1. Introduction
Hospital information systems (HIS) are primarily intended for use in patient care to support the care process and enable legally required documentation. However, the patient data stored in HIS holds more value. Patient data can - if permissible by applicable privacy regulations - be utilized in secondary use scenarios, including, in particular, biomedical research and quality assurance [2].
Translational biomedical research using routine clinical data depends on sufficiently sized, representative data sets. Researchers therefore benefit from accessing data pools from multiple organizational units such as departments, clinics, etc., which, in practical implementations of HIS that comprise multiple subsystems with heterogeneous levels of technical and organizational integration, implies merging data from various technically separate source systems. This can pose challenges even for secondary data usage within single institutions like a university hospital [3]. Effectively utilizing data across institutional boundaries is even more challenging. This challenge has been met in Germany through the Medical Informatics Initiative [1], which is funded by the Federal Ministry of Education and Research. The Medical Informatics Initiative creates national standards for patient consent [4], FHIR-based nationally harmonized clinical data representation [5], and nationally standardized contractual templates and organizational procedures to foster collaborative cross-institutional data exchange and usage [6].
While the nationally standardized interoperability specifications constrain data representation for data exchange via FHIR profiling [7] to specific terminologies or often even value sets defined as subsets of internationally standardized terminologies and ontologies to facilitate plug-and-play interoperability to the extent possible, it is difficult to map specific items from local routine clinical documentation to the relevant terminology/ontology concepts in a way that actually allows cross-institutional data usage.
This is, to a large part, due to the intrinsic ambiguity involved in annotating routine clinical data with terminologies and/or ontologies of sufficient descriptive power and scope to describe available clinical data effectively [
8]. While standardized terminologies and ontologies like SNOMED CT, LOINC and ATC [
9,
10,
11] help to describe the semantics of medical data clearly and unambiguously once annotation has been performed, thus supporting cross-system and cross-institutional data use, the annotation step itself can be extremely resource intensive. Annotation calls for a rare combination of clinical, technical, and terminological expertise since it often requires resolution of ambiguities resulting from, e.g., non-uniqueness of potential mappings to various levels of ontological hierarchies and relational and representational systems. This may require judgment calls that take into account the context of clinical data acquisition, which is typically specific to the contributing site or even organizational sub-unit, the technical consequences of specific choices, and, to the extent possible, the implications of specific choices for the downstream processing and analysis steps in generic secondary use contexts. For example, Kors et al. [
12] annotated biomedical data using a subset of the Unified Medical Language System [
13] to provide a gold-standard of annotated data which future efforts can build upon. While a pre-annotation step could be performed automatically, human annotators were still needed to choose from several possible annotation options and to make the final annotation choices. Automatic annotation alone currently could not provide satisfactory results. Consequently, improving annotation workflows is an active area of research and didactic activities [
14,
15].
In practice, semantic annotation is often performed manually without task-specific tooling, using, e.g. spreadsheet tables in combination with various online resources such as the SNOMED CT browser [
16] or RELMA [
17] to search the terminologies. This exacerbates the challenges outlined above and introduces additional, avoidable sources of error.
Hypothesizing that innovative, task-specific annotation tooling with modern collaborative features could contribute to a more efficient and effective annotation process to support cross-site data usage scenarios, we developed a prototype of a web based, collaborative annotation tool (CoAT) for semantic annotation of medical data and evaluate its usability in intra- and cross-site application scenarios. By proposing this new, collaborative approach and with CoAT providing and evaluating a prototypical implementation, we aim to both provide first evidence for the feasibility and usefulness of this approach and facilitate further research in this direction by making our solution available as open source.
2. Materials and Methods
Prototype conception and development was preceded by a systematic requirements analysis for the annotation tooling. The results from the requirements analysis were formulated as user stories and grouped into “must”, “should” and “could” requirements. Following an agile approach, the development took place in three development cycles. Each cycle focused on a defined set of user stories and produced new requirements for the next development cycle.
The web app was implemented in Java SE 11 using the Spring Boot framework as a fast and easy way to benefit from the advantages of a Spring application. Vaadin 14 was used to develop a modern web graphical user interface with Java code using the framework’s built-in UI components.
We implemented three primary features going beyond the feature set of typical tooling choices for semantic annotation:
Since only metadata, but no patient specific data is to be processed, the implementation poses no risks to patient privacy if used as intended. Risks to staff privacy were minimized by limiting the processing of staff related data items to those strictly necessary to allow for authentication and authorization of users.
The prototype was first evaluated regarding its usability in an intra-site application in Bonn. Three test users were recruited: one physician, one nurse, and one medical documentary. To evaluate the usability, these test users performed think-aloud tests [
23] where they had to complete a given set of tasks in the tool which covered the whole annotation process including usage of the collaborative features. While doing so, the test users verbalized their thoughts and were recorded. The records were transcribed to document where users encountered challenges. The test lead judged the user’s success or failure in the completion of the tasks based on pre-defined expected results.
To evaluate the value of collaborative annotation features and pre-existing semantic annotation data in practice, the system was rolled out to two more sites in the SMITH consortium, Jena and Leipzig. At the Jena University Hospital (JUH), the Collaborative Annotation Tool was evaluated with several real-world value sets from intensive care medicine and laboratory diagnostics. Leipzig University used a real-world scenario from the MII overarching use case POLAR [
24], a feasibility query selecting a number of patient’s characteristics, laboratory values and medications for evaluating the tool. In addition, the tool was also tested with medical informatics students on the same query in order to find out whether specialists without many years of expertise in medical terminology can also make a meaningful initial annotation.
3. Results
3.1. Requirements Analysis
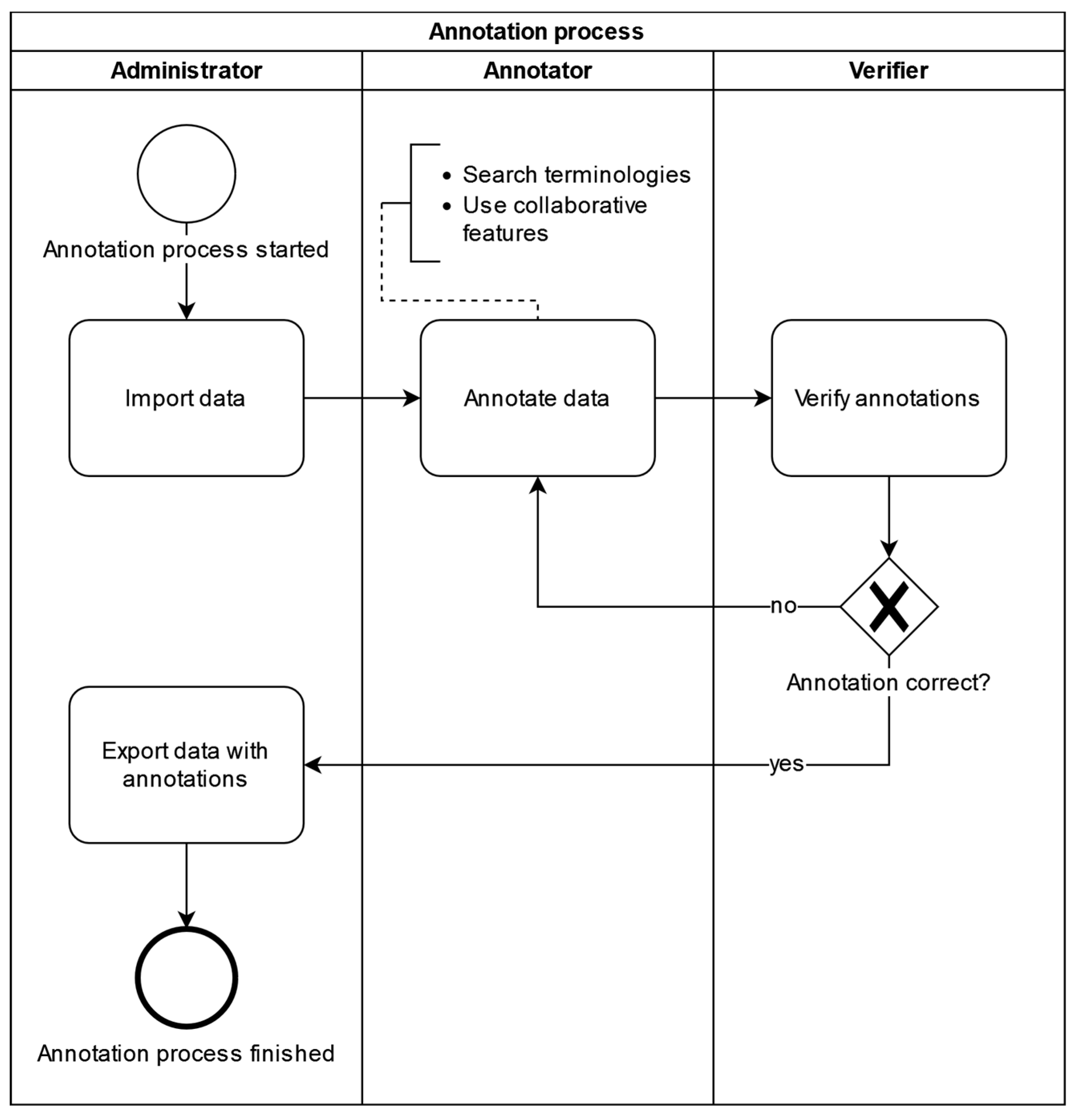
The requirements analysis resulted in functional as well as non-functional (e.g. technical) requirements for the annotation tool. Requirements were formulated and documented as user stories. The requirements analysis also established a data annotation workflow that should be supported by the annotation tool (
Figure 1).
Users are assigned the roles of Administrator, Annotator, or Verifier according to their responsibilities for different parts of the annotation workflow. Each user, along with basic identifying data used for authentication purposes, is assigned a location identifying the site in multicentric collaborative annotation settings like those typical of the MII, where many university hospitals collaborate in and contribute to a national data sharing infrastructure. Administrators start the process by importing medical metadata, typically consisting of local item identifiers uniquely identifying source system and data item, along with any descriptive information available from the source context, into the annotation tool using a CSV file. Imported metadata is tagged with the location of the uploading administrator to enable display of provenance information during the annotation process and support the annotator in assessing applicability and validity of previous annotation choices for the annotation task at hand; in addition, location assignment controls annotator and verifier work allocation - by default, annotators and verifiers are presented only with annotation tasks originating from their location. Annotators can then search terminologies for fitting codes and use collaborative features provided by the tool to annotate data uploaded to their location. Verifiers accept or deny annotations and finally, administrators can then export data with its annotations as a CSV file.
3.2. Implementation Process and Results
Using Spring Boot and Vaadin 14 allowed for a fast implementation of a first prototype. Development of the prototype focuses on the most important features of the application. At the end of the third development cycle all “must”-requirements had been implemented. The application was then deployed to a virtual machine hosted by University Hospital Bonn equipped with 4 vCPUs and 16GB RAM, of which CoAT required less than 1GB. Based on the technologies chosen for implementation, we expect CoAT to run with sufficient performance and scalability on any typical hospital IT infrastructure.
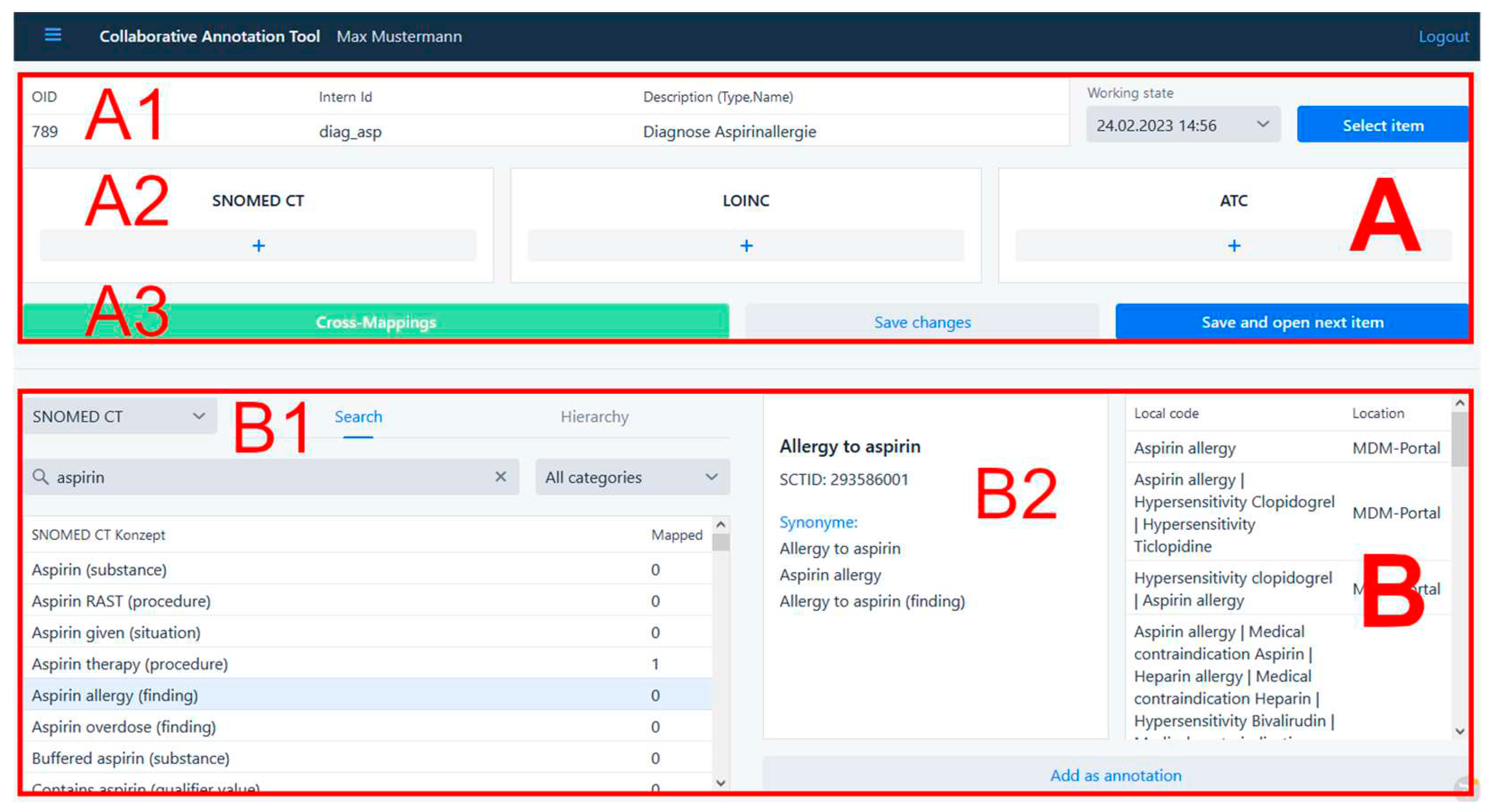
Users with the annotator role can annotate data tagged with their location using the annotation interface (
Figure 2). In the top half of the user interface (A), the actual annotation is performed. At the very top, the data to be annotated is displayed (A1). Each uploaded data item is described by an object identifier, an internal identifier, which typically identifies both source system and data item, and a description. In the three boxes below (A2), codes from the terminologies SNOMED CT, LOINC, and ATC can be added as annotations. Further down is a line of buttons (A3) to use the Cross mapping feature and to save the annotations.
The bottom half of the annotation UI (B) is used to search the terminologies for fitting codes and is further divided into a left and in a right part. On the left (B1), terminologies can be searched by typing in search terms, or, in case of hierarchical terminologies like SNOMED CT, by navigating through the tree structure defined by the hierarchy visualized using the Vaadin Tree Grid widget. Users can click on any search result to obtain detailed information from the terminology, which appears on the right (B2). On the very bottom is a button to directly add the selected search result as an annotation.
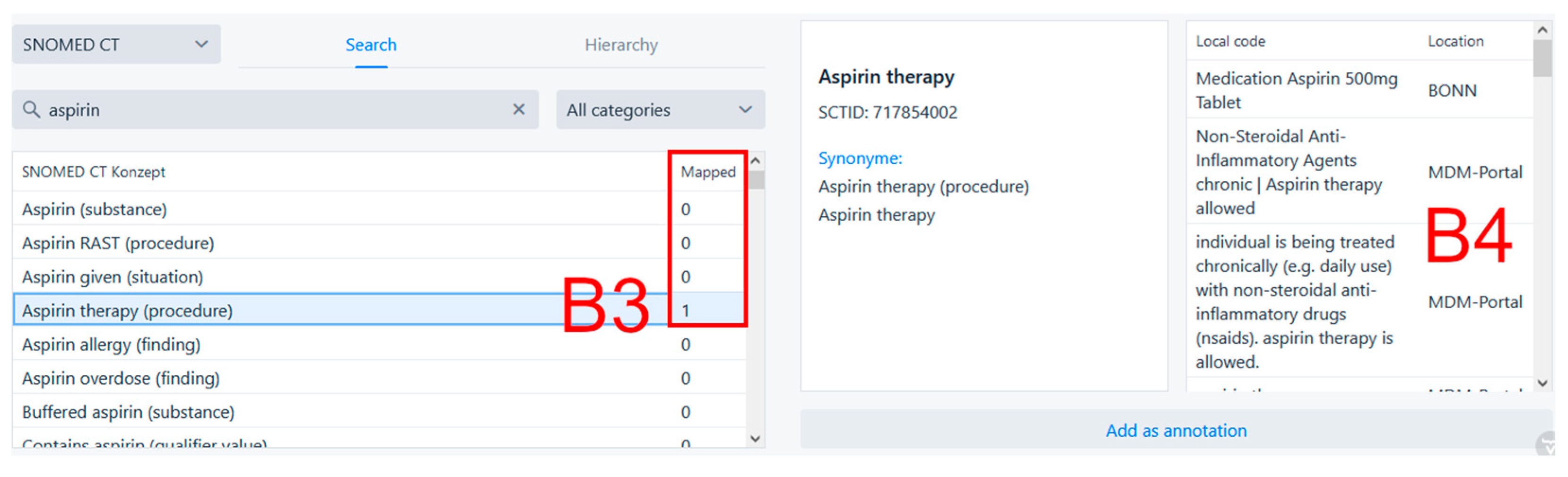
Figure 3 shows the collaborative search feature located in the bottom part of the annotation page. On the left, we can see that searching for “aspirin” in SNOMED CT yielded the result “Aspirin therapy (procedure)”, which has been used to annotate some data once before (
Figure 3, B3) at the Bonn site, as well as in various contexts in the MDM portal. After clicking on the search result, additional information about the search result from the source terminology as well as information about its prior usage in annotation is shown. The table on the very right (
Figure 3, B4) shows what data items in what location were annotated using the selected search result. In this case, the search result was used to annotate the data item “Medication Aspirin 500mg Tablet” at the University Hospital Bonn.
MDM integration via the REST API was achieved as planned thanks to the active support by the MDM operators and provided seamless access from the annotation interface to the more than 500,000 annotated data elements available in MDM. Annotations in the MDM portal are done using UMLS codes. To check for existing annotations in the MDM portal while searching terminologies in the Collaborative Annotation Tool, a selected search result is translated to a set of UMLS codes using the UMLS Metathesaurus. If at least one UMLS code exists, the web service of the MDM portal is called using its RESTful API and provides all data elements annotated using this set of UMLS codes. The annotated data elements from the MDM portal are added to the table with the known annotations as displayed in
Figure 3.
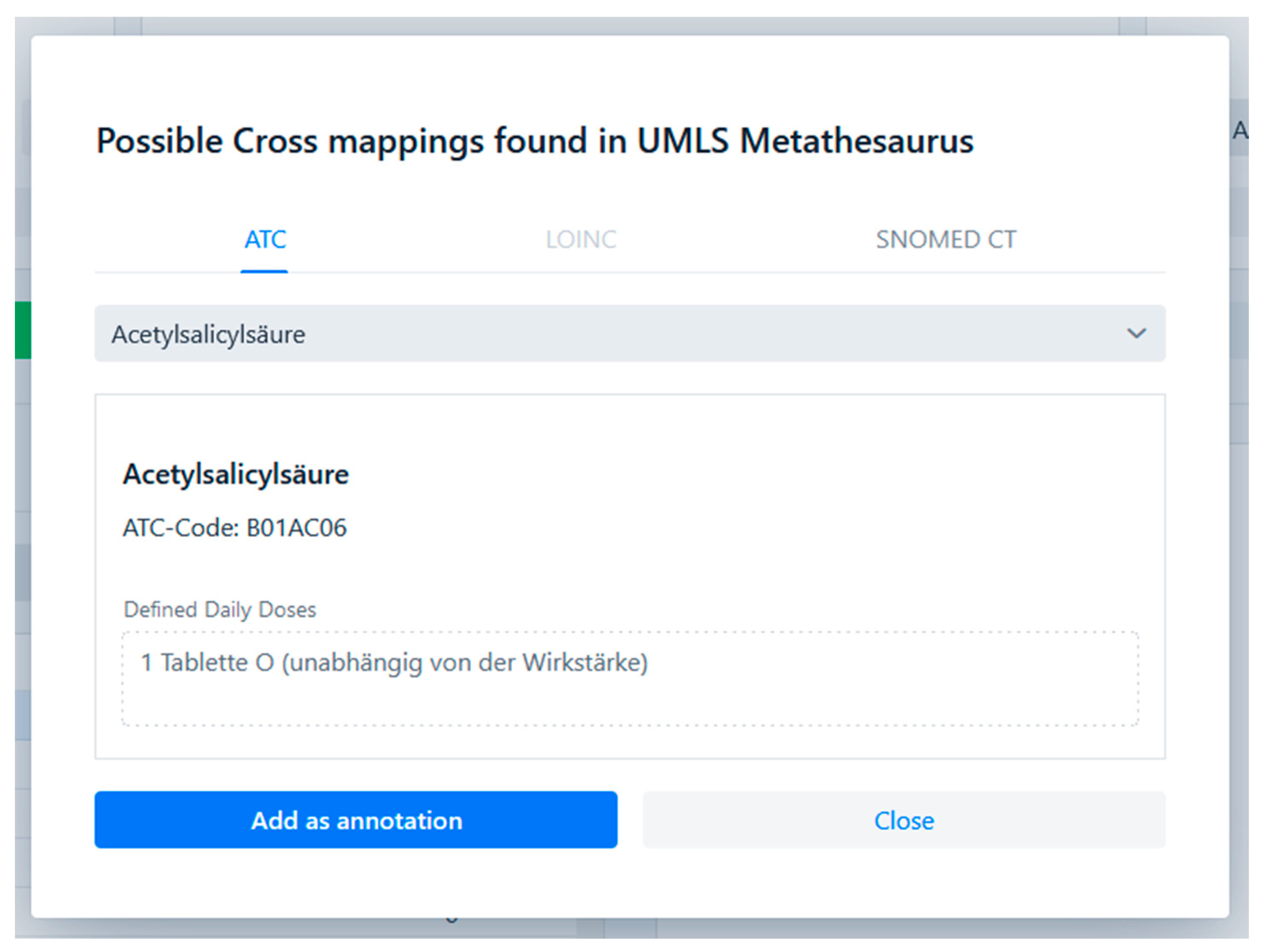
Support for cross-terminological annotation using existing inter-terminology mappings was successfully implemented using the UMLS Metathesaurus as a mapping source. UMLS describes the medical terminology using UMLS concepts and relationships between these. A single UMLS concept describes a single medical term and contains atoms which are codes from different terminologies that are semantically equivalent. Codes that are semantically equivalent to already annotated codes can thus be shown to the user in the CoAT UI. This feature is made accessible to the user with the cross mappings dialogue shown in
Figure 4. When a user annotates a code to a medical data item, the code is looked up in the UMLS Metathesaurus. If a UMLS concept containing this atom and other atoms from the supported terminologies is found, a button to open the cross mapping dialogue is enabled. The dialogue shows all available semantically equivalent codes and allows the user to add them as annotations.
3.3. Usability Evaluation
In
Table 1 the results from the usability evaluation are shown. The tasks the three test users were asked to do cover the key functionalities of the tool. The test users were able to successfully solve most tasks while occasionally encountering problems. The transcripts of the think-aloud tests gave additional insights about the causes of these problems and allowed us to come up with solutions to counter them. To improve the usability of the tool but not over bloat the user interface, additional information to the user was made available by adding small text labels only where necessary and dialog windows that can be opened by clicking a “help” button. Troubled users can also find help in the user guide where all key functionalities are explained.
3.4. Cross-Site Evaluation
3.4.1. Evaluation at Jena University Hospital
Our goal was to map each JUH-internal code from a few example value sets from intensive care medicine and laboratory diagnostics to a LOINC or SNOMED CT code. To achieve this, internal codes were inserted into a CSV file that was uploaded to the tool. The desired LOINC or SNOMED CT codes could be inserted either directly as the numerical code or by searching, e.g., for the SNOMED fully specified name and then choosing the desired code. The extraction of internal codes and the mapping was performed by a semantic interoperability specialist at JUH’s Data Integration Center. The inserted codes had been previously independently validated by a physician from JUH’s department of anesthesiology and intensive care medicine.
Using the export function, the internal codes, each now annotated with a validated international code, were exported into a .csv file. In the example shown in
Table 2, eight JUH-internal codes relating to ECMO and cardiac support procedures were annotated, validated, and exported. The experience with the tool gained at JUH led to systematic feedback to the developers with questions, problem fixes and feature requests, e.g. on provenance of internal codes, categorizing, grouping, and commenting of annotations, export in FHIR ConceptMap format etc.
In summary, with the test at JUH, the basic functionality and usability of the CoAT could be successfully and independently verified. In addition, input from this evaluation contributed to further improvements to CoAT.
3.4.2. Evaluation at the University of Leipzig
We evaluated the tool with three different audiences, each time with the same scenario. The first group consisted of six scientists from our institute, all medical informatics specialists with experience in clinical data and medical terminology. The aim here was to see whether they could be a target group as productive users of the tool, strategically as Verifiers. The second group consisted of 24 Master’s students of medical informatics as part of a course on information management in clinical research in Leipzig. They had minimal prior training in the field of medical documentation and medical terminologies. We wanted to determine how well "untrained" staff can carry out initial annotation and at the same time show the students how medical terminology is used in practice using a realistic example. The third group consisted of 75 participants in a further training course: the TMF School [
25] is an annual training programme run by TMF e.V. and is aimed at experienced researchers and employees in research networks. These include not only medical researchers and medical IT specialists, but also project managers, bioinformaticians, lawyers, etc. In addition to the training effect, it was particularly interesting to see whether there would be any differences in the annotations compared to the students.
The data set to be annotated comprised 19 data elements. It was part of a genuine research project to recognize potentially inadequate medication in multimorbid patients. It is relatively simple in structure and includes demographic data, laboratory values and medication administration. Users were provided with brief documentation on how to use the tool and one value was annotated together as an example. The actual annotation work was scheduled to take 30 minutes.
As a result, users found the tool helpful and illustrative. However, not everyone was able to use it without help. The estimated time window of 30 minutes proved to be far too short. In addition, it was not a formal, systematic evaluation. Some participants obviously used the time to playfully familiarize themselves with the available functions, as the annotations were chosen randomly. Among the "meaningful" annotations, it is noticeable that many users obviously trust the string-based search and sorting, especially for more complex terminologies. For example, the SNOMED CT concept "Age (qualifier value)" is listed prominently in first place for "age" and is therefore selected most frequently, although other concepts like “Current chronological age (observable entity)” appear more suitable. This was not the case for LOINC annotations, where only one person choose “43993-5 | Age at delivery“ instead of “30525-0 | Age“.
4. Discussion
4.1. CoAT Performance Relative to Identified User Needs and Pre-Existent Tooling
Overall, the implementation of identified requirements proved successful. Both in formal usability, where evaluation results were comparable to previously reported evaluation of successful implementation tasks using similar implementation methodology [
26,
27], and cross-site evaluation with very heterogeneous user groups ranging from domain experts with additional deep terminology knowledge and experience to students in a summer school setting with no relevant prior experience, user feedback was positive. The observation that first-time users with no relevant prior experience found the annotation process challenging without further training or support appears unsurprising given the complexity of the task, independent of the tooling [
28]. That even this user group was able to achieve annotation results in a relatively short time frame with multiple target terminologies appears to support the validity of the approach of providing an integrated annotation environment with workflow and cross-terminology mapping features with regard to supporting low-threshold annotation.
While still in an early prototype phase, CoAT provides good usability for complex annotation tasks and proved usable to heterogeneous user groups in a multicentric setting, suggesting that even independently from the innovative collaborative features, further development of CoAT may provide a significant advance over the current state of the art for semantic annotation of medical data.
4.2. Value of Collaborative Features
The collaborative search feature allows annotators to find search results which have previously been used for annotation more efficiently by making previous annotation choices immediately transparent. Additionally, it allows annotators to compare the already annotated data items to the data item that is currently being annotated and thereby support annotators in their judgment whether the search results semantically fit the data item to be annotated. We believe that, compared to approaches where a user actively needs to search pre-existing annotation databases to identify possibly relevant prior work, this seamless and automatic integration of pre-existing knowledge generated by collaboration partners into the annotation workflow realizes a collaboration modality adapted well to high throughput annotation workflows by implementing the communication fundamental to all collaboration implicitly and seamlessly as an integral part of the workflow itself. Nevertheless, additional support for explicit communication with collaborators may be useful and is discussed below (see Future Work).
When annotating data with codes from multiple terminologies, users have to identify (ideally) semantically equivalent codes in these different terminologies. First-time annotation for a medical data item can be challenging since precise identification of the optimal mapping of source item content in its documentation context to a terminology or possibly complex, hierarchical ontology requires both medical and technical knowledge - the relationship between the medical data item and the annotation in effect corresponds to creating new knowledge by the annotator.
However, annotating other semantically equivalent codes from other terminologies once a primary annotation choice has been made in one terminology or ontology could be much easier as the relationships between codes from different terminologies stay the same, are possibly already available and therefore may allow the user to annotate these equivalent codes. This cross-mapping feature should speed up the annotation process as it reduces the time needed to search for equivalent codes and improve the annotation quality.
The collaborative features were found a welcome addition to the tool by the annotators and show promise with regard to improved annotation quality, annotation consistency across sites, and decreased annotation time but require further evaluation.
4.3. Limitations
The usability testing was limited in scope due to resource constraints, while the results presented regarding collaborative cross-site usage are based on the first cross-site practical application of CoAT in a very limited test scenario which, in the case of the evaluation performed in Leipzig, was performed in a teaching context, preventing more detailed statistical analysis of evaluation results. This naturally limits the generalizability of our results, which will have to be verified in larger application scenarios with direct integration into production data provisioning pipelines.
4.4. Future Work
Future research might be directed to extend the evaluation of the tool and verify the results of this paper. Further improvements to the collaborative features or adding new features altogether could create a larger benefit from already existing annotations, including, but not limited to supporting explicit, context-aware communication with collaboration partners to enable discussing specific annotation challenges. As the tool is just a prototype, continued quality assurance and improvement will need to accompany further development to expand on features like supported terminologies and interoperable, bi-directional integration with production ready data use infrastructures [
29] using standardized interfaces like FHIR [
30]. In particular, support for FHIR terminology services would allow the use of central, up-to-date servers provided by the maintainers also from other terminologies than the three ones referenced in this paper. Equally, the feature of exporting standard formats like FHIR Concept Maps or annotating data dictionary templates from EDC systems like REDCap [
31] or LibreClinica [
32] directly would allow to integrate CoAT in routine processes. Systematic comparison of achievable annotation quality and efficiency with CoAT vs. other existing annotation tools may also provide additional insights into the differential contribution of the proposed collaborative features.
5. Conclusions
The idea of supporting semantic annotation of real world data sources by using a combination of cross-site collaborative annotation sharing integrated with both a large portal supplying annotated data sources and a metathesaurus-based cross-terminology mapping feature proved both realizable and useful. Thus, integration of the CoAT implementation with evolving local and national level data provisioning infrastructure promises gains in both annotation efficiency and quality.
Author Contributions
Conceptualization, Daniel Grigutsch and Sven Zenker; Data curation, Thomas Wiktorin, Andrew Heidel, Frank Bloos, Danny Ammon and Matthias Löbe; Formal analysis, Thomas Wiktorin; Funding acquisition, Sven Zenker; Investigation, Thomas Wiktorin, Andrew Heidel, Frank Bloos, Danny Ammon and Matthias Löbe; Methodology, Thomas Wiktorin, Daniel Grigutsch and Sven Zenker; Project administration, Daniel Grigutsch and Sven Zenker; Resources, Sven Zenker; Software, Thomas Wiktorin; Supervision, Daniel Grigutsch, Felix Erdfelder and Sven Zenker; Validation, Daniel Grigutsch and Sven Zenker; Visualization, Thomas Wiktorin; Writing – original draft, Thomas Wiktorin, Andrew Heidel, Frank Bloos, Matthias Löbe and Sven Zenker; Writing – review & editing, Thomas Wiktorin, Andrew Heidel, Matthias Löbe and Sven Zenker. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Federal Ministry of Education and Research (BMBF) grant numbers 01ZZ1803Q, 01ZZ2303G, 01ZZ2303A and 01KX2121 and Deutsche Forschungsgemeinschaft grant number WI 1605/10-2
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Semler, S.C.; Wissing, F.; Heyder, R. German Medical Informatics Initiative. Methods Inf. Med. 2018, 57, e50–e56. [Google Scholar] [CrossRef] [PubMed]
- Meystre, S.M.; Lovis, C.; Bürkle, T.; Tognola, G.; Budrionis, A.; Lehmann, C.U. Clinical Data Reuse or Secondary Use: Current Status and Potential Future Progress. Yearb. Med. Inform. 2017, 26, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, H.T.; Sabroe, S.; Olsen, J. A Framework for Evaluation of Secondary Data Sources for Epidemiological Research. Int. J. Epidemiol. 1996, 25, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Zenker, S.; Strech, D.; Ihrig, K.; Jahns, R.; Müller, G.; Schickhardt, C.; Schmidt, G.; Speer, R.; Winkler, E.; von Kielmansegg, S.G.; et al. Data Protection-Compliant Broad Consent for Secondary Use of Health Care Data and Human Biosamples for (Bio)Medical Research: Towards a New German National Standard. J. Biomed. Inform. 2022, 131, 104096. [Google Scholar] [CrossRef] [PubMed]
- Arbeitsgruppe Interoperabilität | Medizininformatik-Initiative. Available online: https://www.medizininformatik-initiative.de/de/zusammenarbeit/arbeitsgruppe-interoperabilitaet (accessed on 2 November 2023).
- Arbeitsgruppe Data Sharing | Medizininformatik-Initiative. Available online: https://www.medizininformatik-initiative.de/de/zusammenarbeit/arbeitsgruppe-data-sharing (accessed on 2 November 2023).
- Der Kerndatensatz Der Medizininformatik-Initiative | Medizininformatik-Initiative. Available online: https://www.medizininformatik-initiative.de/de/der-kerndatensatz-der-medizininformatik-initiative (accessed on 2 November 2023).
- Pathak, J.; Wang, J.; Kashyap, S.; Basford, M.; Li, R.; Masys, D.R.; Chute, C.G. Mapping Clinical Phenotype Data Elements to Standardized Metadata Repositories and Controlled Terminologies: The eMERGE Network Experience. J. Am. Med. Inform. Assoc. JAMIA 2011, 18, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Aschman, D.J. Snomed® CT: The Fit with Classification in Health. Health Inf. Manag. J. Health Inf. Manag. Assoc. Aust. 2003, 31, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Forrey, A.W.; McDonald, C.J.; DeMoor, G.; Huff, S.M.; Leavelle, D.; Leland, D.; Fiers, T.; Charles, L.; Griffin, B.; Stalling, F.; et al. Logical Observation Identifier Names and Codes (LOINC) Database: A Public Use Set of Codes and Names for Electronic Reporting of Clinical Laboratory Test Results. Clin. Chem. 1996, 42, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Skrbo, A.; Zulić, I.; Hadzić, S.; Gaon, I.D. [Anatomic-therapeutic-chemical classification of drugs]. Med. Arh. 1999, 53, 57–60. [Google Scholar] [PubMed]
- Kors, J.A.; Clematide, S.; Akhondi, S.A.; van Mulligen, E.M.; Rebholz-Schuhmann, D. A Multilingual Gold-Standard Corpus for Biomedical Concept Recognition: The Mantra GSC. J. Am. Med. Inform. Assoc. JAMIA 2015, 22, 948–956. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, C. The Unified Medical Language System (UMLS) of the National Library of Medicine. J. Am. Med. Rec. Assoc. 1990, 61, 40–42. [Google Scholar]
- Krishnan, R.; Rajpurkar, P.; Topol, E.J. Self-Supervised Learning in Medicine and Healthcare. Nat. Biomed. Eng. 2022, 6, 1346–1352. [Google Scholar] [CrossRef] [PubMed]
- Buendía, F.; Gayoso-Cabada, J.; Sierra, J.-L. An Annotation Approach for Radiology Reports Linking Clinical Text and Medical Images with Instructional Purposes. In Proceedings of the Eighth International Conference on Technological Ecosystems for Enhancing Multiculturality; Association for Computing Machinery: New York, NY, USA, 2021; pp. 510–517. [Google Scholar]
- SNOMED CT Browser. Available online: https://browser.ihtsdotools.org/? (accessed on 10 November 2023).
- RELMA. Available online: https://loinc.org/relma/ (accessed on 10 November 2023).
- Hegselmann, S.; Storck, M.; Geßner, S.; Neuhaus, P.; Varghese, J.; Dugas, M. A Web Service to Suggest Semantic Codes Based on the MDM-Portal. Stud. Health Technol. Inform. 2018, 253, 35–39. [Google Scholar] [PubMed]
- Dugas, M.; Neuhaus, P.; Meidt, A.; Doods, J.; Storck, M.; Bruland, P.; Varghese, J. Portal of Medical Data Models: Information Infrastructure for Medical Research and Healthcare. Database J. Biol. Databases Curation 2016, 2016, bav121. [Google Scholar] [CrossRef]
- Dugas, M. Portal für Medizinische Datenmodelle (MDM-Portal). Available online: https://medical-data-models.org (accessed on 26 November 2023).
- Varghese, J.; Bruland, P.; Zenker, S.; Napolitano, G.; Schmid, M.; Ose, C.; Deckert, M.; Jöckel, K.-H.; Böckmann, B.; Müller, M.; et al. Generation of Semantically Annotated Data Landscapes of Four German University Hospitals.; German Medical Science GMS Publishing House, August 29 2017; p. DocAbstr. 203.
- Schuyler, P.L.; Hole, W.T.; Tuttle, M.S.; Sherertz, D.D. The UMLS Metathesaurus: Representing Different Views of Biomedical Concepts. Bull. Med. Libr. Assoc. 1993, 81, 217. [Google Scholar]
- Mcdonald, S.; Edwards, H.; Zhao, T. Exploring Think-Alouds in Usability Testing: An International Survey. IEEE Trans Prof Commun. 2012, 55, 2–19. [Google Scholar] [CrossRef]
- Scherag, A.; Andrikyan, W.; Dreischulte, T.; Dürr, P.; Fromm, M.F.; Gewehr, J.; Jaehde, U.; Kesselmeier, M.; Maas, R.; Thürmann, P.A.; et al. POLAR – POLypharmazie, Arzneimittelwechselwirkungen und Risiken“ – wie können Daten aus der stationären Krankenversorgung zur Beurteilung beitragen? Prävent. Gesundheitsförderung, 2022. [Google Scholar] [CrossRef]
- TMF-School: Fortbildungsreihe für Verbundforschende | TMF e.V. Available online: https://www.tmf-ev.de/veranstaltungen/tmf-akademie/tmf-school (accessed on 29 November 2023).
- Yusupov, I.; Vandermorris, S.; Plunkett, C.; Astell, A.; Rich, J.B.; Troyer, A.K. An Agile Development Cycle of an Online Memory Program for Healthy Older Adults-ERRATUM. Can. J. Aging Rev. Can. Vieil. 2022, 41, 669. [Google Scholar] [CrossRef] [PubMed]
- Feldman, A.G.; Moore, S.; Bull, S.; Morris, M.A.; Wilson, K.; Bell, C.; Collins, M.M.; Denize, K.M.; Kempe, A. A Smartphone App to Increase Immunizations in the Pediatric Solid Organ Transplant Population: Development and Initial Usability Study. JMIR Form. Res. 2022, 6, e32273. [Google Scholar] [CrossRef] [PubMed]
- Jovanović, J.; Bagheri, E. Semantic Annotation in Biomedicine: The Current Landscape. J. Biomed. Semant. 2017, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Erdfelder, F.; Begerau, H.; Meyers, D.; Quast, K.-J.; Schumacher, D.; Brieden, T.; Ihle, R.; Ammon, D.; Kruse, H.M.; Zenker, S. Enhancing Data Protection via Auditable Informational Separation of Powers Between Workflow Engine Based Agents: Conceptualization, Implementation, and First Cross-Institutional Experiences. In Caring is Sharing – Exploiting the Value in Data for Health and Innovation; IOS Press: Amsterdam, The Netherlands, 2023; pp. 317–321. [Google Scholar]
- HL7 FHIR. Available online: https://www.hl7.org/fhir/ (accessed on 28 November 2023).
- REDCap. Available online: https://www.project-redcap.org/ (accessed on 29 November 2023).
- LibreClinica. Available online: https://www.libreclinica.org/ (accessed on 29 November 2023).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).