Submitted:
03 January 2024
Posted:
17 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
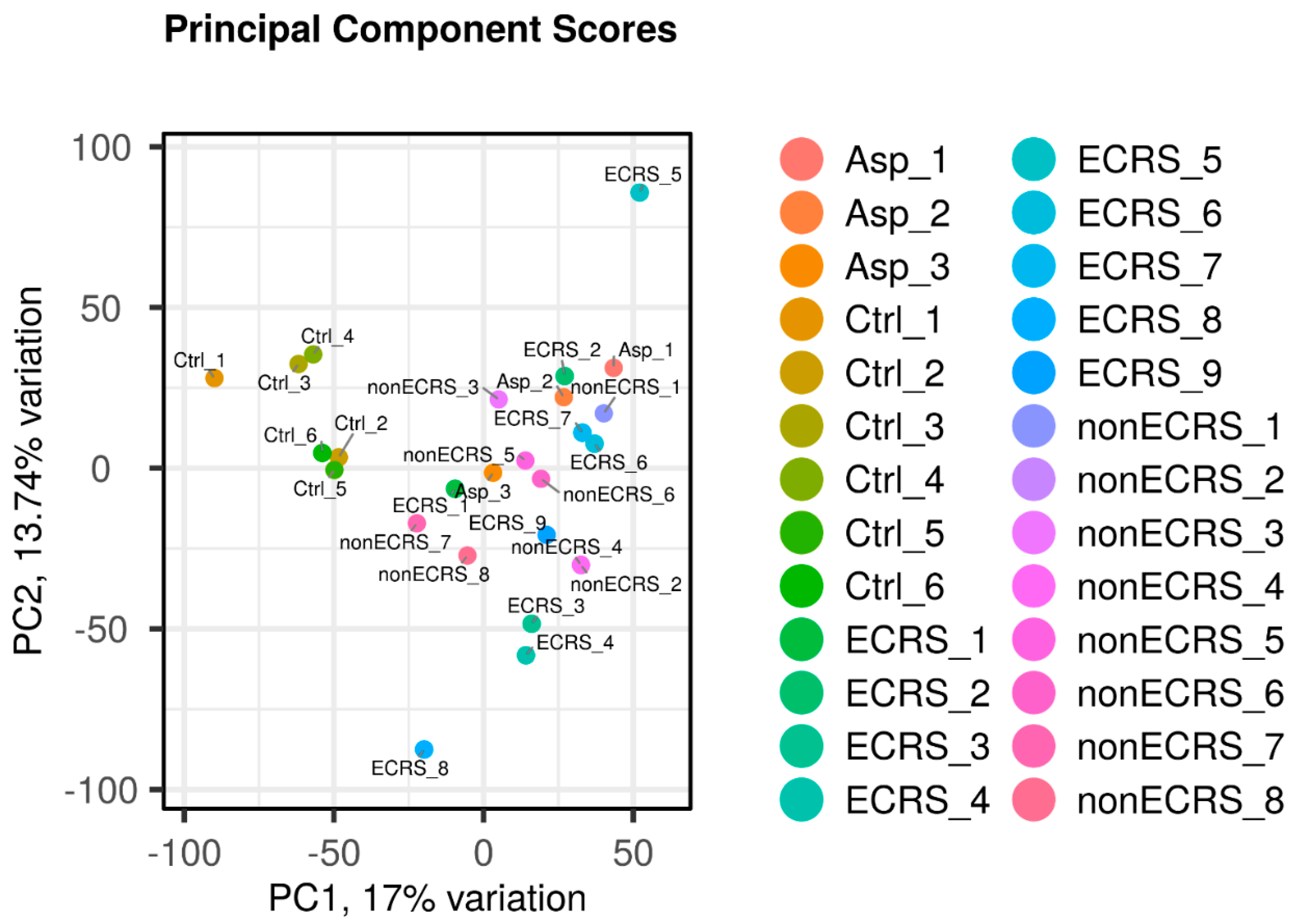
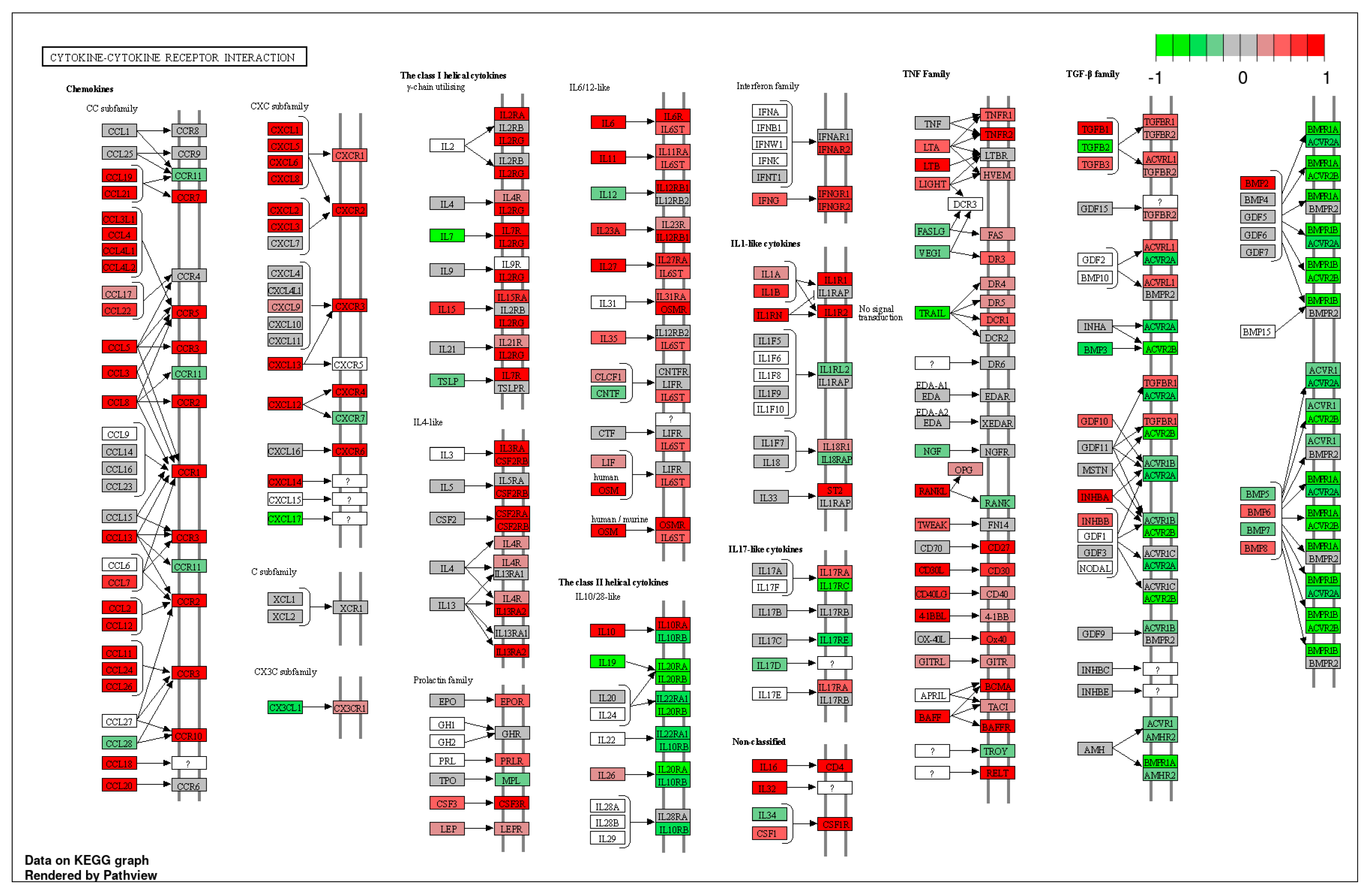
2.1. Gene expression profiles of NPs between ECRS including Asp and nonECRS.
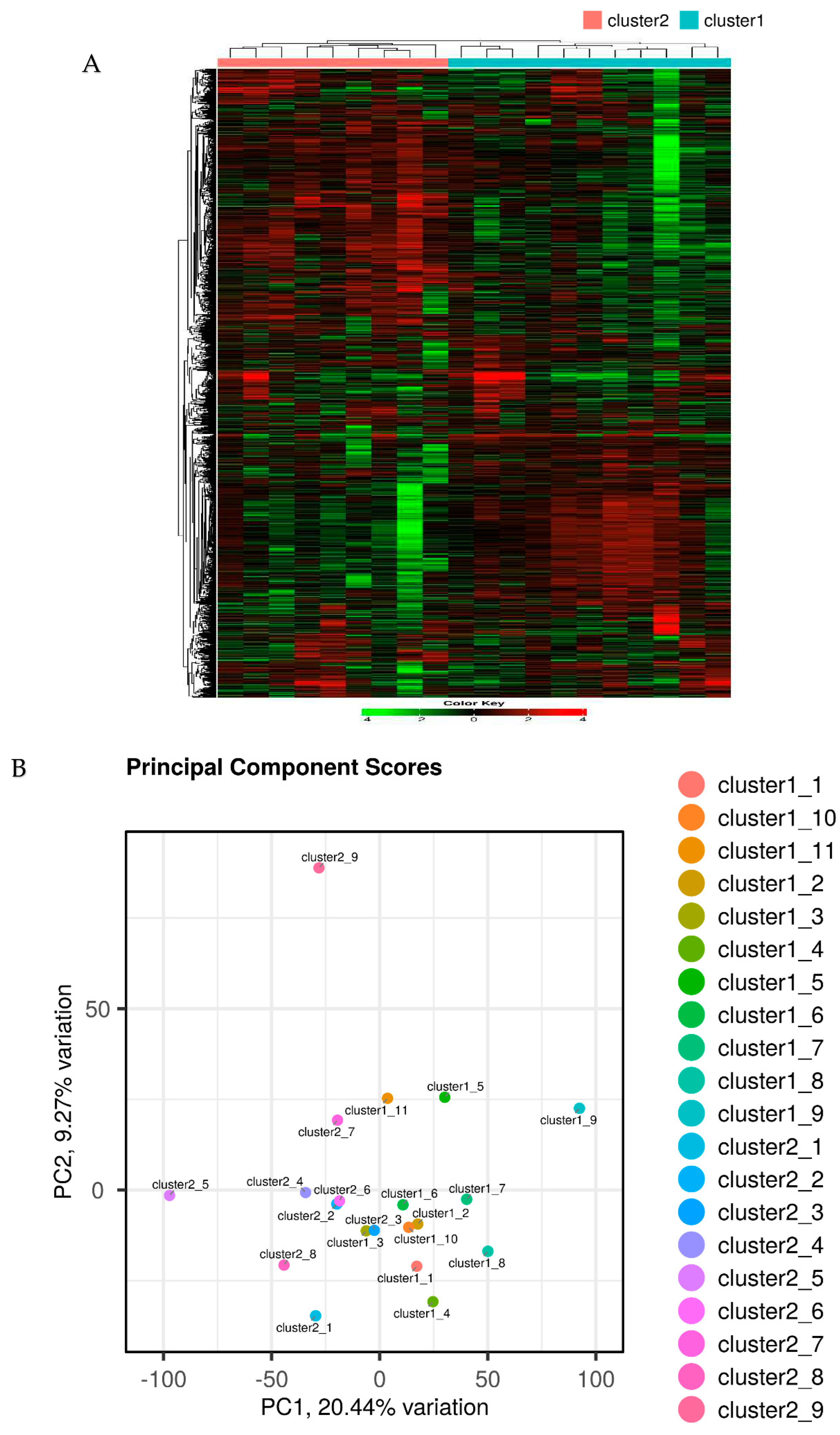
2.2. Segregation of all CRSwNPs into two clusters using hierarchical cluster analysis
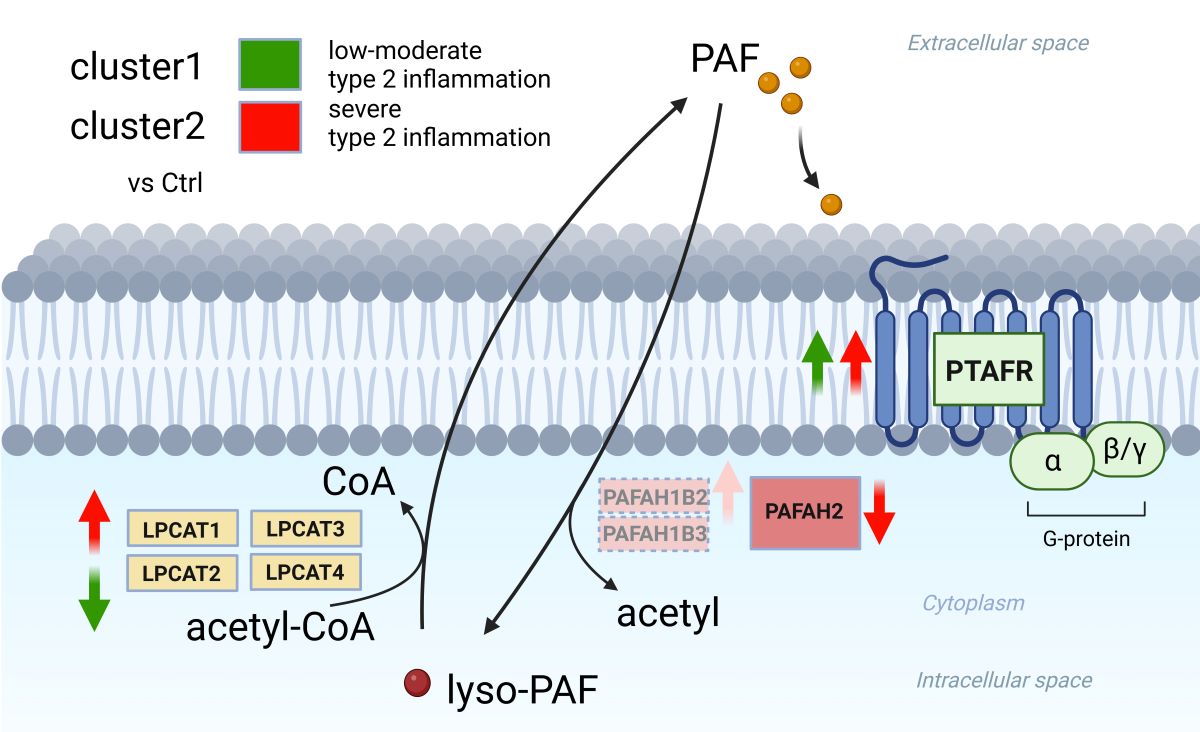
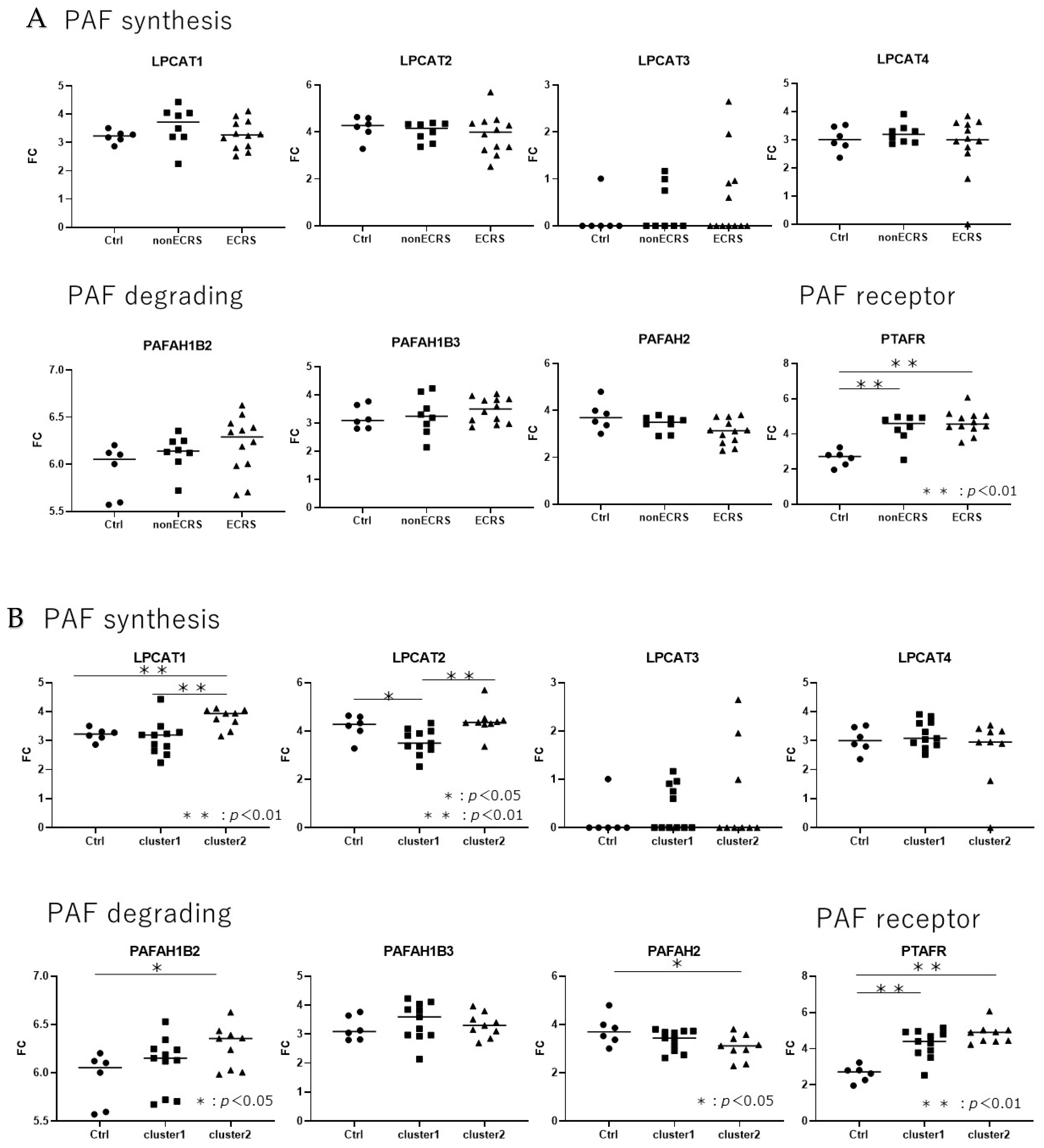
2.3. Gene expression analysis about PAF-metabolism associated gene based on the JESREC-based clinical classification and clusters reflecting the severity of type 2 inflammation.
2.4. Gene expression analysis between PAF-metabolism associated gene and clinical features.
3. Discussion
4. Materials and Methods
4.1. Patient recruitment
 |
4.2. RNA-seq using BRB-seq
4.3. Data processing of BRB-seq
4.4. Data analysis for PAF metabolism associated gene expression
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tokunaga, T.; Sakashita, M.; Haruna, T.; Asaka, D.; Takeno, S.; Ikeda, H.; Nakayama, T.; Seki, N.; Ito, S.; Murata, J.; et al. Novel scoring system and algorithm for classifying chronic rhinosinusitis: the JESREC Study. Allergy. 2015, 70, 995–1003. [Google Scholar] [CrossRef]
- Grayson, JW.; Hopkins, C.; Mori, E.; Senior, B.; Harvey, RJ. Contemporary Classification of Chronic Rhinosinusitis Beyond Polyps vs No Polyps: A Review. JAMA. Otolaryngol. Head. Neck. Surg. 2020, 146, 831–838. [Google Scholar] [CrossRef]
- Matsuwaki, Y.; Ookushi, T.; Asaka, D.; Mori, E.; Nakajima, T.; Yoshida, T.; Kojima, J.; Chiba, S.; Ootori, N.; Moriyama, H. Chronic rhinosinusitis: risk factors for the recurrence of chronic rhinosinusitis based on 5-year follow-up after endoscopic sinus surgery. Int. Arch. Allergy. Immunol. 2008, 146 (Suppl), 77–81. [Google Scholar] [CrossRef]
- Walgama, ES.; Hwang, PH. Aspirin-exacerbated respiratory disease. Otolaryngol. Clin. North. Am. 2017, 50, 83–94. [Google Scholar] [CrossRef]
- Laidlaw, TM.; Boyce, JA. Pathogenesis of aspirin-exacerbated respiratory disease and reactions. Immunol. Allergy. Clinics. North. America. 2013, 33, 195–210. [Google Scholar] [CrossRef]
- Min, YG. The pathophysiology, diagnosis and treatment of allergic rhinitis. Allergy Asthma Immunol Res. 2010, 2, 65–76. https://doi.org/10.4168/aair.2010.2.2.65. Epub 2010 Mar 24. [PubMed]
- Sin, B.; Togias. A. Pathophysiology of allergic and nonallergic rhinitis. Proc. Am. Thorac. Soc. 2011, 8:106–14. 1: 8. [CrossRef] [PubMed]
- Small P, Keith PK, Kim H. Allergic rhinitis. Allergy asthma. Clin. Immunol. 2018,14(s2),51. [CrossRef] [PubMed]
- Muñoz-Cano, RM.; Casas-Saucedo, R.; Valero Santiago. A.; Bobolea, I.; Ribó, P.; Mullol, J. Platelet-Activating Factor (PAF) in allergic rhinitis: clinical and therapeutic implications. J. Clin. Med. 2019,8,1338. [CrossRef] [PubMed]
- Furukawa, M.; Ogura, M.; Tsutsumi, T.; Tsuji, H.; Yamashita, T. Presence of platelet-activating factor in nasal polyps and eosinophils. Acta. Otolaryngol. 2002, 122, 872–876. [Google Scholar] [CrossRef] [PubMed]
- Leggieri, E.; Tedeschi, A.; Lorini, M.; Bianco, A.; Miadonna, A. Study of the effects of paf-acether on human nasal airways. Allergy. 1991, 46, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, V. Role of histamine and platelet-activating factor in allergic rhinitis. J. Physiol. Biochem. 2004, 60, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Cano, R.; Valero, A.; Roca-Ferrer, J.; Bartra, J.; Sanchez-Lopez, J.; Mullol, J. Platelet-activating factor nasal challenge induces nasal congestion and reduces nasal volume in both healthy volunteers and allergic rhinitis patients. Am. J. Rhinol. Allergy. 2013,27:e48-52. [CrossRef] [PubMed]
- Shindou, H.; Hishikawa, D.; Nakanishi, H.; Harayama, T.; Ishii, S.; Taguchi, R. A single enzyme catalyzes both platelet-activating factor production and membrane biogenesis of inflammatory cells. Cloning and characterization of acetyl-CoA:LYSO-PAF acetyltransferase. J. Biol. Chem. 2007,282,6532–6539. https://doi.org/10.1074/jbc.M609641200. Epub 2006 Dec 20. [PubMed]
- Harayama, T.; Shindou, H.; Shimizu, T. Biosynthesis of phosphatidylcholine by human lysophosphatidylcholine acyltransferase 1. J. Lipid. Res. 2009,50,1824–1831. https://doi.org/10.1194/jlr.M800500-JLR200. Epub 2009 Apr 21. [PubMed] [PubMed Central]
- Saito. RF.; Rangel, MC.; Halman, JR.; Chandler, M.; de Sousa Andrade, LN.; Odete-Bustos, S.; Furuya, TK.; Carrasco, AGM.; Chaves-Filho, AB.; Yoshinaga, MY.; Miyamoto, S.; Afonin, KA.; Chammas, R. Simultaneous silencing of lysophosphatidylcholine acyltransferases 1-4 by nucleic acid nanoparticles (NANPs) improves radiation response of melanoma cells. Nanomedicine. 2021,36,102418. https://doi.org/10.1016/j.nano.2021.102418. Epub 2021 Jun 24. [PubMed]
- Palgan, K.; Bartuzi, Z. Platelet activating factor in allergies. Int. J. Immunopathol. Pharmacol. 2015,28,584–589. https://doi.org/10.1177/0394632015600598. Epub 2015 Oct 20. [PubMed]
- Vadas, P.; Gold, M.; Perelman, B.; Liss, GM.; Lack, G.; Blyth, T. Platelet-activating factor, PAF acetylhydrolase, and severe anaphylaxis. N. Engl. J. Med. 2008, 358, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Upton, JEM.; Grunebaum, E.; Sussman, G.; Vadas, P. Platelet Activating Factor (PAF): A Mediator of Inflammation. Biofactors. 2022,48,1189-1202. https://doi.org/10.1002/biof.1883. Epub 2022 Aug 27. [PubMed]
- Karasawa, K.; Harada, A.; Satoh, N.; Inoue, K.; Setaka, M. Plasma platelet activating factor-acetylhydrolase (PAF-AH). Prog Lipid. Res. 2003, 42, 93–114. [Google Scholar] [CrossRef] [PubMed]
- Honda, Z.; Ishii, S.; Shimizu, T. Platelet-activating factor receptor. J. Biochem. 2002,131,773–779. [CrossRef] [PubMed]
- Howard, KM. Platelet-activating factor receptor. In Encyclopedia of Biological Chemistry II. 2nd ed; Lennarz, WJ., Lane, MD., Eds.; Academic Press.: San Diego, CA, 2013; pp. 533–537. [Google Scholar]
- Vadas, P.; Perelman, B. Effect of epinephrine on platelet-activating factorstimulated human vascular smooth muscle cells. J. Allergy. Clin. Immunol 2012,129,1329-1333. https://doi.org/10.1016/j.jaci.2012.02.027. Epub 2012 Mar 27. [PubMed]
- Thivierge, M.; Parent, JL.; Stankova, J.; Rola-Pleszczynski, M. Modulation of human platelet-activating factor receptor gene expression by protein kinase C activation. J. Immunol. 1996, 157, 4681–4687. [Google Scholar] [CrossRef] [PubMed]
- Moritoki, H.; Hisayama, T.; Takeuchi, S.; Miyano, H.; Kondoh, W. Involvement of nitric oxide pathway in the PAF-induced relaxation of rat thoracic aorta. Br. J. Pharmacol. 1992, 107, 196–201. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Teo, CWL.; Png, SJY.; Ung, YW.; Yap, WN. Therapeutic effects of intranasal tocotrienol-rich fraction on rhinitis symptoms in platelet-activating factor induced allergic rhinitis. Allergy. Asthma. Clin. Immunol. 2022,18,52. [CrossRef] [PubMed]
- Roca-Ferrer, J.; Pérez-González, M.; Alobid, I.; Tubita, V.; Fuentes, M.; Bantulà, M.; Muñoz-Cano, R.; Valero, A.; Izquierdo, I.; Mullol, J. Upregulation of Platelet-Activating Factor Receptor Expression and Lyso-Platelet-Activating Factor Isoforms in Human Nasal Polyp Tissues. J. Clin. Med. 2023, 12, 7357. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ishino, T.; Takeno, S.; Takemoto, K.; Yamato, K.; Oda, T.; Nishida, M.; Horibe, Y.; Chikuie, N.; Kono, T.; Taruya, T.; Hamamoto, T.; Ueda, T. Distinct Gene Set Enrichment Profiles in Eosinophilic and Non-Eosinophilic Chronic Rhinosinusitis with Nasal Polyps by Bulk RNA Barcoding and Sequencing. Int. J. Mol. Sci. 2022, 23, 5653. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kohnz, RA.; Mulvihill, MM.; Chang, JW.; Hsu, KL.; Sorrentino, A.; Cravatt, BF.; Bandyopadhyay, S.; Goga, A.; Nomura, DK. Activity-Based Protein Profiling of Oncogene-Driven Changes in Metabolism Reveals Broad Dysregulation of PAFAH1B2 and 1B3 in Cancer. ACS. Chem. Biol. 2015,10,1624-1630. https://doi.org/10.1021/acschembio.5b00053. Epub 2015 May 7. [PubMed] [PubMed Central]
- Fulkerson, PC.; Rothenberg, ME. Targeting eosinophils in allergy, inflammation and beyond. Nat. Rev. Drug. Discov. 2013,12,117-129. https://doi.org/10.1038/nrd3838. Epub 2013 Jan 21. [PubMed] [PubMed Central]
- Ricciardolo, FL. Multiple roles of nitric oxide in the airways. Thorax. 2003, 58, 175–182. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alpern, D.; Gardeux, V.; Russeil, J.; Mangeat, B.; Meireles-Filho, ACA.; Breysse, R.; Hacker, D.; Deplancke, B. BRB-seq: ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing.Genome Biol 2019,20, 71. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





