a crucial nutrient reserve for the embryo, serves as an indicator of germination potential.
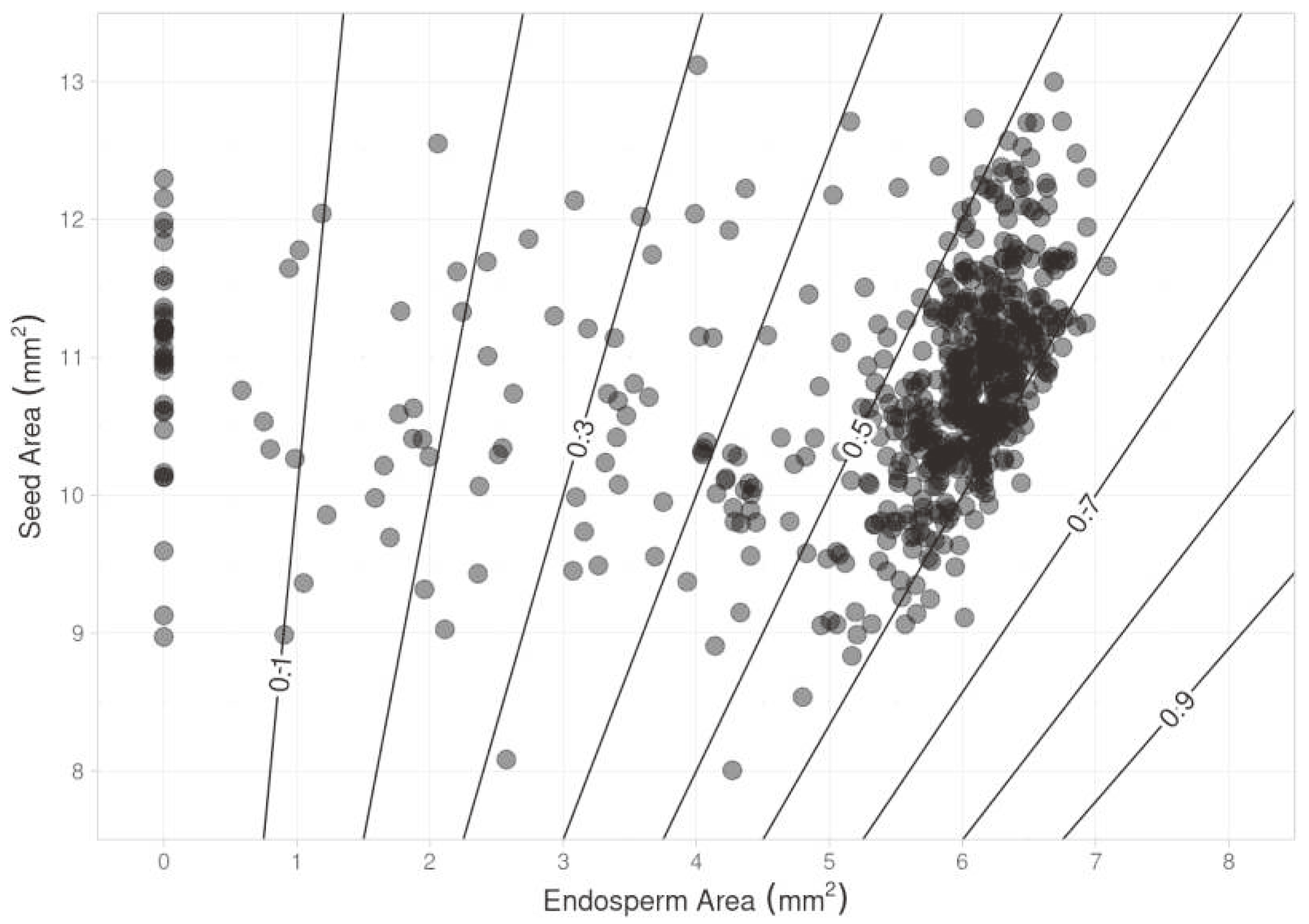
In
Figure 6, we depict the relationship between segmented seed and endosperm areas, with contour levels representing endosperm/seed area ratios—a potential indicator of seed quality. The majority of points fall within the 50-60% ratio range. Notably, no points surpass 70%, establishing an initial benchmark for high vigor classification.
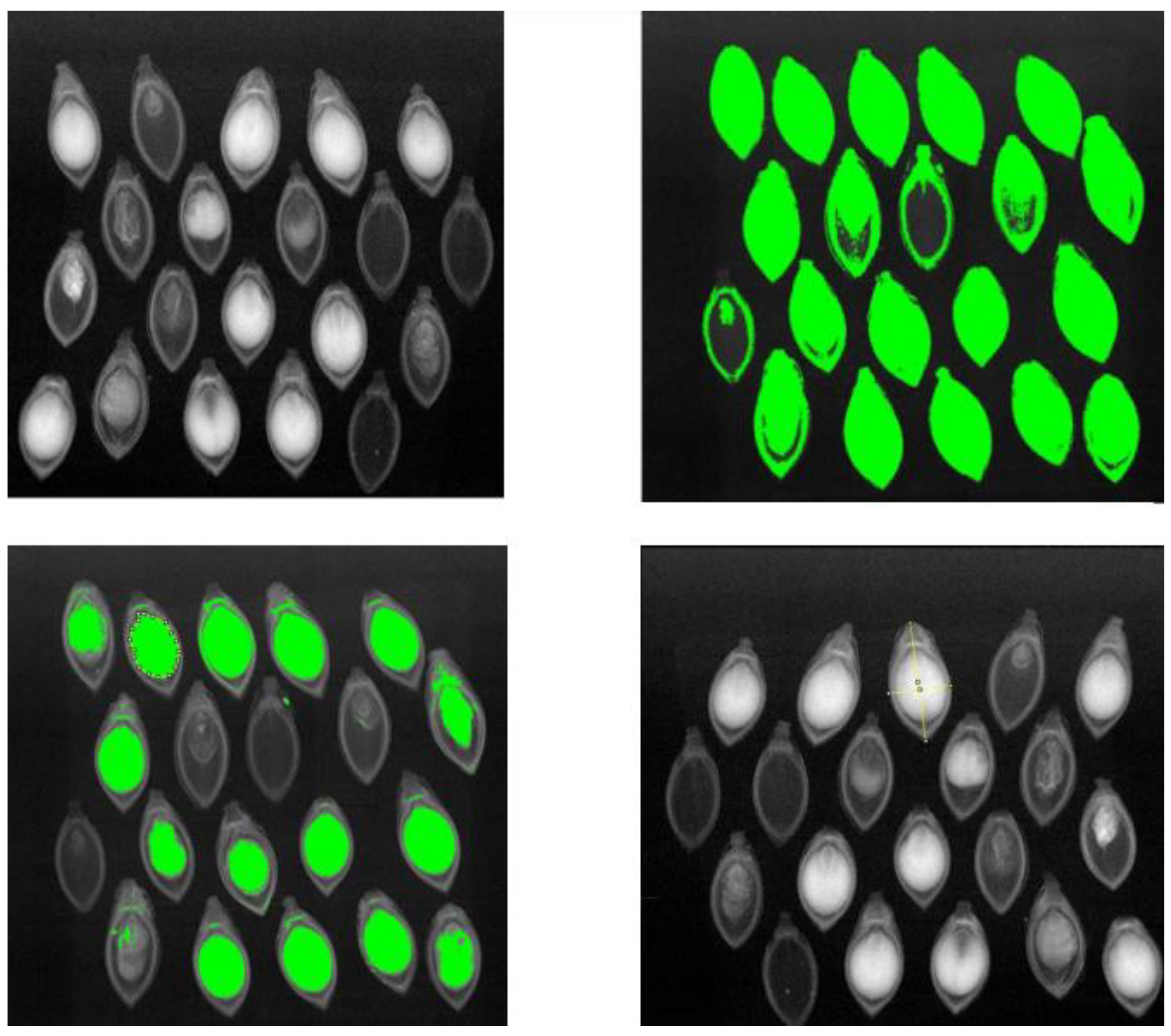
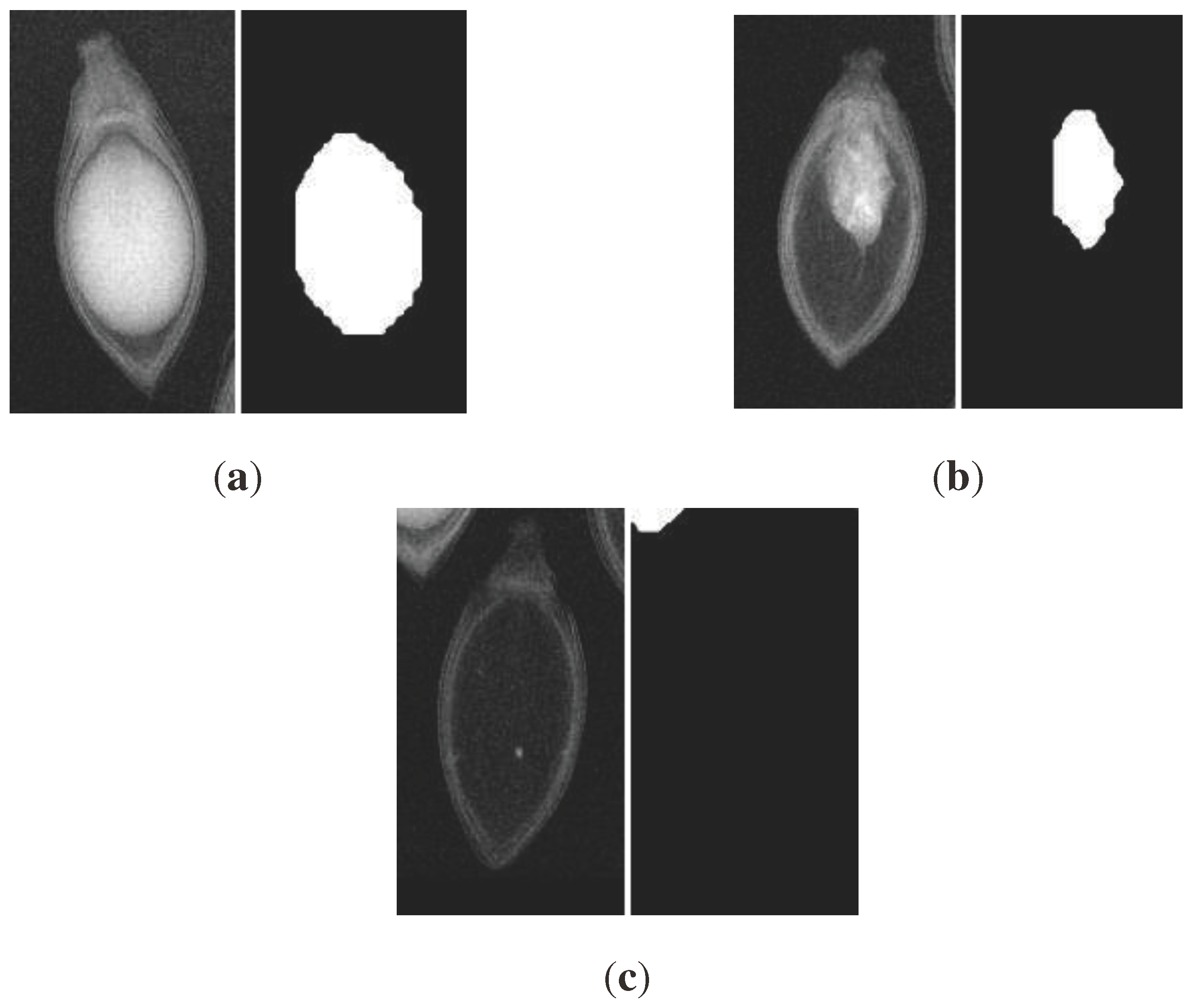
Upon qualitative visual inspection (refer to
Figure 7), a clear and accurate alignment is evident between true seed/endosperm boundaries in X-ray images and segmentation masks predicted by YOLOv8. The model demonstrates proficiency in handling high appearance variation arising from internal structural ambiguities, rotations, and varying endosperm densities.
4. Discussion
The YOLOv8 model showcases impressive segmentation and classification performance even with limited dataset sizes. Given the challenges of compiling specialized agricultural data, the statistical similarity between 500, 1500, and 3000 training epochs suggests robust models that avoid overfitting. Notably, segmentation still reached 97.2% Average Precision (AP) when trained on just 5 seed X-ray images (refer to
Table 1).
This finding is comparable to a computer vision approach used to assess the viability of guavira treated seeds with tetrazolium salt [15], which achieved better precision with 97.90% correct recognition for mucilage and 96.71% for lime. However, it relied on destructive tests. In a vigor test for rice seeds using computer vision techniques [16], a new prediction model was introduced for non-destructive germination forecasting, achieving a high accuracy of 94.17%. This demonstrates that our results outperform other deep learning methods.
The 50-60% endosperm/seed area ratio range, encompassing over 50% of samples (see
Figure 6), offers a clear indication for assessing the viability of seed lots. Although further physiochemical testing can refine category boundaries, this morphological indicator facilitates quick sorting and selective harvesting.
Deep learning, by automatically extracting spatial features predictive of germination, circumvents the need for extensive manual measurement while enhancing consistency compared to subjective human visual assessments.
Table 2 highlights the accuracy of endosperm identification and segmentation, reaching a maximum of 95.6% in 1,500 epochs with 15 training images. The automatic extraction of interpretable morphological features from X-ray scans through deep learning enables swift and reproducible seed sorting without requiring specialized image analysis expertise. This method generalizes well across varying appearances, orientations, and shapes compared to template-matching approaches. Additionally, easy retraining allows for updates in biological classifications as expert knowledge evolves.
5. Conclusion
We suggest using YOLOv8 for studying the internal structure of Brachiaria Brizantha cv. Xaraés seeds through X-ray images to evaluate seed vigor. YOLOv8, with an added post-segmentation module, helps in obtaining quality descriptors for seed batches based on their internal morphology. This process automates the analysis of segmented images, replicating human visual analysis. We used image augmentation to create more training images artificially, employing the ImageDataGenerator API in the Keras library.
Our findings show the here proposed model performs well in segmenting and classifying despite having a relatively small dataset. It achieved up to 95.6% accuracy in identifying and segmenting the endosperm over 1,500 epochs with just 15 training images. The endosperm/seed area ratio, specifically in the 50-60% range, which covers over 50% of the samples, offers a meaningful measure for assessing the viability of seed batches.
As a future step, we plan to develop a user-friendly web application. This application aims to be a valuable tool in Agricultural Engineering post-harvest processes. It will assist seed production companies by automatically categorizing seed batches, providing the market with options based on cost-effectiveness.
References
- Kopittke, P.M.; Menzies, N.W.; Wang, P.; McKenna, B. A.; Lombi, E. Soil and the intensification of agriculture for global food security. Environment International, 2019; 132, 105078. [CrossRef]
- Abud, H. F.; Cícero, S. M.; Gomes Junior, F. G. Radiographic images and relationship of the internal morphology and physiological potential of broccoli seeds. Acta Scientiarum. Agronomy, 2018; 40(1), 34950. [CrossRef]
- Simak, M. Testing of forest tree and shrub seeds by X-radiography. p. 1-28. In: Gordon, A.G.; Gosling, P.; Wang, B.S.P. Tree and shrub seed handbook. ISTA, Zurich, Switzerland, 1991.
- de Freitas, M. N., Dias, M. A. N., Gomes-Junior, F. G., Abud, H. F., de Araújo, L. B., & de Moraes, T. F. (2021). Discrimination of Urochloa seed genotypes through image analysis: Morphological features. Agronomy Journal, 113(6), 4930–4944. [CrossRef]
- Domingues, R.C.; Fruet, G.; Abud, H.F.; Gomes, D.G (2023). Imagens de Raios X e YOLOv8 para Avaliação Automatizada, Precisa e Não Destrutiva da Qualidade de Sementes Braquiária (Urochloa brizantha). In: Congresso Brasileiro de Agroinformática, 2023, Anais do XIV Congresso Brasileiro de Agroinformática (SBIAGRO 2023). https://sol.sbc.org.br/index.php/sbiagro/article/view/26555.
- Bianchini, V.D.J.M.; Mascarin, G.M.; Silva, L.C.A.S.; Arthur, V.; Carstensen, J.M.; Boelt, B.; da Silva, C.B. Multispectral and X-ray images for characterization of Jatropha curcas L. seed quality. Plant Methods, 2021; 17(1). [CrossRef]
- Rahman, A.; Cho, B.K. Assessment of seed quality using non-destructive measurement techniques: a review. Seed Science Research, 2016; 26(4), 285–305. [CrossRef]
- de Oliveira, G. R.F; Mastrangelo, C.B.; Hirai, W.Y.; Batista, T.B.; Sudki, J.M.; Petronilio, A.C.P.; Crusciol, C.A.C.; da Silva, E.A.A. An Approach Using Emerging Optical Technologies and Artificial Intelligence Brings New Markers to Evaluate Peanut Seed Quality. Frontiers in Plant Science, 2022; 13. [CrossRef]
- Glenn J. Ultralytics YOLOv8 (2023) https://github.com/ultralytics/ultralytics.
- Joseph O'Rourke, Alok Aggarwal, Sanjeev Maddila, and Michael Baldwin. An optimal algorithm for finding minimal enclosing triangles. Journal of Algorithms, 7(2):258–269, 1986.
- Victor Klee and Michael C Laskowski. Finding the smallest triangles containing a given convex polygon. Journal of Algorithms, 6(3):359–375, 1985.
- Shapiro, S. S., and M. B. Wilk. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 52, no. 3/4: 591–611, 1965.
- O'Neill, M.E. and Mathews, K. Theory & Methods: A Weighted Least Squares Approach to Levene's Test of Homogeneity of Variance. Australian & New Zealand Journal of Statistics, 42: 81-100, 2000.
- Stuart, Alan L. and William J. Conover. Practical Nonparametric Statistics. International Statistical Review/Revue Internationale de Statistique 40, no. 3: 393–393, 1972.
- Nucci, H.H.P., de Azevedo, R.G., Nogueira, M.C., Costa, C.S., de Oliveira Guilherme, D., Hirokawa Higa, G.T. and Pistori, H. (2023). Use of computer vision to verify the viability of guavira seeds treated with tetrazolium salt. Smart Agricultural Technology, [online] 5, p.100239. [CrossRef]
- Qiao, J., Liao, Y., Yin, C., Yang, X., Tú, H.M., Wang, W. and Liu, Y. (2023). Vigour testing for the rice seed with computer vision-based techniques. Frontiers in Plant Science, [online] 14, p.1194701. [CrossRef]
- Silva, Daniel de Amaral da; Bomfim, Isac Gabriel Abrahão; Braga, Antonio Rafael; Gomes, Danielo G. (2023). Applying Computer Vision Models to Detect in Real Time the Pollen Flow at the Input of Honeybee Hives (Apis mellifera L.). In: Workshop de Computação Aplicada à Gestão do Meio Ambiente e Recursos Naturais (WCAMA), 14. , 2023, p. 21-30. ISSN 2595-6124. [CrossRef]
- Fruet, Gabriel Vasconcelos; Bonfim, Isac Gabriel Abrahão; Domingues, Rafael Capelo; Braga, Antonio Rafael; Gomes, Danielo G. (2023). ApisFlow: a Real-Time Automated Tool to Detect, Classify and Count Honey Bees Castes at the Hive Entrance. In: Workshop de Computação Aplicada à Gestão do Meio Ambiente e Recursos Naturais (WCAMA), 14. , 2023, João Pessoa/PB. p. 1-10. ISSN 2595-6124. [CrossRef]
- Wang, C., Bochkovskiy, A., & Liao, H.M. (2022). YOLOv7: Trainable Bag-of-Freebies Sets New State-of-the-Art for Real-Time Object Detectors. 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 7464-7475. [CrossRef]
- Montgomery, D.C., Peck, E.A., & Vining, G.G. (2013). Introduction to Linear Regression Analysis. Wiley Series in Probability and Statistics. Wiley. pp. 172-175.
- Yang, W., Wu, J., Zhang, J, Gao, K., Du, R., Wu, Z., Firkat, E., Li, D. (2023) Deformable convolution and coordinate attention for fast cattle detection. Computers and Electronics in Agriculture, Volume 211, 108006, ISSN 0168-1699. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).