Submitted:
16 January 2024
Posted:
17 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
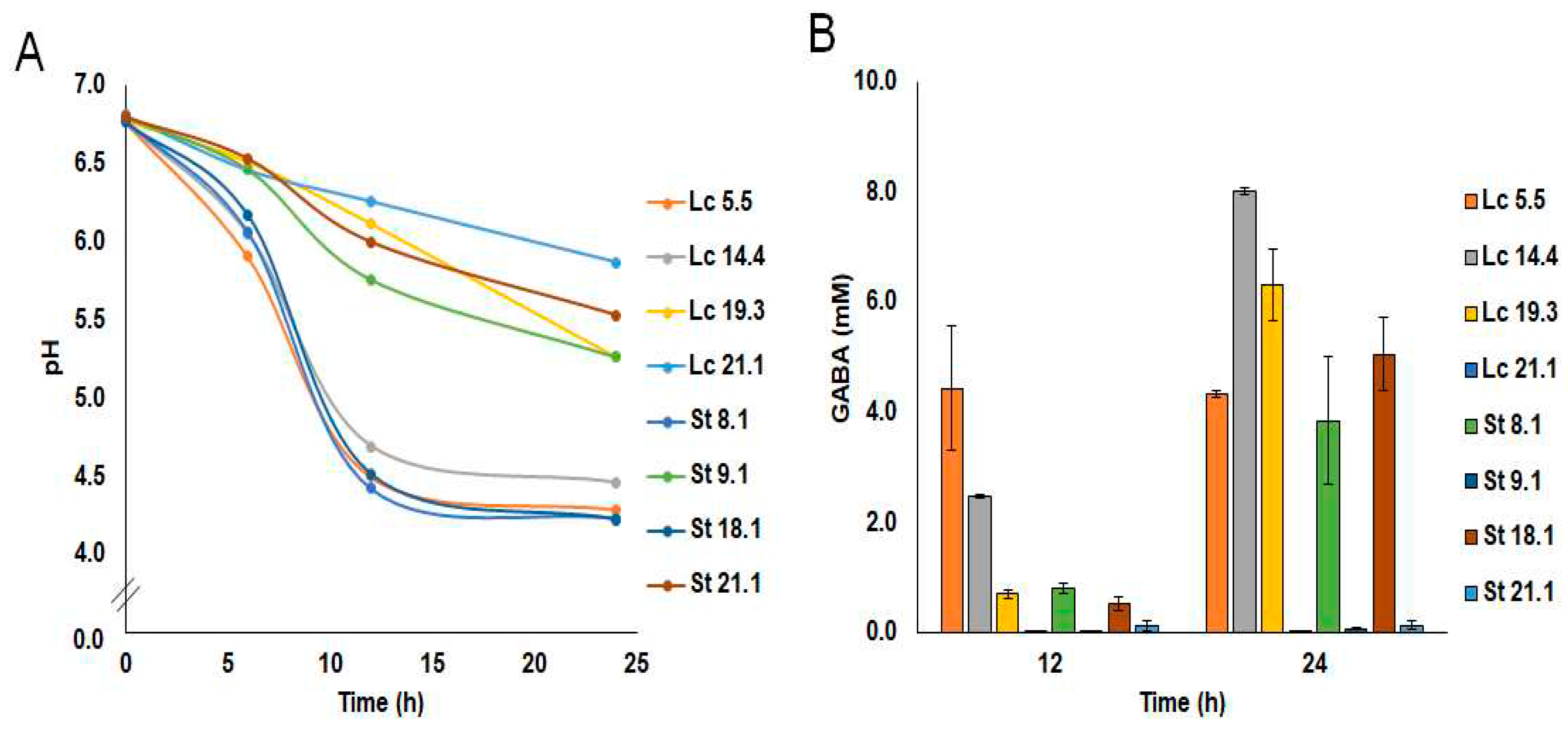
2.1. Molecular Identification and GABA Production
2.2. Biochemical and Technological Characterization
2.3. Safety Evaluation
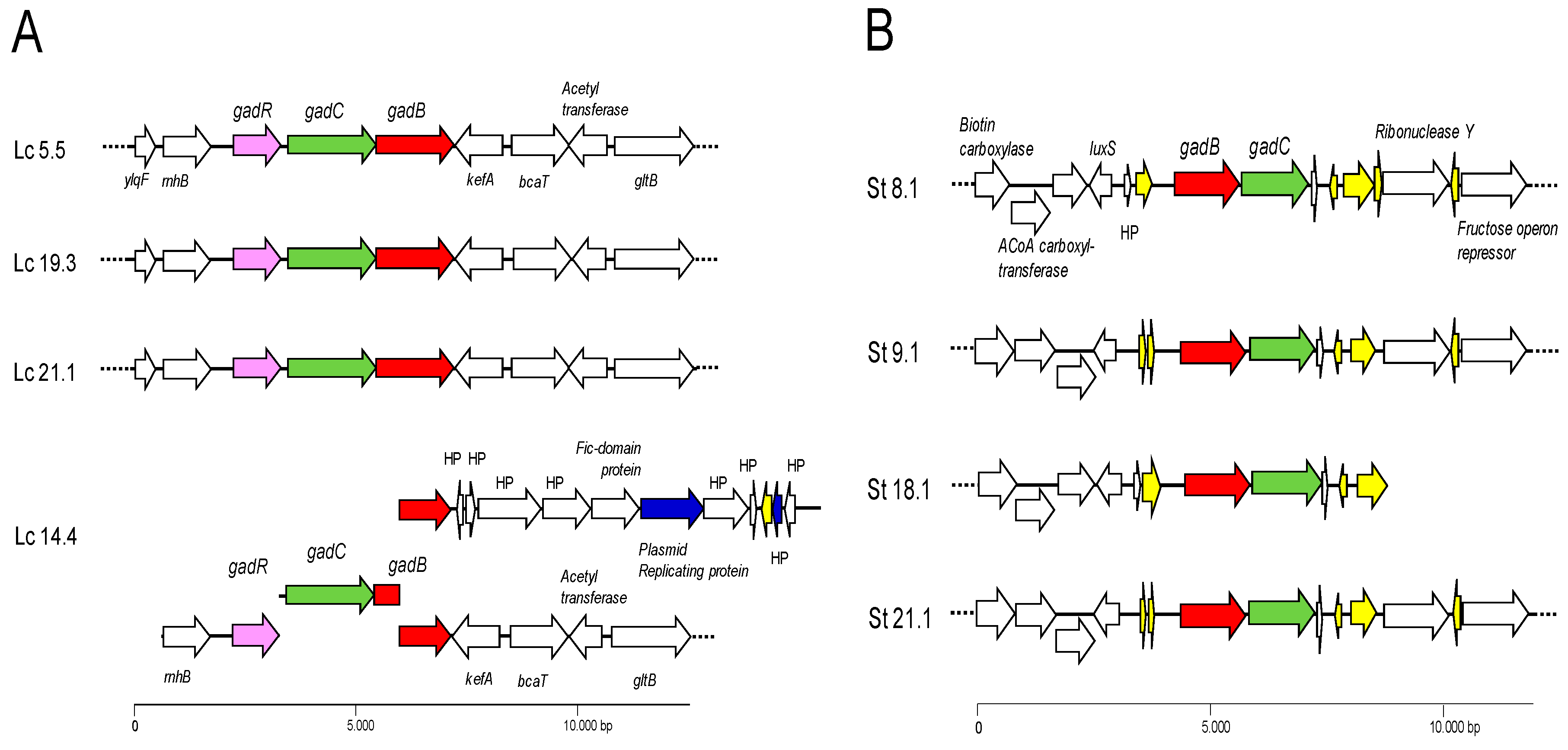
2.4. Genome Sequence and Analysis
3. Discussion
4. Materials and Methods
4.1. Milk Sampling and Bacterial Culture Conditions
4.2. Identification and Typing
4.3. GABA Production and Quantification
2.4. Phenotypic, Technological and Safety Profiles of GABA-Producing Strains
2.4.1. Phenotype Characterization
2.4.2. Growth in Milk and Production of Organic Acids and Volatile Compounds
2.4.3. GABA Production in Milk and Feces
2.4.4. Production of Biogenic Amines
2.4.5. Antibiotic Resistance
4.4.6. Production of Bacteriocins
2.5. Whole Genome Sequencing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hepsomali, P.; Groeger, J.A.; Nishihira, J.; Scholey, A. Effects of oral gamma-aminobutyric acid (GABA) administration on stress and sleep in humans: A systematic review. Front. Neurosci. 2020, 14, 923. [Google Scholar] [CrossRef] [PubMed]
- Ngo, D.H.; Vo, T.S. An updated review on pharmaceutical properties of gamma-aminobutyric acid. Molecules 2019, 24, 2678. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Shirai, T.; Ochiai, H.; Kasao, M.; Hayakawa, K.; Kimura, M.; Sansawa, H. Blood-pressure-lowering effect of a novel fermented milk containing gamma-aminobutyric acid (GABA) in mild hypertensives. Eur. J. Clin. Nutr. 2003, 57, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Pouliot-Mathieu, K.; Gardner-Fortier, C.; Lemieux, S.; St-Gelais, D.; Champagne, C.P.; Vuillemard, J.C. Effect of cheese containing gamma-aminobutyric acid-producing lactic acid bacteria on blood pressure in men. Pharma Nutr. 2013, 1, 141–148. [Google Scholar] [CrossRef]
- Shimada, M.; Hasegawa, T.; Nishimura, C.; Kan, H.; Kanno, T.; Nakamura, T.; Matsubayashi, T. Anti-hypertensive effect of gamma-aminobutyric acid (GABA)-rich Chlorella on high-normal blood pressure and borderline hypertension in placebo-controlled double blind study. Clin. Exp. Hypertens. 2009, 31, 342–354. [Google Scholar] [CrossRef]
- Essa, M.M.; Bishir, M.; Bhat, A.; Chidambaram, S.B.; Al-Balushi, B.; Hamdan, H.; Govindarajan, N.; Freidland, R.P.; Qoronfleh, M.W. Functional foods and their impact on health. J. Food Sci. Technol. 2023, 60, 820–834. [Google Scholar] [CrossRef] [PubMed]
- Yogeswara, I.B.A.; Maneerat, S.; Haltrich, D. Glutamate decarboxylase from lactic acid bacteria-A key enzyme in GABA synthesis. Microorganisms 2020, 8, 1923. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, R.; Bajpai, V.K.; Baek, K.H. Production of GABA (γ-aminobutyric acid) by microorganisms: A review. Braz. J. Microbiol. 2012, 43, 1230–1241. [Google Scholar] [CrossRef]
- Franciosi, E.; Carafa, I.; Nardin, T.; Schiavon, S.; Poznanski, E.; Cavazza, A.; Larcher, R. Tuohy, K.M. Biodiversity and γ-aminobutyric acid production by lactic acid bacteria isolated from traditional alpine raw cow’s milk cheeses. BioMed Res. Inter. 2015, 2015, 25740. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Kimoto, H.; Someya, Y.; Fukurawa, S.; Suzuki, I. Production of γ-aminobutyric acid by cheese starters during cheese ripening. Dairy Foods 1998, 81, 1486–1491. [Google Scholar] [CrossRef] [PubMed]
- Diana, M.; Tres, A.; Llombart, M.; Rafecas, M. Spanish cheese screening and selection of lactic acid bacteria with high gamma-aminobutyric acid production. LWT-Food Sci. Technol. 2014, 56, 351–355. [Google Scholar] [CrossRef]
- Linares, D.M.; O’Callaghan, T.F.; O’Connor, P.M.; Ross, R.P.; Stanton, C. Streptococcus thermophilus APC151 strain is suitable for the manufacture of naturally GABA-enriched bioactive yogurt. Front. Microbiol. 2016, 7, 1876. [Google Scholar] [CrossRef]
- Siragusa, S.; De Angelis, M.; Di Cagno, R.; Rizzello, C.G.; Coda, R.; Gobbetti, M. Synthesis of γ-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl. Environ. Microbiol. 2007, 73, 7283–7290. [Google Scholar] [CrossRef] [PubMed]
- Renes, E.; Linares, D.M.; González, L.; Fresno, J.M.; Tornadijo, M.E.; Stanton, C. Production of conjugated linoleic acid and gamma-aminobutyric acid by autochthonous lactic acid bacteria and detection of the genes involved. J. Funct. Foods 2017, 34, 340–346. [Google Scholar] [CrossRef]
- Sezgin, E.; Tekin, B. Molecular evolution and population genetics of glutamate decarboxylase acid resistance pathway in lactic acid bacteria. Front. Genet. 2023, 14, 1027156. [Google Scholar] [CrossRef] [PubMed]
- Su, M.S.; Schlicht, S.; Gänzle, M.G. Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb. Cell Fact. 2011, 10, S8. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, T.; Hayashi, H.; Abe, K. Exchange of glutamate and gamma-aminobutyrate in a Lactobacillus strain. J. Bacteriol. 1997, 179, 3362–3364. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.; Guinee, T.P.; Cogan, T.M.; and McSweeney, P.L.H. Starter cultures. In Fundamentals of Cheese Science; Fox, P., Guinee, T.P., Cogan, T.M., and McSweeney, P.L.H., Eds.; Springer Nature: New York, USA, 2017; pp. 121–183. [Google Scholar]
- Valenzuela, J.A.; Flórez, A.B.; Vázquez, L.; Vasek, O.M.; Mayo, B. Production of γ-aminobutyric acid (GABA) by lactic acid bacteria strains isolated from traditional, starter-free dairy products made of raw milk. Benef. Microbes 2019, 10, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Nakajima, I.; Fujita, Y.; Kobayashi, M.; Kimoto, H.; Suzuki, I.; Aso, H. Lactococcus lactis contains only one glutamate decarboxylase gene. Microbiol. 2000, 145, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.W.; Leenhouts, K.; Burghoorn, J.; Brands, J.R.; Venema, G.; Kok, J. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol. 1998, 27, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.X.; Xie, C.; Gu, Z.X. Optimisation of fermentative parameters for GABA enrichment by Lactococcus lactis. Czech J. Food Sci. 2009, 27, 433–442. [Google Scholar] [CrossRef]
- Somkuti, G.A.; Renye, J.A., Jr.; Steinberg, D.H. Molecular analysis of the glutamate decarboxylase locus in Streptococcus thermophilus ST110. J. Ind. Microbiol. Biotechnol. 2012, 39, 957–963. [Google Scholar] [CrossRef]
- Pereira, G.V.M.; De Carvalho Neto, D.P.; Junqueira, A.C.D.O.; Karp, S.G.; Letti, L.A.; Magalhães Júnior, A.I.; Soccol, C.R. A review of selection criteria for starter culture development in the food fermentation industry. Food Rev. Int. 2020, 36, 135–167. [Google Scholar] [CrossRef]
- Iskandar, C.F.; Cailliez-Grimal, C.; Borges, F.; Revoll-Junelles, A.M. Review of lactose and galactose metabolism in lactic acid bacteria dedicated to expert genomic annotation. Trends Food Sci. Technol. 2019, 88, 121–132. [Google Scholar] [CrossRef]
- Flórez, A.B.; Vázquez, L.; Rodríguez, J.; Mayo, B. Phenotypic and safety assessment of the cheese strain Lactiplantibacillus plantarum LL441, and sequence analysis of its complete genome and plasmidome. Int. J. Mol. Sci. 2023, 24, 605. [Google Scholar] [CrossRef]
- Hyink, O.; Balakrishnan, M.; Tagg, J.R. Streptococcus rattus strain BHT produces both a class I two-component lantibiotic and a class II bacteriocin. FEMS Microbiol. Lett. 2005, 252, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, A.H.; Bakkes, P.J.; Lubelski, J.; Agustiandari, H.; Kuipers, O.P.; Driessen, A.J. The ABC-type multidrug resistance transporter LmrCD is responsible for an extrusion-based mechanism of bile acid resistance in Lactococcus lactis. J. Bacteriol. 2008, 190, 7357–7366. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic acid bacteria in raw-milk cheeses: From starter cultures to probiotic functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef] [PubMed]
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef] [PubMed]
- Couderc, C.; Laroute, V.; Coddeville, M.; Caillaud, M.A.; Jard, G.; Raynaud, C.; Cocaign-Bousquet, M.; Tormo, H.; Daveran-Mingot, M.-L. Harnessing diversity of Lactococcus lactis from raw goat milk: Design of an indigenous starter for the production of Rocamadour, a French PDO cheese. Int. J. Food Microbiol. 2022, 379, 109837. [Google Scholar] [CrossRef] [PubMed]
- Fusieger, A.; Martins, M.C.F.; de Freitas, R.; Nero, L.A.; de Carvalho, A.F. Technological properties of Lactococcus lactis subsp. lactis bv. diacetylactis obtained from dairy and non-dairy niches. Braz. J. Microbiol. 2020, 51, 313–321. [Google Scholar] [CrossRef]
- Monnet, C.; Nardi, M.; Hols, P.; Gulea, M.; Corrieu, G.; Monnet, V. Regulation of branched-chain amino acid biosynthesis by alpha-acetolactate decarboxylase in Streptococcus thermophilus. Lett. Appl. Microbiol. 2003, 36, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Redruello, B.; Saidi, Y.; Sampedro, L.; Ladero, V.; Del Rio, B.; Álvarez, M.A. GABA-Producing Lactococcus lactis strains isolated from camel’s milk as starters for the production of GABA-enriched cheese. Foods 2021, 10, 633. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.X.; Chen, Z.; Gu, Z.X.; Han, Y. Isolation of γ-aminobutyric acid-producing bacteria and optimization of fermentative medium. Biochem. Eng. J. 2008, 41, 48–52. [Google Scholar] [CrossRef]
- Laroute, V.; Aubry, N.; Audonnet, M.; Mercier-Bonin, M.; Daveran-Mingot, M.L.; Cocaign-Bousquet, M. Natural diversity of lactococci in γ-aminobutyric acid (GABA) production and genetic and phenotypic determinants. Microb. Cell Fact. 2023, 22, 178. [Google Scholar] [CrossRef] [PubMed]
- Nardi, M.; Renault, P.; Monnet, V. Duplication of the pepF gene and shuffling of DNA fragments on the lactose plasmid of Lactococcus lactis. J. Bacteriol. 1997, 179, 4164–4171. [Google Scholar] [CrossRef] [PubMed]
- Barrett, E.; Ross, R.P.; O’Toole, P.W.; Fitzgerald, G.F; Stanton, C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012, 113, 411–417. [Google Scholar] [CrossRef]
- Duranti, S.; Ruiz, L.; Lugli, G.A.; Tames, H.; Milani, C.; Mancabelli, L.; Mancino, W.; Longhi, G.; Carnevali, L.; Sgoifo, A.; Margolles, A.; Ventura, M.; Ruas-Madiedo, P.; Turroni, F. Bifidobacterium adolescentis as a key member of the human gut microbiota in the production of GABA. Sci. Rep. 2020, 10, 14112. [Google Scholar] [CrossRef] [PubMed]
- Bruinenberg, P.G.; De Vos, W.M.; Siezen, R.J. Deletion of various carboxy-terminal domains of Lactococcus lactis SK11 proteinase: Effects on activity, specificity, and stability of the truncated enzyme. Appl. Environ. Microbiol. 2000, 66, 2859–2865. [Google Scholar] [CrossRef] [PubMed]
- Dandoy, D.; Fremaux, C.; de Frahan, M.H.; Horvath, P.; Boyaval, P.; Hols, P.; Fontaine, L. The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS. Microb. Cell Fact. 2011, 10, S21. [Google Scholar] [CrossRef] [PubMed]
- Hayes, F.; Caplice, E.; McSweeney, A.; Fitzgerald, G.F.; Daly, C. pAMbeta1-associated mobilization of proteinase plasmids from Lactococcus lactis subsp. lactis UC317 and L. lactis subsp. cremoris UC205. Appl. Environ. Microbiol. 1990, 56, 195–201. [Google Scholar] [CrossRef]
- Vos, P.; van Asseldonk, M.; van Jeveren, F.; Siezen, R.; Simons, G.; de Vos, W.M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J. Bacteriol. 1989, 171, 2795–2802. [Google Scholar] [CrossRef] [PubMed]
- Juillard, V.; Furlan, S.; Foucaud, C.; Richard, J. Mixed cultures of proteinase-positive and proteinase-negative strains of Lactococcus lactis in milk. J. Dairy Sci. 1996, 79, 964–970. [Google Scholar] [CrossRef]
- Bushin, L.B.; Covington, B.C.; Rued, B.E.; Federle, M.J.; Seyedsayamdost, M.R. Discovery and biosynthesis of streptosactin, a sactipeptide with an alternative topology encoded by commensal bacteria in the human microbiome. J. Am. Chem. Soc. 2020, 142, 16265–16275. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Barba, J.L.; Caballero-Guerrero, B.; Maldonado-Barragán, A.; Jiménez-Díaz, R. Coculture with specific bacteria enhances survival of Lactobacillus plantarum NC8, an autoinducer-regulated bacteriocin producer, in olive fermentations. Food Microbiol. 2010, 27, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Bukowski, M.; Wladyka, B.; Dubin, G. Exfoliative toxins of Staphylococcus aureus. Toxins (Basel) 2010, 2, 1148–1165. [Google Scholar] [CrossRef]
- Lunde, M.; Aastveit, A.H.; Blatny, J.M.; Nes, I.F. Effects of diverse environmental conditions on ΦLC3 prophage stability in Lactococcus lactis. Appl. Environ, Microbiol. 2005, 71, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Desiere, F.; Lucchini, S.; Canchaya, C.; Ventura, M.; and Brüssow, H. Comparative genomics of phages and prophages in lactic acid bacteria. Antonie van Leeuwenhoek 2002, 82, 73–91. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.; González-Guerra, A.; Vázquez, L.; Fernández-López, R.; Flórez, A.B.; de la Cruz, F.; Mayo, B. Isolation and phenotypic and genomic characterization of Tetragenococcus spp. from two Spanish traditional blue-veined cheeses made of raw milk. Int. J. Food Microbiol. 2022, 371, 109670. [Google Scholar] [CrossRef]
- Lanyi, B. Classical and rapid identification: Methods for medically important bacteria. In Methods in Microbiology, Vol. 19; Colwell, R.R., Grigorova, R., Eds.; Academic Press: New York, USA, 1987; pp. 1–67. [Google Scholar]
- Alegría, A.; González, P.; Delgado, S.; Flórez, A.B. Hernández-Barranco, A.; Rodríguez, A. Mayo, B. Characterisation of the technological behaviour of mixtures of mesophilic lactic acid bacteria isolated from traditional cheeses made of raw milk without added starters. Int. J. Dairy Technol. 2016, 69, 1–13. [Google Scholar] [CrossRef]
- Ziadi, M.; Wathelet, J.P.; Marlier, M.; Hamdi, M.; Thonart, P. Analysis of volatile compounds produced by 2 strains of Lactococcus lactis isolated from leben (Tunisian fermented milk) using solid-phase microextraction-gas chromatography. J. Food Sci. 2008, 73, S247–S252. [Google Scholar] [CrossRef]
- Redruello, B.; Ladero, V.; Cuesta, I.; Álvarez-Buylla, J.R.; Martín, M.C.; Fernández, M.; Álvarez, M.A. A fast, reliable, ultra-high performance liquid chromatography method for the simultaneous determination of amino acids, biogenic amines and ammonium ions in cheese, using diethyl ethoxymethylenemalonate as a derivatising agent. Food Chem. 2013, 139, 1029–1035. [Google Scholar] [CrossRef]
- EFSA FEEDAP Panel. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.M.; Kwon, S.J.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
| Species/strain | Organic acid sugar a | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lactose | Glucose | Galactose | Orotic acid | Citric acid | Pyruvic acid | Succinic acid | Lactic acid | Formic acid | Acetic acid | Uric acid | Hippuric acid | |
| L. lactis | ||||||||||||
| Lc 5.5 | 6.68±2.3 | 2.3±0.1 | 44.4±12.2 | 9.2±2.5 | 272.5±86.4 | 7.5±1.7 | 2.1±3.0 | 938.2±224.4 | 7.3±1.4 | - | 3.1±1.0 | - |
| Lc 14.4 | 4.49±0.2 | 1.8±0.1 | 51.4±1.5 | 6.0±0.4 | - | 7.4±2.9 | 2.0±2.8 | 826.3±68.1 | 9.8±3.3 | 73.7±19.1 | 2.3±0.1 | 2.3±0.0 |
| Lc 19.3 | 5.36±1.2 | 1.9±0.1 | 59.7±22.0 | 9.4±2.5 | 218.4±47.7 | 2.8±0.8 | - | 405.7±132.6 | - | 6.6±9.4 | 2.5±0.7 | 0.6±0.3 |
| Lc 21.1 | 7.12±2.5 | 2.5±1.3 | 18.4±5.8 | 10.7±2.9 | 21.5±8.3 | 15.1±2.9 | - | 281.5±62.9 | - | 74.2±13.9 | 2.4±0.6 | 3.2±0.9 |
| S. thermophilus | ||||||||||||
| St 8.1 | 5.05±1.1 | 9.1±2.1 | 982.4±129.1 | 10.5±1.1 | 263.2±43.7 | 8.0±0.3 | - | 1173.3±55.5 | - | - | 2.9±0.4 | 2.6±0.5 |
| St 9.1 | 5.84±1.5 | 32.7±7.4 | 391.9±105.2 | 10.2±2.6 | 251.4±65.4 | 1.0±0.1 | - | 464.0±116.2 | - | - | 2.6±0.7 | 2.1±0.4 |
| St 18.1 | 5.07±3.0 | 9.0±5.4 | 972.2±547.8 | 10.8±5.8 | 265.5±147.3 | 8.3±3.4 | - | 1203.6±617.5 | - | - | 3.0±1.7 | 2.6±1.3 |
| St 21.1 | 6.84±2.3 | 77.0±24 | 365.5±105.8 | 11.2±3.1 | 287.7±92.2 | 1.6±0.1 | - | 375.6±90.4 | - | - | 2.6±0.6 | 2.0±0.5 |
| Milk (control) | 6.53±0.3 | 42.2±8.2 | 282.7±152.4 | 8.5±0.1 | 191.8±1.4 | 1.4±1.0 | - | 313.1±184.2 | - | - | 1.8±0.2 | 1.9±0.3 |
| Compounda | Control milk | L. lactis | S. thermophilus | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Lc 5.5 | Lc 14.4 | Lc 19.3 | Lc 21.1 | St 8.1 | St 9.1 | St 18.1 | St 21.1 | ||
| Alcohols | |||||||||
| 2-Phenyl ethanol | - | 19.13 | 33.26 | - | - | - | - | - | - |
| Isoamyl alcohol | - | 100.86 | 164.49 | - | - | - | - | - | - |
| Carboxylic acids | |||||||||
| Acetic acid | - | 3.63 | 14.36 | 3.28 | 6.2 | 1.39 | 0.84 | 1.63 | 0.64 |
| Benzoic acid | - | 13.44 | 0.86 | 12.26 | 0.57 | 2.55 | 5.19 | 0.95 | 4.24 |
| Butanoic acid | 0.26 | 4.43 | 3.85 | 4.67 | 2.39 | 6.09 | 5.09 | 6.38 | 4.89 |
| Hexanoic acid | 1.07 | 33.09 | 31.86 | 33.35 | 20.27 | 38.75 | 35.79 | 39.55 | 34.24 |
| Heptanoic acid | - | 0.98 | 1.01 | 1.04 | - | 0.85 | 0.81 | 0.94 | 0.81 |
| Octanoic acid | 1.87 | 35.99 | 37.83 | 36.51 | 30.9 | 41.25 | 40.64 | 42.48 | 39.72 |
| n-Decanoic acid | 2.62 | 20.69 | 21.14 | 19.67 | 18.07 | 21.6 | 20.34 | 22.58 | 20.23 |
| 9-Decenoic acid | - | 2.27 | 2.37 | 2.32 | 1.82 | 2.8 | 2.54 | 2.8 | 2.47 |
| Nonanoic acid | - | 0.48 | 0.71 | 0.7 | 0.56 | 0.63 | 0.76 | 0.84 | 0.73 |
| Dodecanoic acid | 0.52 | 3.62 | 3.48 | 3.11 | 2.73 | 4.38 | 3.64 | 4.09 | 4.05 |
| Tetradecanoic acid | - | - | - | - | - | 1.04 | 0.82 | 0.95 | 0.88 |
| Isovaleric acid | - | 14.99 | 31.97 | 7.96 | - | - | - | - | - |
| Ketones | |||||||||
| Acetoinb | - | - | 17.02 | - | 25.54 | 13.58 | 8.31 | 12.81 | 9.82 |
| 2-Heptanone | 2.67 | - | - | - | - | 3.86 | 3.26 | 4.42 | 3.38 |
| 2-Nonanone | 1.85 | 2.12 | 1.9 | 2.05 | 3.06 | 2.56 | 2.6 | 2.96 | 2.52 |
| 2-Pentadecanone | - | - | 0.55 | - | - | - | - | - | - |
| 2-Tridecanone | 0.37 | 0.52 | 0.58 | 0.51 | 0.47 | 0.56 | 0.65 | 0.66 | 0.65 |
| 2-Undecanone | 0.86 | 1.09 | 1.17 | 1.28 | 1.2 | 1.45 | 1.41 | 1.54 | 1.44 |
| 4-Octanone | - | 40.7 | 79.5 | 20.59 | - | - | - | - | - |
| 2,5-Dimethyl-3-hexanone | - | - | 3.99 | 0.67 | - | - | - | - | - |
| Lactones | |||||||||
| δ-Decalactone | 0.35 | 0.66 | 0.7 | 0.66 | 0.62 | 0.73 | 0.64 | 0.71 | 0.61 |
| γ-Dodecalactone | 0.35 | 0.45 | 0.43 | 0.4 | 0.4 | 0.47 | 0.66 | 0.48 | 0.5 |
| δ-Dodecalactone | 0.44 | 0.69 | 0.71 | 0.63 | 0.61 | 0.83 | 0.74 | 0.78 | 0.76 |
| Other | |||||||||
| Benzaldehyde | 0.60 | 1.49 | 1.63 | 1.35 | 0.72 | 0.77 | - | 0.81 | - |
| Phenyl acetaldehyde | 0.34 | 11.12 | 11.73 | 4.56 | - | - | - | - | |
| 2,3-Heptanedione | - | - | 1.72 | 0.89 | - | - | - | - | - |
| 5-Methyl-2-phenyl-2-hexenal | - | 1.45 | 1.62 | - | - | - | - | - | - |
| Property/protein-encoding genes | Species and strains | |||||||
|---|---|---|---|---|---|---|---|---|
| L. lactis | S. thermophilus | |||||||
| Lc 5.5 | Lc 14.4 | Lc 19.3 | Lc 21.1 | St 8.1 | St 9.1 | St 18.1 | St 21.1 | |
| Genome size (bp) | 2,718,046 | 2,652,084 | 2,575,310 | 2,433,628 | 1,806,293 | 1,761,434 | 1,807,628 | 1,794,253 |
| G+C content | 35.17 | 35.16 | 35.17 | 34.79 | 39.77 | 39.40 | 39.89 | 39.47 |
| No. of contigs | 95 | 73 | 84 | 104 | 46 | 49 | 54 | 45 |
| No. of coding sequences | 2,882 | 2,762 | 2,721 | 2,558 | 2,043 | 1,989 | 2,043 | 2,041 |
| No. of BV-BRC subsystems | 207 | 207 | 208 | 204 | 202 | 202 | 202 | 202 |
| Penicillin binding proteins | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Efflux-related proteins | 28 | 24 | 29 | 27 | 27 | 34 | 28 | 34 |
| Virulence Factors (VFDB/Victors) | 2/8 | 2/9 | 1/8 | 1/7 | 1/39 | 1/41 | 1/39 | 1/41 |
| Antibiotic Resistance (BV-BRC/CARD) | 26/2 | 26/2 | 27/2 | 26/2 | 24/0 | 24/0 | 24/0 | 24/0 |
| Resistance to heavy metals | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 2 |
| rRNA sequences (23S+16S+5S) | 4 (1+1+2) | 4 (1+1+2) | 4 (1+1+2) | 4 (1+1+2) | 3 (1+1+1) | 2 (1+1+0) | 3 (1+1+1) | 2 (1+1+0) |
| tRNA molecules | 52 | 50 | 50 | 51 | 37 | 30 | 35 | 30 |
| Proteases | 12 | 15 | 12 | 15 | 11 | 11 | 11 | 11 |
| Peptidases | 26 | 27 | 27 | 26 | 25 | 23 | 26 | 23 |
| Transposases/integrases/excisionases | 27 | 18 | 24 | 24 | 25 | 24 | 24 | 26 |
| Phage-derived proteins | 333 | 245 | 252 | 225 | 7 | 5 | 6 | 61 |
| Plasmid replication proteins | 13 | 11 | 14 | 12 | 1 | 1 (pUB110-type) | 1 | - |
| CRISPR-Cas loci | 0 | 0 | 0 | 0 | 3 | 3 | 3 | 3 |
| Bacteriocins | Lactococcin B and Q, sactipeptides | Enterolysin A, lactococcin A, B, and Q, sactipetides | 2 x lactococcin B, linaridin | Lactococcin B, sactipeptides | BhtR, BlpD, BlpK, BlpU, streptide | BlpD, BlpK, BlpU, streptide | BlpD, BlpK, BlpU, streptide | BlpD, BlpK, BlpU, streptide |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





