1. Introduction
Skin cancer is a prevalent and potentially life-threatening disease that affects millions of people worldwide. The harmful effects of skin cancer can be severe and detrimental. The effects of skin cancer can vary depending on the type and stage of the disease. It can result in disfigurement, significant medical expenses, and even mortality if not diagnosed and treated at an early stage. Current estimates are that one in five Americans will develop skin cancer in their lifetime. It is estimated that approximately 9,500 people in the U.S. are diagnosed with skin cancer every day [
1]. In addition, skin cancer can also cause physical and emotional distress, as it may require invasive treatments and leave visible scars.
Skin cancer [
2] is a prevalent malignancy arising from uncontrolled growth of skin cells, primarily attributed to prolonged exposure to ultraviolet (UV) radiation, emanating from the sun or artificial sources, like tanning beds. This radiation inflicts DNA damage, culminating in genetic mutations that promote abnormal cell proliferation. Individuals with fair skin, light-colored eyes, and light hair are particularly vulnerable due to reduced melanin, a natural UV protectant. Those with a history of sunburns, particularly in childhood, face heightened risk. Genetic predisposition is a factor, especially in cases of familial skin cancer, with melanoma carrying the highest risk. Inherited conditions like xeroderma pigmentosum and dysplastic nevus syndrome also elevate susceptibility. Advancing age compounds risk as cumulative UV exposure increases DNA damage. Immune system suppression, as seen in transplant recipients or HIV/AIDS patients, compromises the body’s ability to detect and eliminate abnormal cells, further heightening the risk. Certain chemical exposures, such as arsenic and industrial compounds, contribute to the risk profile. Individuals with a prior history of skin cancer are at an increased risk of recurrence, necessitating vigilant follow-upand skin checks.
The ISIC 2019 dataset [
3] is a significant compilation within the International Skin Imaging Collaboration (ISIC) series, specifically curated for advancing research in dermatology, particularly in the field of computer-aided diagnosis (CAD) for skin cancer detection and classification. This dataset [
3], released in 2019, is a continuation of the effort to provide a comprehensive collection of high-quality dermoscopic images accompanied by annotations and metadata. It consists of thousands of images showcasing various skin lesions, including melanomas, nevi, and other types of benign and malignant skin conditions.
One of the primary objectives of the ISIC 2019 dataset is to facilitate the development and evaluation of machine learning algorithms, computer vision models, and artificial intelligence systems geared towards accurate and early detection of skin cancers. Researchers, data scientists, and developers leverage this dataset to train, validate, and test their algorithms for automated skin lesion analysis, classification, and diagnosis. The availability of annotated images within the ISIC 2019 dataset [
3] allows for supervised learning approaches, enabling algorithms to learn patterns and features associated with different types of skin lesions. By utilizing this dataset, researchers aim to improve the accuracy and efficiency of diagnostic tools, potentially aiding dermatologists and healthcare professionals in making more precise and timely diagnoses.
In recent years, deep learning [
4] has brought about a transformative revolution in the field of machine learning. It stands out as the most advanced subfield, centering on artificial neural network algorithms inspired by the structure and function of the human brain. Deep learning techniques find extensive application in diverse domains, including but not limited to speech recognition, pattern recognition, and bioinformatics. Notably, in comparison to traditional machine learning methods, deep learning systems have demonstrated remarkable achievements in these domains. Recent years have witnessed the adoption of various deep learning strategies for computer-based skin cancer detection. This paper delves comprehensively into the examination and evaluation of deep learning-based skin cancer classification techniques.
Our approach incorporates state-of-the-art deep learning models, including ImageNet ConvNets [
5] and Vision Transformer (ViT) [
6], through techniques like transfer learning, and fine-tuning. Evaluation encompasses quantitative assessments using confusion matrices, classification reports, and visual evaluations using tools like Score-CAM [
7].
The integration of "Naturalize" techniques, as referenced in [
8], alongside these strides represents significant headway in automating the analysis of skin cancer classification. This paper extensively explores the methodologies and outcomes derived from these state-of-the-art approaches, shedding light on their transformative capacity within the realm of skin cancer research
After this introduction, the rest of the paper will continue as follows:
Section 2 highlights the relevant literature related to the detection and classification of skin cancer using pre-trained CNNs, and
Section 3 describes the methodology used in this study. In addition,
Section 4 presents the experimental results obtained using pre-trained models and Google ViT for the skin cancer classification an in-depth analysis of results is made. Finally, the paper is concluded in
Section 5.
2. Related Works
Recent advancements in deep learning models for skin lesion classification have showcased significant progress. This review consolidates findings from notable studies employing diverse convolutional neural network (CNN) architectures for this purpose. These studies explore methodologies and performances using the ISIC2019 dataset.
Kassem et al. [
9] utilized a GoogleNet (Inception V1) model with transfer learning on the ISIC2019 dataset, achieving 94.92% accuracy. They demonstrated commendable performance in recall (79.80%), precision (80.36%), and F1-score (80.07%).
In 2022, Li et al. [
10] introduced the Quantum Inception-ResNet-V1, achieving 98.76% accuracy on the same ISIC2019 dataset. Their model exhibited substantial improvements in recall (98.26%), precision (98.40%), and F1-score (98.33%), signifying a significant leap in accuracy.
Mane et al. [
11] leveraged MobileNet with transfer learning, achieving an accuracy of 83% on the ISIC2019 dataset. Despite relatively lower results compared to other models, their consistent performance across recall, precision, and F1-score at 83% highlighted robust classification.
Hoang et al. [
12] introduced the Wide-ShuffleNet combined with segmentation techniques, achieving an accuracy of 84.80%. However, their model showed comparatively lower metrics for recall (70.71%), precision (75.15%), and F1-score (72.61%) than prior studies.
In 2023, Fofanah et al. [
13] presented a Four-layer DCNN model, achieving 84.80% accuracy on a modified dataset split. Their model demonstrated balanced performance across recall (83.80%), precision (80.50%), and F1-score (81.60%).
Similarly, Alsahaf et al. [
14] proposed a Residual Deep CNN model in the same year, achieving 94.65% accuracy on a different dataset split. They maintained balanced metrics for recall (70.78%), precision (72.56%), and F1-score (71.33%).
These studies collectively showcase the evolution of skin lesion classification models, indicating significant progress in accuracy and performance metrics. Comparative analysis highlights the strengths and weaknesses of each model, laying the groundwork for further advancements in dermatological image classification.
The literature review focuses on a series of studies (
Table 1) concentrating on automating skin cancer classification using the ISIC2019 dataset, offering a summarized view of these endeavors.
Our groundbreaking research presents the novel augmentation technique "Naturalize," specifically designed to tackle the challenges posed by data scarcity and class imbalance within deep learning. Through the implementation of "Naturalize," we have successfully overcome these hurdles, achieving an unprecedented 100% average testing accuracy, precision, recall, and F1-Score in our skin cancer classification model. This groundbreaking technique revolutionizes the landscape of deep learning, offering a solution that not only elevates classification performance but also redefines the potential for accurate and reliable diagnosis across various imbalanced skin cancer classes.
3. Materials and Methods
This section provides a detailed explanation of our advanced approach aimed at achieving highly precise skin cancer image classification on the challenging ISIC2019 dataset. Our classification efforts utilized state-of-the-art Deep Learning (DL) methodologies, prominently featuring the "ImageNet ConvNets" and "Keras Vision Transformers (ViT)." To enhance performance, we incorporated the innovative "Naturalize" augmentation technique to address data insufficiencies in specific classes like (AK, DER, VAS, SCC) while simultaneously increasing the available image data in other classes. In pursuit of excellence, we equalized the number of images across classes by reducing three classes to 2.4k images, aligning them with the BKL class. This adjustment aimed to eliminate the imbalance in image numbers between these three types (MEL, NV, BCC), which ranged from 10.4k to 1k images.
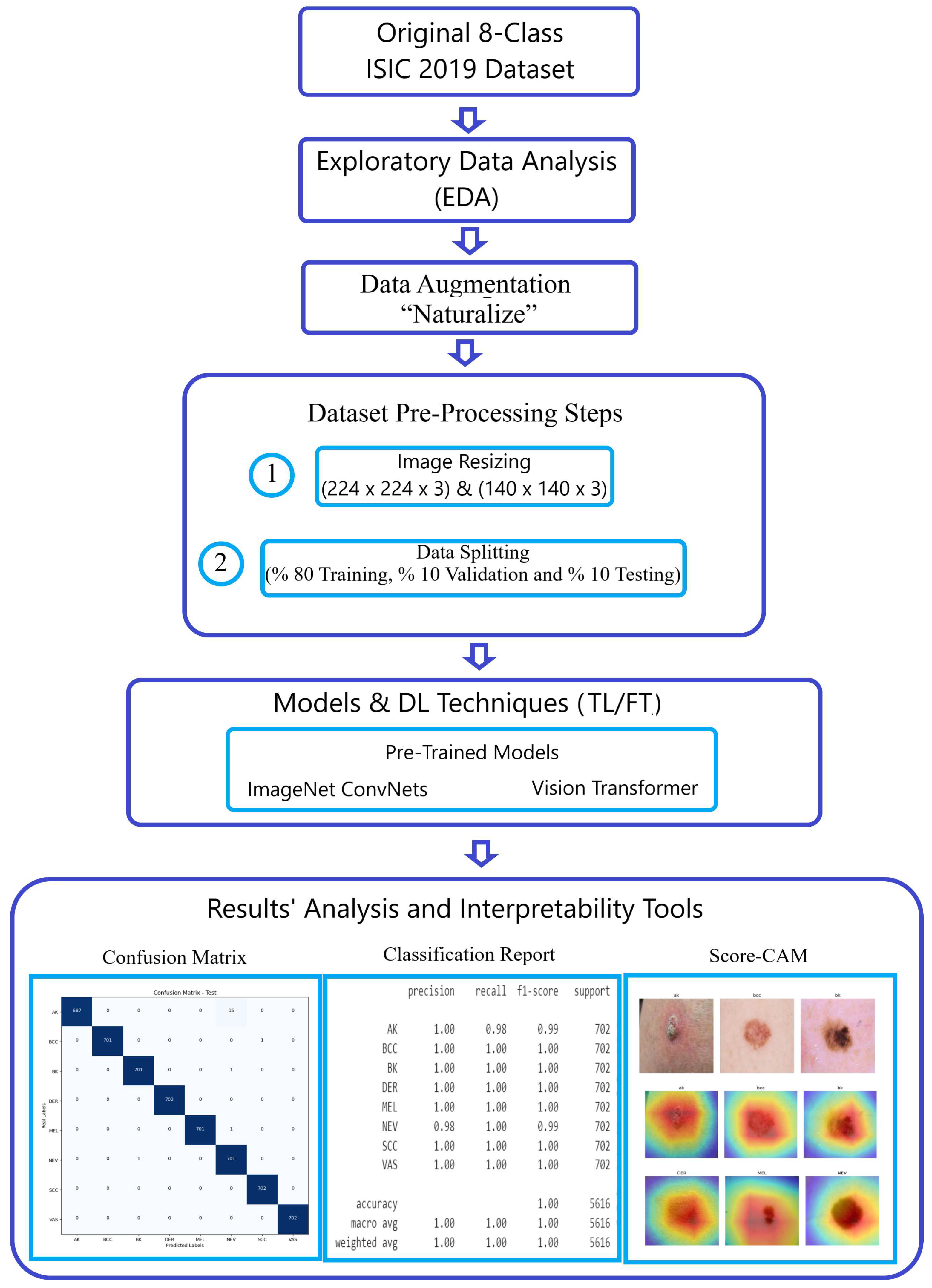
Figure 1 visually illustrates the breadth and sophistication of our methodology, serving as a testament to our steadfast commitment to advancing the field of image classification.
3.0.1. Original 8-Class ISIC 2019 dataset
3.1. ISIC-2019 Dataset
3.1.1. Original 8-Class ISIC 2019 dataset
The initial ISIC 2019 dataset [
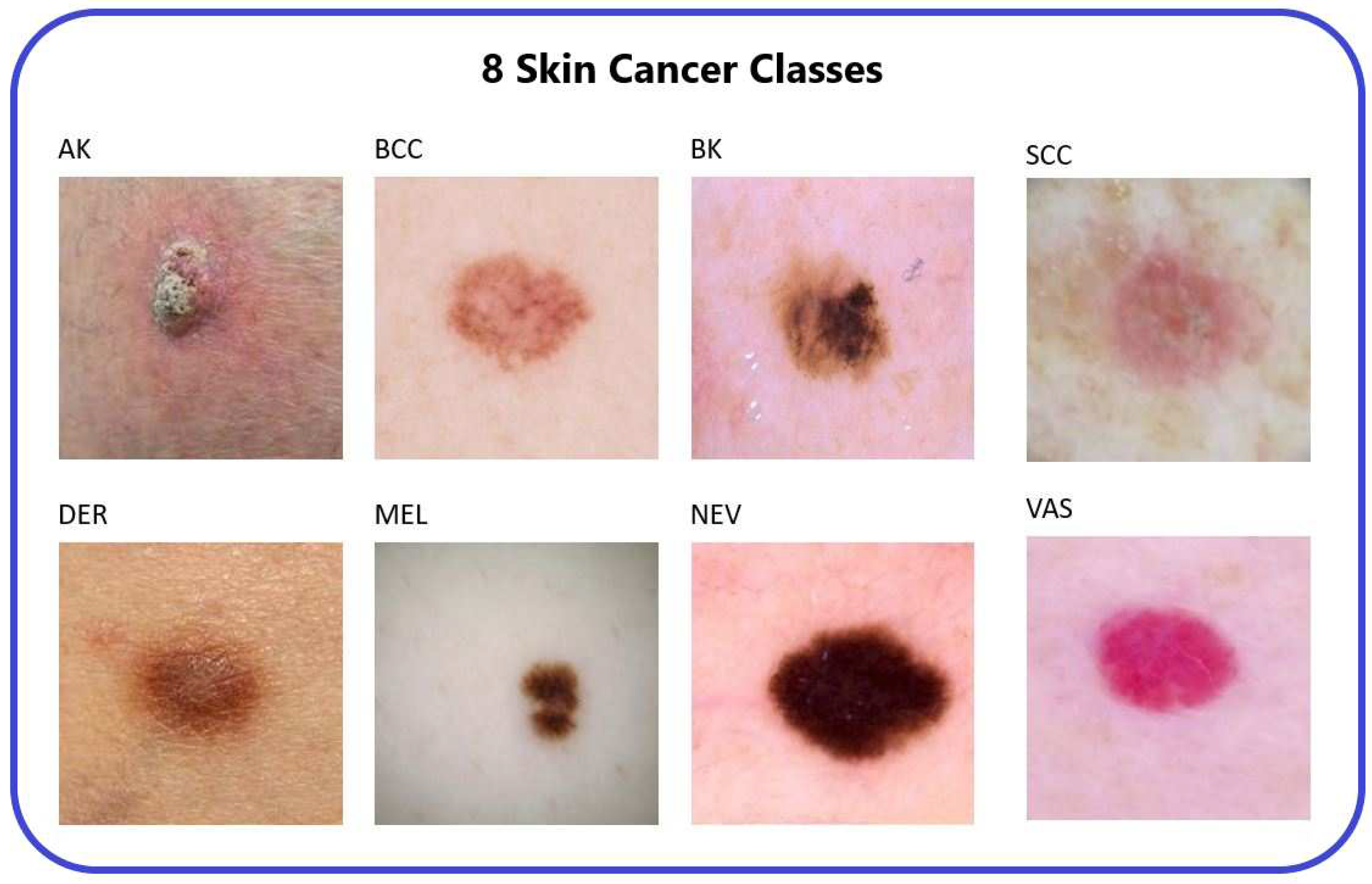
3], obtained from an online repository, consists of 25,331 images categorized into eight distinct classes representing different types of skin cancer. These classes are Actinic Keratosis (AK), Basal Cell Carcinoma (BCC), Benign Keratosis (BK), Dermatofibroma (DER), Melanocytic Nevi (NEV), Melanoma (MEL), Vascular Skin Lesion (VAS), and Squamous Cell Carcinoma (SCC).
To address the unbalanced distribution of images within the original ISIC-2019 dataset, we modified it by reducing the number of images for three types of skin cancer (MEL, NV, BCC) to 2.4k, aligning them with the existing count of 2.4k images for the BK type. This adjustment was made to achieve balance among the different cancer types. We applied the Naturalize Augmentation technique during this process. Consequently, the updated dataset now comprises 19,200 balanced images across the eight types of skin cancer.
Table 2 [
14] provides an overview of the distribution of the eight skin cancer classes within the original ISIC 2019 dataset.
The images in the ISIC dataset adhere to a standard size of 1024x1024 pixels [
14], which needs to be resized into “224 × 224” and “140 × 140” to make the use of it more flexible in the work.
3.1.2. Pruned 2.4K ISIC2019
Due to substantial variations in the quantity of available images, it was necessary to reduce the number of photos in specific categories. This adjustment aimed to alleviate the pronounced differences among various types of skin cancer.
Table 3 summarizes the distribution of the Pruned 2.4K ISIC2019 dataset in the 8 classes.
3.1.3. Naturalized 2.4k and 7.2k ISIC2019 datasets
Our goal was to achieve an equal number of photos across all eight types of skin cancer. The Naturalize augmentation is employed to achieve this target. Two balanced updated version of ISIC2019 are created using the Naturalize augmentation technique: Naturalized 2.4K ISIC2019 and Naturalized 7.2K ISIC2019 datasets.
Table 4 summarizes the distribution of the Naturalized 2.4K ISIC2019 dataset in the 8 classes.
Table 5 summarizes the distribution of the Naturalized 7.2K ISIC2019 dataset in the 8 classes.
The Naturalize augmentation technique can generate any number of skin cancer images with unique content and quality resembling the original ISIC2019 dataset. This is achieved through the benefit from the power of randomness of the addition of segmented skin cancer images with different skin backgrounds.
3.2. Exploratory Data Analysis (EDA)
Exploratory Data Analysis (EDA) was conducted to gain insights into the nature of the dataset. This involved training and testing pre-trained ImageNet models with the original ISIC 2019 dataset, analyzing the confusion matrix, and generating a classification report.
The primary observation from the exploratory data analysis (EDA) reveals a significant influence stemming from extreme class imbalances, notably in categories such as DER and NEV, within the ISIC 2019 dataset. This imbalance markedly impacts the overall performance metrics (average accuracy, precision, recall, and F1-score).
Addressing this issue is where the "Naturalize" augmentation technique comes into play. This technique involves generating new images for classes that have insufficient representation, maintaining a quality that mirrors the original images. As a result, "Naturalize" effectively resolves the pronounced imbalances among classes while preserving image quality.
3.3. Data Augmentation “Naturalize”
The pseudocode shown in Algorithm 1 demonstrates the principle behind the “Naturalize” augmentation technique and how it works.
|
Algorithm 1:Naturalize Algorithm |
- 1:
# Imports and Paths - 2:
Import os, random, image_processing, SAM_model - 3:
Define file paths and import essential libraries - 4:
- 5:
# Load SAM_model and ISIC 2019 Dataset - 6:
Mount Google_drive - 7:
Load Skin Cancer images from ISIC 2019 dataset - 8:
SAM = load_model(SAM_model) - 9:
- 10:
# Segment ISIC 2019 Dataset Using SAM - 11:
Segment ISIC 2019 images using SAM into segmented “Cancer” images - 12:
Save segmented "Cancer" images into “Skin Cancer” dataset on Google_drive - 13:
- 14:
# Random Selection from Skin Background dataset - 15:
Select randomly Skin Background image from Skin Background dataset - 16:
- 17:
# Composite Image Creation - 18:
for i in range(num_images) do - 19:
Load Skin Background image - 20:
Select randomly "Cancer" image from “Skin Cancer” dataset - 21:
Rotate randomly "Cancer" image - 22:
Add "Cancer" image at random position to Skin Background image - 23:
Save the composite image on Google_drive - 24:
end for |
The “Naturalize” augmentation technique consists of two primary steps:
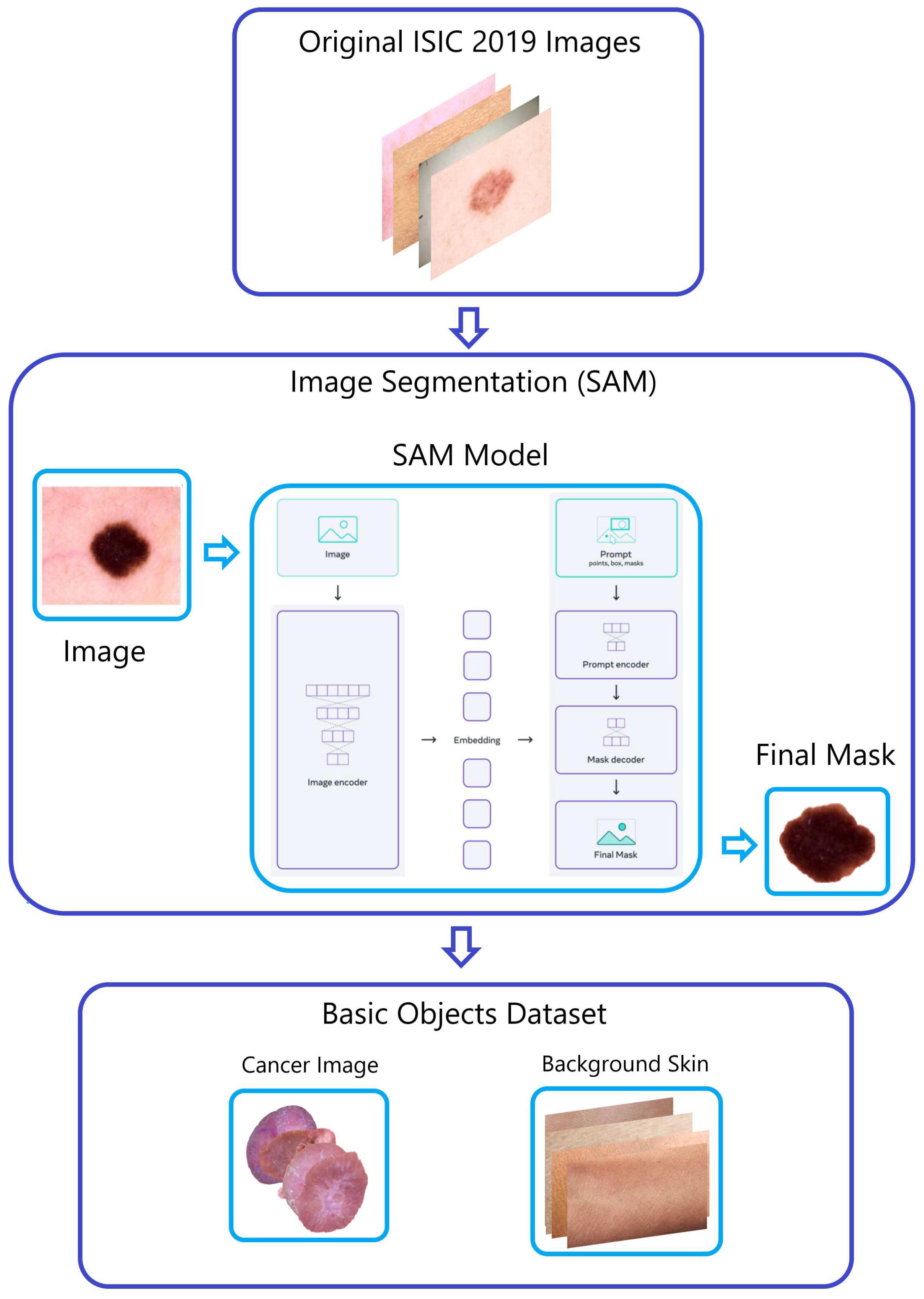
-
Within the ISIC 2019 dataset, images depicting four different types of skin cancer were divided into smaller sets through the application of the "Segment Anything Model (SAM)" developed by Meta AI [
15]. This process produced segments for AK, DER, VAS, and SCC. The inclusion of these new images in the classes positively influenced the accuracy of classification, as evidenced by the performance metrics and classification report from the prior EDA analysis.
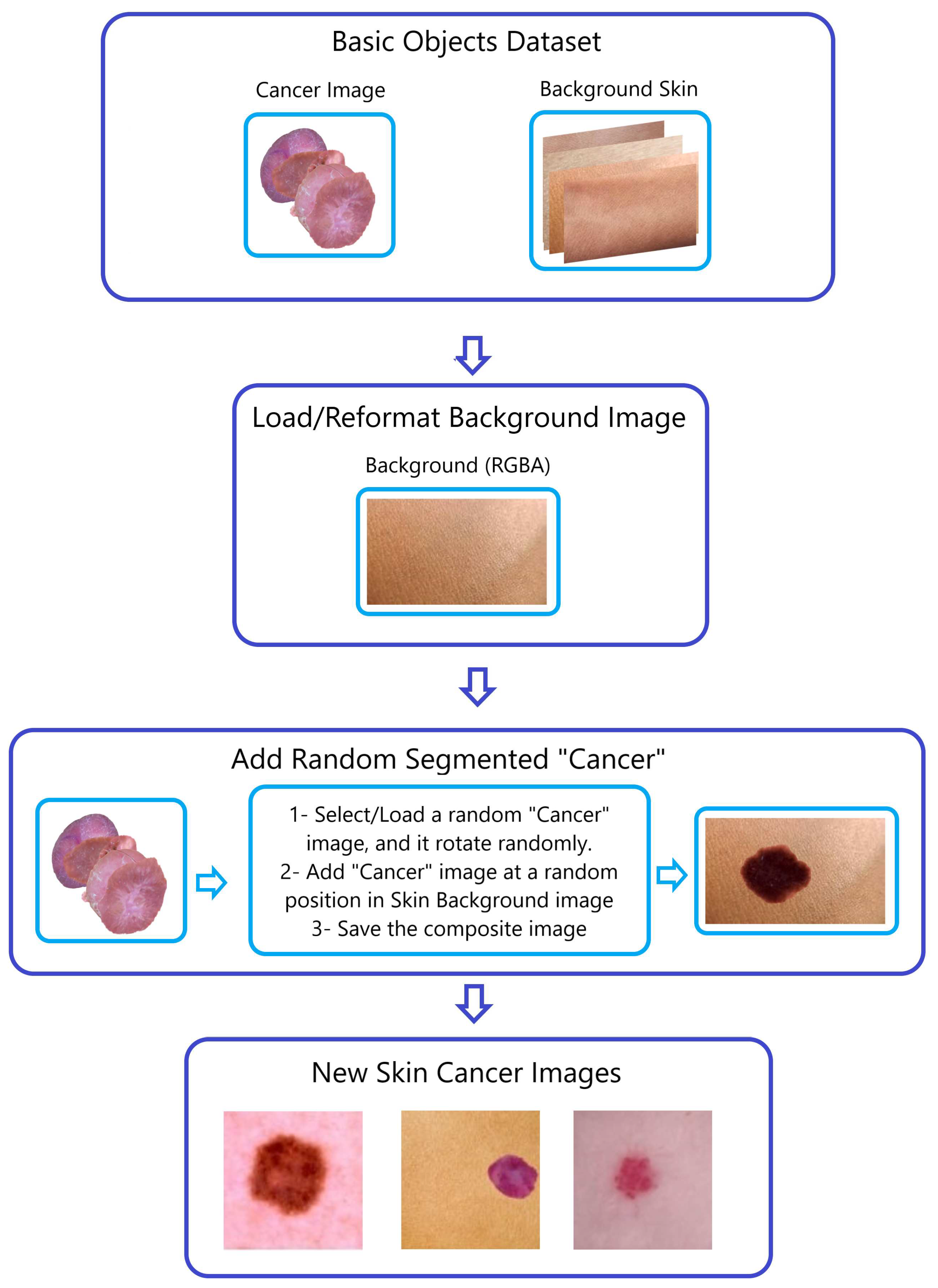
-
Step 2—Generating Composite Images (
Figure 4):
To produce composite images, we merged the four SAM-segmented categories with randomly chosen photographs of healthy skin within the respective sub-datasets (AK, DER, VAS, and SCC). This procedure is visually demonstrated in
Figure 3 and
Figure 4, using the creation of composite skin cancer images as an example.
Guided by our meticulous exploratory data analysis (EDA), we have judiciously pruned select images from multiple classes, embodying our unwavering commitment to data quality. Following the incorporation of images, the quantity of skin cancer images within the initial 8-class ISIC2019 dataset saw a significant rise. Through the initial utilization of the "Naturalize" technique, the number of skin cancer images surged from 1,987 images to a substantial 9,600 images. TABLE IV effectively portrays the remarkable evolution of the original 1,987-skin cancer images referring to (AK, DER, VAS, SCC) in ISIC2019 dataset into 9,600 skin cancer images spanning four different types (AK, DER, VAS, and SCC).
The dataset experienced significant growth due to the implementation of the "Naturalize" augmentation method, resulting in the development of the ISIC2019 dataset with approximately 9.6K images. This expansion was achieved by adding between 1,500 to 2,000 images to each of the four sub-datasets representing the following skin cancer classes: AK, DER, VAS, and SCC.
The choice to exclusively incorporate the skin cancer categories "AK, DER, VAS, and SCC" in the "Naturalize" applications stems from the findings in the classification report. This decision is driven by the goal of enhancing the overall precision, recall, F1-Score, and accuracy averages.
3.4. Comparison between Naturalize and Conventional Augmentation Techniques
Conventional methods of image augmentation typically rely on basic transformations like rotation, flipping, and color adjustments to diversify datasets. These techniques aim to enhance general image variability. In contrast, the "Naturalize" augmentation method is more intricate and tailored. It leverages a targeted segmentation process using the "Segment Anything Model" to isolate specific classes of skin cancer within the dataset. This segmentation strategy is rooted in previous data analyses, emphasizing the improved performance of certain classes.
The segmentation process using the SAM model yields a notably substantial number of segmented skin cancer instances from the original ISIC 2019 dataset. Integrating these segmented objects (cancer images) at random results in an extensive array of unique and authentic replicas of the initial dataset.
Moreover, "Naturalize" grants precision control over the image quality by enabling the random rotation of segmented skin cancer and their placement within randomly selected skin images from the "Background Skin" dataset. This level of control is uncommon in traditional augmentation methods, ensuring enhanced precision in the generated images.
Beyond medical imaging, the versatility of "Naturalize" is evident. By segmenting all objects within the original images and reintegrating them into background images, this technique can be applied across various domains, not confined solely to medical image augmentation. This adaptability underscores its potential for widespread utilization, showcasing its applicability beyond the medical field.
3.5. Naturalized 2.4K ISIC2019 and Naturalized 7.2K ISIC2019 Datasets Preprocessing
The preprocessing of the Naturalized 2.4K ISIC2019 and Naturalized 7.2K ISIC2019 datasets involved two primary steps:
Step 1—Image Resizing: The images were resized to match the standard “224 × 224” image input size required by pre-trained ImageNet ConvNets and ViT models. Additionally, the images were resized to dimensions of "140 × 140," aiming to optimize computational resources, especially with a sizable dataset like the Naturalized 7.2K ISIC2019 dataset.
Step 2—Data Splitting: The Naturalized 2.4K ISIC2019 and Naturalized 7.2K ISIC2019 datasets were split into three subsets: an 80% training set, a 10% validation set, and a 10% testing set.
3.6. Models and DL Techniques (TL/FT)
Two types of model architectures were utilized in this study: pre-trained ImageNet ConvNets, and pre-trained Vision Transformers (ViT). Additionally, two DL techniques [
16] were employed to train pre-trained models: Transfer Learning (TL), Fine-Tuning (FT).
3.6.1. Pre-Trained ImageNet ConvNets
Pre-trained ImageNet models are an explicit example of ConvNets, which are trained on a large dataset.
For this study, pre-trained ImageNet models formed the core of the research. Notable models utilized in this investigation included ConvNexTBase and ConvNeXtLarge [
17], DenseNet-201 [
18], EfficientNetV2 B0 [
19], InceptionResNet [
20], Xception [
21], VGG16 [
22], and VGG-19 [
22].
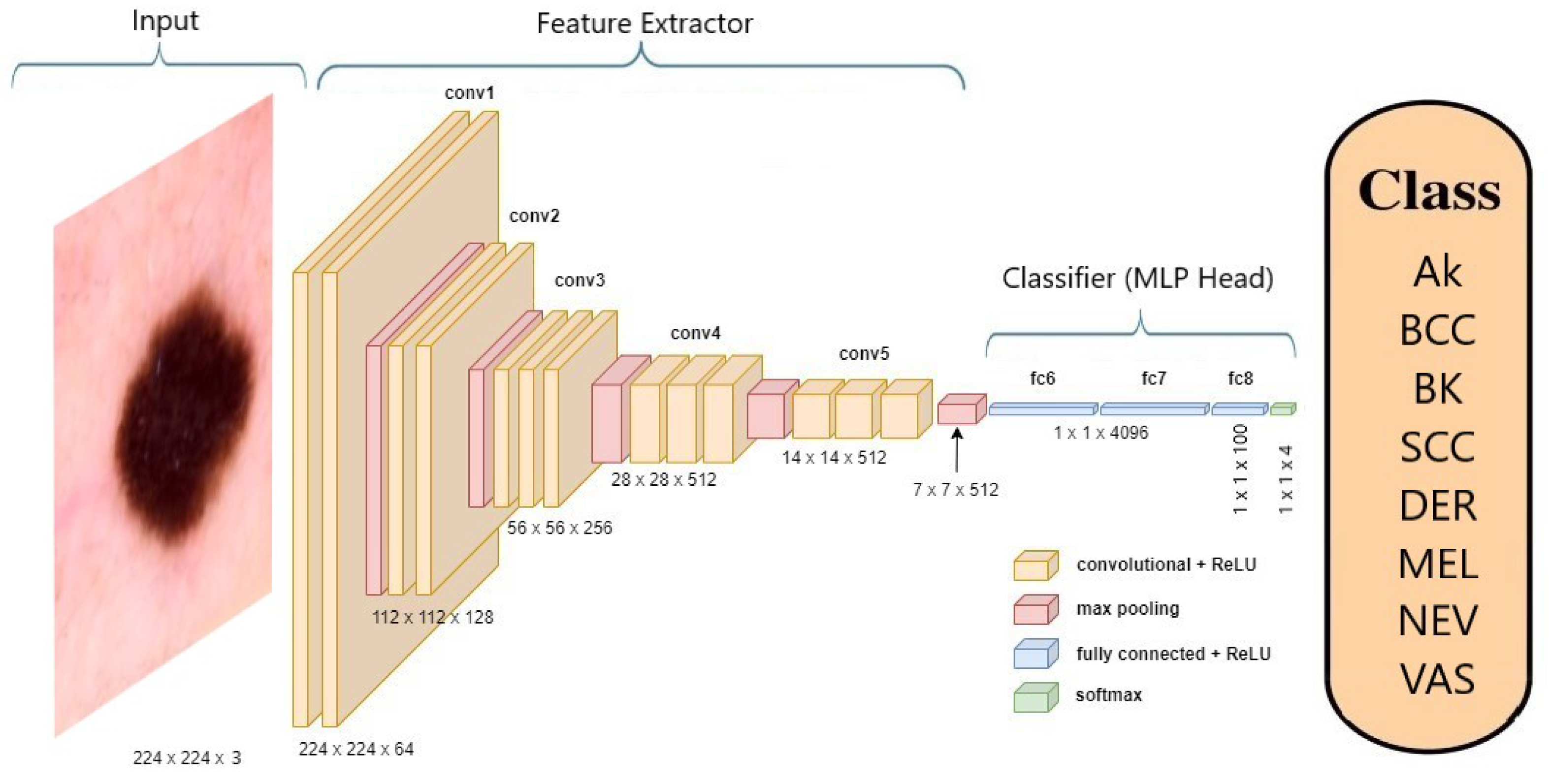
Figure 5 offers an illustration of the VGG-19 [
22] model’s architecture when applied to skin cancer classification.
3.6.2. Pre-Trained Vision Transformer (ViT)
The study employed the Vision Transformer (ViT) [
6] architecture, which is derived from the transformer architecture frequently utilized in Natural Language Processing (NLP). This approach entailed dividing input images into smaller patches and subjecting each patch to processing through a transformer encoder. In contrast to conventional convolutional layers, ViT employed self-attention mechanisms to extract features from the input images, enabling the model to analyze the entire image simultaneously. The research utilized the "ViT" configuration with 12 encoder blocks, and
Figure 6 demonstrates its use in classifying skin cancer [
6].
3.6.3. DL Techniques (TL/FT)
A pre-trained ImageNet model comprises a Convolutional Base, responsible for extracting features, and a Classifier, which is a Multi-Layer Perceptron (MLP) head. In the context of transfer learning (TL) [
23], the process involves replacing the MLP head with a new one and then retraining the model on a specific dataset. During this transfer learning phase, the Convolutional Base remains fixed and not trainable.
When fine-tuning (FT) [
24] is applied, both the Convolutional Base and the MLP head undergo further training, adjusting their parameters to suit a new learning task.
To achieve an optimal Deep Learning skin cancer tool, this work employs two deep learning techniques, namely Transfer Learning (TL) and Fine-Tuning (FT).
3.7. Results’ Analysis and Interpretability Tools
Apart from the accuracy metrics, which include accuracy and loss, three tools for analyzing and interpreting results are employed. These tools consist of the confusion matrix, classification reports, and Score-CAM
3.7.1. Confusion Matrix
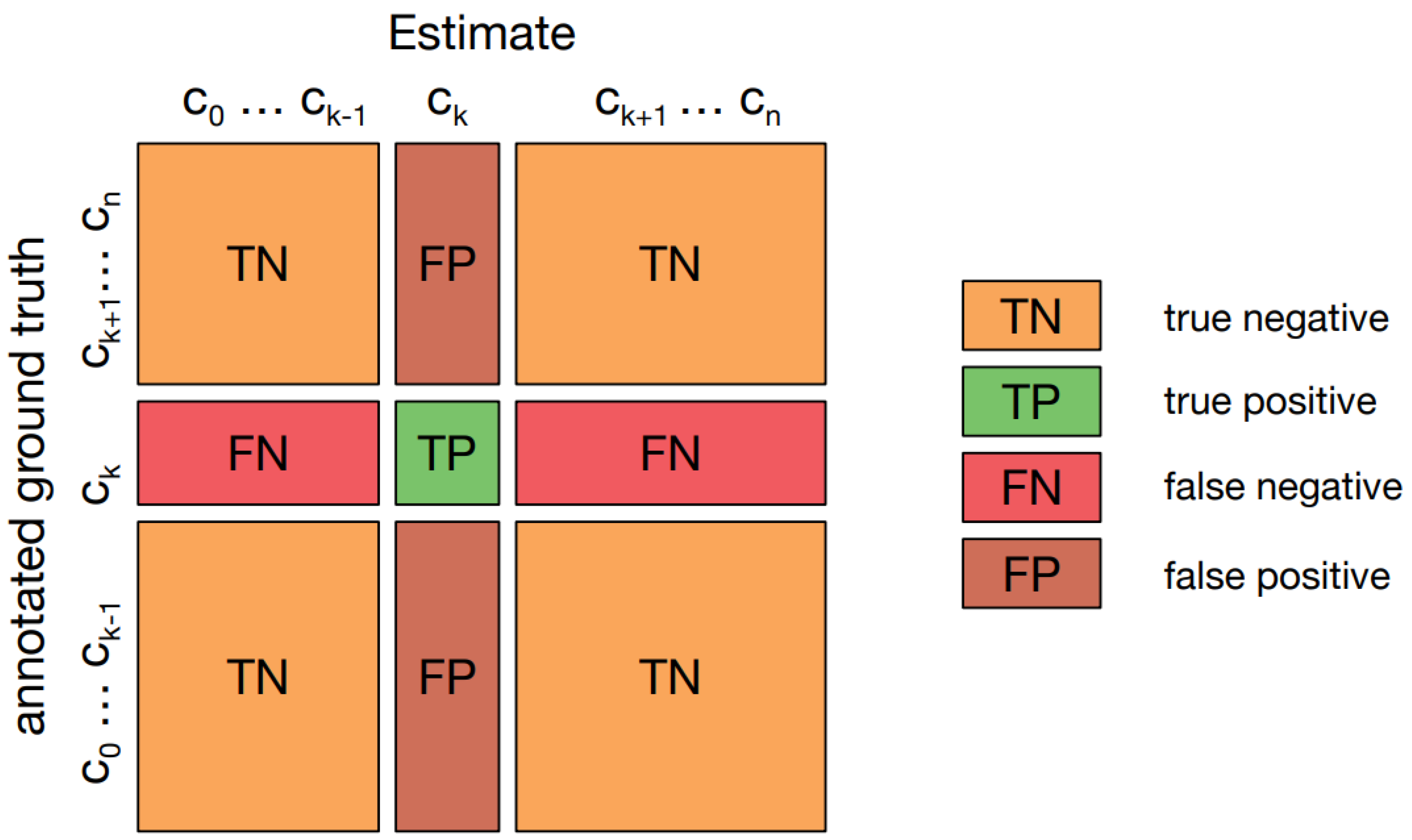
A confusion matrix [
25], also known as an error matrix, provides a visual representation of how well an algorithm performs, particularly in supervised learning scenarios. It presents actual classes in the rows and predicted classes in the columns.
Figure 7 illustrates such a matrix in a multi-class classification context, highlighting "TN and TP" for correctly identified negative and positive cases, and "FN and FP" for cases that were incorrectly classified.
3.7.2. Classification Report
In the assessment, the evaluation of prediction quality relies on metrics such as precision, recall, and F1-score for individual classes. Additionally, it includes macro and weighted average accuracies to gauge overall performance.
Accuracy, computed as a percentage of correct predictions, is determined by Equation (
1) [
25]:
Precision measures the quality of a positive prediction made by the model and the Equation (
2) [
25] demonstrates its computational process:
Recall measures how many of the true positives (TPs) were recalled (found) and calculated using the Equation (
3) [
25]:
F1-Score is the harmonic mean of precision and recall and can be calculated using the Equation (
4) [
25]:
3.7.3. Score-CAM
A Score-CAM, as described in reference [
7], is a visual explanation technique that assigns weights to scores using class activation mapping (CAM) in CNN models. It serves the purpose of providing insights into the inner workings of CNN models.
4. Results and Discussion
4.1. Results
This section offers a comprehensive summary of experiments focused on the 8-class classification of skin cancer. Various models, such as pre-trained ImageNet (ConvNextBase, ConvNeXtLarge, DenseNets, InceptionResNet V2, EfficientNetB0, VGG-19, VGG16, and Xception) and ViT models, were employed. The experiments were carried out using the challenging ISIC2019 dataset. To address class imbalance, the "Naturalize" augmentation technique was introduced, leading to the creation of two new balanced datasets named Naturalized 2.4K ISIC2019 and Naturalized 7.2K ISIC2019. The performance of the models was assessed quantitatively through confusion matrices and classification reports, and visually using Score-CAM on four types of datasets: original ISIC2019, updated ISIC2019, and Naturalized 2.4K ISIC2019 and Naturalized 7.2K ISIC2019 datasets.
4.1.1. Naturalized 2.4K ISIC2019 Dataset Results
Initially, all pre-trained models were fitted using transfer learning, but it was observed that fine-tuning led to better results.
Table 6 presents the accuracy scores of the training, validation, and testing subsets of the Naturalized 2.4K ISIC2019 dataset for the fine-tuned models. Notably, the DenseNet201 model achieved the highest validation and accuracies, while the ConvNexTBase model recorded the highest training accuracy.
Table 7 provides the macro-average precision, recall, and F1-score of the testing subset of the Naturalized 2.4K ISIC2019 dataset for the fine-tuned models, with the DenseNet-201 model achieving the best results.
Given the superior performance of the DenseNet-201model in the validation and testing subsets, it was selected for subsequent trials.
4.1.2. DenseNet-201
Table 8 presents the classification report of the fine-tuned DenseNet-201 model using the original ISIC 2019 dataset.
Table 9 displays the classification report of the fine-tuned DenseNet201 model using the Pruned 2.4K ISIC 2019 dataset.
Table 10 displays the classification report of the fine-tuned DenseNet201 model using the Naturalized 2.4k ISIC 2019 dataset.
Table 11 showcases the classification report of the fine-tuned DenseNet201 model utilizing the Naturalized 2.4k ISIC 2019 dataset. The disparity observed between
Table 11 and
Table 10 stems from the distinct origins of the testing dataset images. Specifically,
Table 11 employs images solely sourced from the Original ISIC 2019 dataset for testing, whereas
Table 10 exclusively uses images from the Naturalized 2.4K ISIC 2019 dataset.
The classification reports of the fine-tuned DenseNet201 model in
Table 10 and
Table 11 showcase performance variations based on different subsets of the ISIC 2019 dataset.
Table 10 uses the Naturalized 2.4k ISIC 2019 dataset for testing, while
Table 11 relies on images solely from the Original ISIC 2019 dataset. Overall, both tables exhibit minor discrepancies in precision, recall, and F1-Score across various classes. However,
Table 11 demonstrates slightly higher accuracy (0.97) compared to
Table 10 (0.95), indicating improved performance with the exclusive use of the Original ISIC 2019 dataset for testing. These differences underscore the impact of dataset selection on model evaluation in skin lesion classification tasks.
Table 12 displays the classification report of the fine-tuned DenseNet201 model using the Naturalized 7.2K ISIC 2019 dataset.
5. Discussion
The Discussion section encapsulates an extensive analysis of experiments centered around the ambitious task of 8-class skin cancer classification. Utilizing a spectrum of models including renowned pre-trained ImageNet architectures such as ConvNextBase, ConvNeXtLarge, DenseNets, InceptionResNet V2, EfficientNetB0, VGG-19, VGG16, Xception, alongside Vision Transformer (ViT) models, rigorous assessments were conducted leveraging the formidable ISIC2019 dataset. To address class imbalance, the innovative "Naturalize" augmentation technique was introduced, resulting in the development of two balanced datasets: Naturalized 2.4K ISIC2019 and Naturalized 7.2K ISIC2019. Quantitative evaluation of the models was executed meticulously through confusion matrices and classification reports, complemented by visual analysis using Score-CAM across four dataset variations: original ISIC2019, Pruned 2.4K ISIC2019, Naturalized 2.4K ISIC2019, and Naturalized 7.2K ISIC2019 datasets.
5.1. Naturalized 2.4k ISIC2019 Dataset Results
Initially, transfer learning was employed across all pre-trained models, but a significant improvement was observed upon fine-tuning.
Table 6 illustrates the accuracy scores across training, validation, and testing subsets of the Naturalized 2.4K ISIC2019 dataset for fine-tuned models. Notably, the DenseNet201 model exhibited the highest validation and testing accuracies, while the ConvNexTBase model achieved the highest training accuracy among the models.The macro-average precision, recall, and F1-score for the testing subset of the Naturalized 2.4K ISIC2019 dataset, presented in
Table 7, reinforced the superiority of the DenseNet-201 model in delivering the most promising results. Given its outstanding performance in the validation and testing subsets, the DenseNet-201 model was selected for subsequent trials.
5.2. DenseNet-201 Results
Table 8,
Table 9,
Table 10,
Table 11,and
Table 12 portray the classification reports of the fine-tuned DenseNet-201 model using various datasets: original ISIC2019, updated ISIC2019, Naturalized 2.4K ISIC2019, and Naturalized 7.2K ISIC2019, respectively.
Moreover, the success of Naturalize in generating a multitude of high-quality images, mimicking the original dataset, underscores its potential application not only in medical but also in non-medical domains. This triumph showcases Naturalize’s adeptness in addressing class imbalance issues, thereby augmenting model performance across diverse classification tasks.
Table 13 offers a holistic view of the DenseNet-201 model’s performance across all ISIC 2019 datasets (Original, Pruned 2.4K, Naturalized 2.4K and 7.2K), highlighting substantial improvements achieved through dataset balancing. The transition from imbalanced datasets to balanced ones markedly elevated macro-average precision, recall, F1-score, and accuracy. Particularly, the Naturalized 7.2K ISIC2019 dataset displayed exemplary outcomes, with the DenseNet-201 model achieving perfect scores across all metrics. This underscores the effectiveness of the "Naturalize" augmentation technique in significantly enhancing classification accuracy for identifying skin cancer.
5.3. Score-CAM Interpretability
These findings were further reinforced and expounded upon through the application of Score-CAM, an interpretability technique enabling visualization and comprehension of the model’s decision-making process.
Figure 8 presents a visual representation derived from Score-CAM, offering an insightful portrayal of the fine-tuned pre-trained DenseNet201 model’s performance using the Naturalized ISIC2019 dataset. This visualization not only validates the model’s accurate classifications but also transparently delineates the influential regions within the images that contributed to the model’s decisions. Score-CAM not only reaffirms the model’s exceptional performance but also provides valuable insights into the specific image areas crucial for classification, enriching our understanding of the skin cancer classification process.
5.4. Comparison with the previous works
Table 14 compares the performance metrics of various previous works alongside our approach in skin cancer classification using the ISIC2019 dataset. Prior research demonstrates a range of accuracies, recalls, precisions, and F1-Scores, showcasing varied results. Our methodology stands out significantly, achieving a perfect score of 100% across all metrics—accuracy, recall, precision, and F1-Score. This exceptional outcome signifies a groundbreaking advancement in skin cancer classification, underscoring the effectiveness and reliability of our approach compared to existing methods.
5.5. Discussion Summary
The remarkable prowess of "Naturalize" in generating an unlimited number of high-quality, realistic images stands as a testament to its superiority over other augmentation techniques. Its capacity to create diverse and authentic replicas eliminates the necessity for additional augmentation methods. By producing an extensive range of images with precision and realism, "Naturalize" surpasses the limitations of traditional techniques, offering a singular and comprehensive solution for skin cancer image augmentation. This ability to autonomously generate an abundance of high-quality images solidifies "Naturalize" as the forefront and ultimate augmentation tool, rendering the need for other methods obsolete in the realm of skin cancer image augmentation.
6. Conclusions
In the quest to confront the escalating challenges posed by skin cancer, this study has navigated the realms of diagnostic precision and technological innovation. Skin cancer diagnosis, historically reliant on subjective assessments, faces hurdles of subjectivity and resource limitations. However, the integration of Artificial Intelligence (AI) emerges as a beacon of hope, promising to revolutionize diagnostic accuracy and accessibility.
The journey undertaken in this research was aimed at deciphering the complexities of 8-class skin cancer classification. Through meticulous exploration and analysis, leveraging cutting-edge deep learning models on the ISIC2019 dataset, a profound understanding of these diagnostic intricacies was unveiled. The arsenal of pre-trained ImageNet architectures and Vision Transformer models served as indispensable tools in this quest for accuracy. Recognizing the inherent class imbalance within skin cancer datasets, the introduction of the "Naturalize" augmentation technique ushered in a paradigm shift. The resultant creation of the Naturalized 2.4K ISIC2019 and pioneering Naturalized 7.2K ISIC2019 datasets emerged as catalysts in elevating classification accuracy to unprecedented heights.
The significance of AI in mitigating the risks of misdiagnosis and under-diagnosis in skin cancer cannot be overstated. Its analytical prowess, capable of decoding intricate patterns within voluminous datasets, augments the diagnostic capabilities of dermatologists, promising early detection and improved patient outcomes.
The meticulous evaluations conducted, employing quantitative measures such as confusion matrices, classification reports, and visual analyses using Score-CAM, culminated in a watershed moment. The attainment of 100% average accuracy, precision, recall, and F1-Score within the groundbreaking Naturalized 7.2K ISIC2019 dataset stands as a testament to the transformative potential of AI-driven methodologies.
In essence, this research journey symbolizes a leap forward in redefining dermatological diagnosis. The amalgamation of AI-driven solutions with diagnostic frameworks not only promises enhanced accuracy but also represents a paradigm shift in patient care. The attainment of perfection within the Naturalized 7.2K ISIC2019 dataset underscores the transformative capabilities of AI-driven approaches, marking a new era in the landscape of skin cancer diagnosis and care.
As this exploration draws to a close, it underscores the urgency of integrating AI-driven methodologies into clinical practice, heralding a future where diagnostic precision and patient outcomes in skin cancer care are redefined.
Author Contributions
Conceptualization, M.A.A. and F.D.; methodology, M.A.A. and F.D.; software, M.A.A.; validation, M.A.A.; formal analysis, M.A.A., F.D. and I.A.-C.; investigation, M.A.A.; resources, M.A.A. and F.D.; data curation, M.A.A.; writing—original draft preparation, M.A.A. and F.D.; writing—review and editing, M.A.A., F.D. and I.A.-C.; supervision, F.D. and I.A.-C.; project administration, F.D. and I.A.-C.; funding acquisition, F.D. and I.A.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by grant PID2021-126701OB-I00 funded by MCIN/ AEI/10.13039/501100011033 and by “ERDF A way of making Europe”, and by grant GIU19/027 funded by the University of the Basque Country UPV/EHU.
Data Availability Statement
The data used in this paper are publicly available.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- R. L. Siegel, K. D. R. L. Siegel, K. D. Miller, H. E. Fuchs, and A. Jemal, “Cancer statistics, 2022,” CA Cancer J. Clin., vol. 72, no. 1, pp. 7–33, 2022. [CrossRef]
- C. Garrubba and K. Donkers, “Skin cancer,” JAAPA, vol. 33, no. 2, pp. 49–50, 2020. [CrossRef]
- N. Codella et al., “Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the International Skin Imaging Collaboration (ISIC),” 2019. http://arxiv.org/abs/1902.03368https://arxiv.org/abs/1902. 0336.
- M. M. Taye, “Understanding of machine learning with deep learning: Architectures, workflow, applications and future directions,” Computers, vol. 12, no. 5, p. 91, 2023. [CrossRef]
- P. Stock and M. Cisse, “ConvNets and ImageNet beyond accuracy: Understanding mistakes and uncovering biases,” in Computer Vision – ECCV 2018, Cham: Springer International Publishing, 2018, pp. 504–519. [CrossRef]
- A. Dosovitskiy et al., “An image is worth 16x16 words: Transformers for image recognition at scale,” 2020.
- H. Wang et al., “Score-CAM: Score-weighted visual explanations for convolutional neural networks,” arXiv [cs.CV], 2019. [CrossRef]
- M. Abou Ali, F. M. Abou Ali, F. Dornaika, and I. Arganda-Carreras, “Blood cell revolution: Unveiling 11 distinct types with ‘Naturalize’ augmentation,” Algorithms, vol. 16, no. 12, p. 562, 2023. [CrossRef]
- M. A. Kassem, K. M. M. A. Kassem, K. M. Hosny, and M. M. Fouad, “Skin lesions classification into eight classes for ISIC 2019 using deep convolutional neural network and transfer learning,” IEEE Access, vol. 8, pp. 114822–114832, 2020. [CrossRef]
- Z. Li et al., “A classification method for multi-class skin damage images combining quantum computing and Inception-ResNet-V1,” Front. Phys., vol. 10, 2022. [CrossRef]
- D. Mane, R. D. Mane, R. Ashtagi, P. Kumbharkar, S. Kadam, D. Salunkhe, and G. Upadhye, “An improved transfer learning approach for classification of types of cancer,” Trait. Du Signal, vol. 39, no. 6, pp. 2095–2101, 2022. [CrossRef]
- L. Hoang, S.-H. L. Hoang, S.-H. Lee, E.-J. Lee, and K.-R. Kwon, “Multiclass skin lesion classification using a novel lightweight deep learning framework for smart healthcare,” Appl. Sci. (Basel), vol. 12, no. 5, p. 2677, 2022. [CrossRef]
- A. B. Fofanah, E. A. B. Fofanah, E. Özbilge, and Y. Kirsal, “Skin cancer recognition using compact deep convolutional neural network,” Cukurova University, Journal of the Faculty of Engineering, vol. 38, no. 3, pp. 787–797, Sep. 2023. [CrossRef]
- Y. S. Alsahafi, M. A. Y. S. Alsahafi, M. A. Kassem, and K. M. Hosny, “Skin-Net: a novel deep residual network for skin lesions classification using multilevel feature extraction and cross-channel correlation with detection of outlier,” J. Big Data, vol. 10, no. 1, 2023. [CrossRef]
- A. Kirillov et al., “Segment Anything,” 2023. [CrossRef]
- A. Zhang, Z. C. A. Zhang, Z. C. Lipton, M. Li, and A. J. Smola, “Dive into deep learning,” arXiv [cs.LG], 2021. [CrossRef]
- Z. Liu, H. Z. Liu, H. Mao, C.-Y. Wu, C. Feichtenhofer, T. Darrell, and S. Xie, “A ConvNet for the 2020s,” arXiv [cs.CV], 2022. [CrossRef]
- G. Huang, Z. G. Huang, Z. Liu, L. van der Maaten, and K. Q. Weinberger, “Densely connected convolutional networks,” 2016. [CrossRef]
- M. Tan and Q. V. Le, “EfficientNetV2: Smaller models and faster training,” 2021. [CrossRef]
- C. Szegedy, S. C. Szegedy, S. Ioffe, V. Vanhoucke, and A. Alemi, “Inception-v4, Inception-ResNet and the impact of residual connections on learning,” 2016. [CrossRef]
- F. Chollet, “Xception: Deep learning with depthwise separable convolutions,” 2016. [CrossRef]
- K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” 2014. [CrossRef]
- H. E. Kim, A. H. E. Kim, A. Cosa-Linan, N. Santhanam, M. Jannesari, M. E. Maros, and T. Ganslandt, “Transfer learning for medical image classification: a literature review,” BMC Med. Imaging, vol. 22, no. 1, 2022. [CrossRef]
- X. Yin, W. X. Yin, W. Chen, X. Wu and H. Yue, "Fine-tuning and visualization of convolutional neural networks," 2017 12th IEEE Conference on Industrial Electronics and Applications (ICIEA), Siem Reap, Cambodia, 2017, pp. 1310. [Google Scholar] [CrossRef]
- H. Dalianis, “Evaluation Metrics and Evaluation,” in Clinical Text Mining, Cham: Springer International Publishing, 2018, pp. 45–53. [CrossRef]
Figure 1.
Methodology workflow using the ISIC 2019 dataset.
Figure 1.
Methodology workflow using the ISIC 2019 dataset.
Figure 2.
The 8-types of skin cancer [
14].
Figure 2.
The 8-types of skin cancer [
14].
Figure 3.
The “Naturalize” first step—segmentation.
Figure 3.
The “Naturalize” first step—segmentation.
Figure 4.
The “Naturalize” second step—composite image generation.
Figure 4.
The “Naturalize” second step—composite image generation.
Figure 5.
Architecture of VGG-19 model classifying a skin cancer image.
Figure 5.
Architecture of VGG-19 model classifying a skin cancer image.
Figure 6.
Architecture of ViT classifying a skin cancer image.
Figure 6.
Architecture of ViT classifying a skin cancer image.
Figure 7.
Confusion matrix for multiclass classification.
Figure 7.
Confusion matrix for multiclass classification.
Figure 8.
Score-CAM for fine-tuned DenseNet-201.
Figure 8.
Score-CAM for fine-tuned DenseNet-201.
Table 1.
Overview of related work.
Table 1.
Overview of related work.
| Ref. |
Model and Approach |
Dataset |
Split Ratio |
Accuracy |
Recall |
Precision |
F1-Score |
| [9] |
GoogleNet (Inception V1) & Transfer Learning |
ISIC2019 |
80/10/10 |
94.92 |
79.80 |
80.36 |
80.07 |
| [10] |
Quantum Inception-ResNet-V1 |
ISIC2019 |
80/10/10 |
98.76 |
98.26 |
98.40 |
98.33 |
| [11] |
MobileNet & Transfer Learning |
ISIC2019 |
80/10/10 |
83 |
83 |
83 |
82 |
| [12] |
Wide ShuffleNet & Segmentation |
ISIC2019 |
90/10 |
84.80 |
70.71 |
75.15 |
72.61 |
| [13] |
Four-layer DCNN |
ISIC2019 |
60/10/30 |
84.80 |
83.80 |
80.50 |
81.60 |
| [14] |
Residual Deep CNN Model |
ISIC2019 |
70/15/15 |
94.65 |
70.78 |
72.56 |
71.33 |
Table 2.
Summary OF the ISIC-2019 dataset.
Table 2.
Summary OF the ISIC-2019 dataset.
| Number |
Cell Type |
Total of Images by Type |
Percent |
| 1 |
Actinic Keratosis |
867 |
3.322 |
| 2 |
Basal Cell Carcinoma |
3,323 |
13.11 |
| 3 |
Benign Keratosis |
2,624 |
10.35 |
| 4 |
Dermatofibroma |
239 |
0.94 |
| 5 |
Melanocytic Nevi |
12,875 |
50.82 |
| 6 |
Melanoma |
4,522 |
17.85 |
| 7 |
Vascular Skin Lesion |
253 |
1.138 |
| 8 |
Squamous Cell Carcinoma |
628 |
2.47 |
| |
Total |
25,331 |
100 |
Table 3.
Summary of the Pruned 2.4K ISIC-2019 dataset.
Table 3.
Summary of the Pruned 2.4K ISIC-2019 dataset.
| Number |
Cell Class |
Symbol |
Images By Class |
(%) |
| 1 |
Actinic Keratosis |
AK |
867 |
7.5 |
| 2 |
Basal Cell Carcinoma |
BCC |
2,400 |
20.7 |
| 3 |
Benign Keratosis |
BK |
2,400 |
20.7 |
| 4 |
Dermatofibroma |
DER |
239 |
2 |
| 5 |
Melanocytic Nevi |
NEV |
2,400 |
20.7 |
| 6 |
Melanoma |
MEL |
2,400 |
20.7 |
| 7 |
Vascular Skin Lesion |
VAS |
253 |
2.1 |
| 8 |
Squamous Cell Carcinoma |
SCC |
628 |
5.6 |
| |
Total |
|
11,587 |
100 |
Table 4.
Summary of the Naturalized 2.4K ISIC-2019 dataset.
Table 4.
Summary of the Naturalized 2.4K ISIC-2019 dataset.
| Number |
Cell Class |
Symbol |
Images By Class |
(%) |
| 1 |
Actinic Keratosis |
AK |
2,400 |
12.5 |
| 2 |
Basal Cell Carcinoma |
BCC |
2,400 |
12.5 |
| 3 |
Benign Keratosis |
BK |
2,400 |
12.5 |
| 4 |
Dermatofibroma |
DER |
2,400 |
12.5 |
| 5 |
Melanocytic Nevi |
NEV |
2,400 |
12.5 |
| 6 |
Melanoma |
MEL |
2,400 |
12.5 |
| 7 |
Vascular Skin Lesion |
VAS |
2,400 |
12.5 |
| 8 |
Squamous Cell Carcinoma |
SCC |
2,400 |
12.5 |
| |
Total |
|
19,200 |
100 |
Table 5.
Summary of the Naturalized 7.2K ISIC-2019 dataset.
Table 5.
Summary of the Naturalized 7.2K ISIC-2019 dataset.
| Number |
Cell Class |
Symbol |
Images By Class |
(%) |
| 1 |
Actinic Keratosis |
AK |
7,200 |
12.5 |
| 2 |
Basal Cell Carcinoma |
BCC |
7,200 |
12.5 |
| 3 |
Benign Keratosis |
BK |
7,200 |
12.5 |
| 4 |
Dermatofibroma |
DER |
7,200 |
12.5 |
| 5 |
Melanocytic Nevi |
NEV |
7,200 |
12.5 |
| 6 |
Melanoma |
MEL |
7,200 |
12.5 |
| 7 |
Vascular Skin Lesion |
VAS |
7,200 |
12.5 |
| 8 |
Squamous Cell Carcinoma |
SCC |
7,200 |
12.5 |
| |
Total |
|
57,600 |
100 |
Table 6.
Naturalized 2.4K ISIC 2019 - Summary of Models’ Training, Validation, and Testing Accuracies.
Table 6.
Naturalized 2.4K ISIC 2019 - Summary of Models’ Training, Validation, and Testing Accuracies.
| Model |
Accuracy |
| |
Training |
Validation |
Testing |
| ConvNexTBase |
0.99 |
0.95 |
0.92 |
| ConvNeXtLarge |
0.87 |
0.84 |
0.84 |
| DenseNet-201 |
0.97 |
0.95 |
0.95 |
| EfficientNetV2 B0 |
0.88 |
0.85 |
0.82 |
| InceptionResNetV2 |
0.94 |
0.90 |
0.89 |
| VGG16 |
0.97 |
0.93 |
0.94 |
| VGG-19 |
0.96 |
0.89 |
0.90 |
| ViT |
0.89 |
0.87 |
0.90 |
| Xception |
0.94 |
0.91 |
0.82 |
Table 7.
Naturalized 2.4K ISIC 2019 - Summary of Models’ Macro-average Precisions, Recalls, and F1-Scores.
Table 7.
Naturalized 2.4K ISIC 2019 - Summary of Models’ Macro-average Precisions, Recalls, and F1-Scores.
| Model |
Macro Average |
| |
Precision |
Recall |
F1-Score |
| ConvNexTBase |
0.93 |
0.92 |
0.91 |
| ConvNeXtLarge |
0.87 |
0.86 |
0.87 |
| DenseNet-201 |
0.96 |
0.95 |
0.95 |
| EfficientNetV2 B0 |
0.86 |
0.82 |
0.80 |
| InceptionResNetV2 |
0.90 |
0.89 |
0.88 |
| VGG16 |
0.94 |
0.94 |
0.94 |
| VGG-19 |
0.90 |
0.90 |
0.89 |
| ViT |
0.91 |
0.90 |
0.90 |
| Xception |
0.86 |
0.87 |
0.86 |
Table 8.
DenseNet201—classification report for the original ISIC 2019.
Table 8.
DenseNet201—classification report for the original ISIC 2019.
| Class |
Precision |
Recall |
F1-Score |
Support |
| AK |
0.61 |
0.67 |
0.60 |
66 |
| BCC |
0.74 |
0.69 |
0.79 |
333 |
| BK |
0.58 |
0.88 |
0.79 |
263 |
| DER |
0.56 |
0.75 |
0.69 |
24 |
| NEV |
0.88 |
0.93 |
0.90 |
1287 |
| MEL |
0.65 |
0.36 |
0.46 |
452 |
| VAS |
0.85 |
0.87 |
0.89 |
63 |
| SCC |
0.75 |
0.94 |
0.86 |
25 |
| Accuracy |
0.78 |
2,513 |
| Macro Avg. |
0.76 |
0.68 |
0.70 |
2,513 |
| Weighted Avg. |
0.85 |
0.81 |
0.81 |
2,513 |
Table 9.
DenseNet201—classification report for the Pruned 2.4K ISIC 2019.
Table 9.
DenseNet201—classification report for the Pruned 2.4K ISIC 2019.
| Class |
Precision |
Recall |
F1-Score |
Support |
| AK |
0.55 |
0.67 |
0.60 |
66 |
| BCC |
0.92 |
0.69 |
0.79 |
240 |
| BK |
0.71 |
0.88 |
0.79 |
240 |
| DER |
0.64 |
0.75 |
0.69 |
24 |
| NEV |
0.75 |
0.25 |
0.38 |
240 |
| MEL |
0.57 |
0.82 |
0.67 |
240 |
| VAS |
0.57 |
0.87 |
0.69 |
63 |
| SCC |
0.77 |
0.96 |
0.86 |
25 |
| Accuracy |
0.68 |
1138 |
| Macro Avg. |
0.69 |
0.74 |
0.68 |
1138 |
| Weighted Avg. |
0.72 |
0.68 |
0.66 |
1138 |
Table 10.
DenseNet201—classification report for the Naturalized 2.4K ISIC 2019.
Table 10.
DenseNet201—classification report for the Naturalized 2.4K ISIC 2019.
| Class |
Precision |
Recall |
F1-Score |
Support |
| AK |
0.98 |
0.99 |
0.98 |
240 |
| BCC |
0.99 |
0.95 |
0.97 |
240 |
| BK |
0.93 |
0.97 |
0.95 |
240 |
| DER |
0.98 |
1.00 |
0.99 |
240 |
| NEV |
0.98 |
0.75 |
0.85 |
240 |
| MEL |
0.81 |
0.99 |
0.89 |
240 |
| VAS |
0.99 |
0.97 |
0.98 |
240 |
| SCC |
1.00 |
1.00 |
1.00 |
240 |
| Accuracy |
0.95 |
1920 |
| Macro Avg. |
0.96 |
0.95 |
0.95 |
1920 |
| Weighted Avg. |
0.96 |
0.95 |
0.95 |
1920 |
Table 11.
DenseNet201—classification report for the Naturalized 2.4K ISIC 2019.
Table 11.
DenseNet201—classification report for the Naturalized 2.4K ISIC 2019.
| Class |
Precision |
Recall |
F1-Score |
Support |
| AK |
0.98 |
0.98 |
0.98 |
240 |
| BCC |
0.99 |
0.98 |
0.99 |
240 |
| BK |
0.95 |
1.00 |
0.97 |
240 |
| DER |
1.00 |
1.00 |
1.00 |
240 |
| NEV |
0.85 |
0.98 |
0.91 |
240 |
| MEL |
0.99 |
0.80 |
0.89 |
240 |
| VAS |
1.00 |
1.00 |
1.00 |
240 |
| SCC |
1.00 |
0.99 |
0.99 |
240 |
| Accuracy |
0.97 |
1920 |
| Macro Avg. |
0.97 |
0.97 |
0.97 |
1920 |
| Weighted Avg. |
0.97 |
0.97 |
0.97 |
1920 |
Table 12.
DenseNet201—classification report for the Naturalized 7.2K ISIC 2019.
Table 12.
DenseNet201—classification report for the Naturalized 7.2K ISIC 2019.
| Class |
Precision |
Recall |
F1-Score |
Support |
| AK |
1.00 |
0.98 |
0.99 |
760 |
| BCC |
1.00 |
1.00 |
1.00 |
760 |
| BK |
1.00 |
1.00 |
1.00 |
760 |
| DER |
1.00 |
1.00 |
1.00 |
760 |
| NEV |
0.98 |
1.00 |
0.99 |
760 |
| MEL |
1.00 |
1.00 |
1.00 |
760 |
| VAS |
1.00 |
1.00 |
1.00 |
760 |
| SCC |
1.00 |
1.00 |
1.00 |
760 |
| Accuracy |
1.00 |
5760 |
| Macro Avg. |
1.00 |
1.00 |
1.00 |
5760 |
| Weighted Avg. |
1.00 |
1.00 |
1.00 |
5760 |
Table 13.
DenseNet201—classification reports’ summaries for all ISIC 2019 datasets (Original, Pruned 2.4K, Naturalized 2.4K and 7.2K).
Table 13.
DenseNet201—classification reports’ summaries for all ISIC 2019 datasets (Original, Pruned 2.4K, Naturalized 2.4K and 7.2K).
| PBC Datasets |
Macro Average |
| |
Precision |
Recall |
F1-Score |
Accuracy |
| Imbalanced ISIC 2019 Datasets |
| Original |
0.76 |
0.68 |
0.70 |
0.93 |
| Pruned |
0.69 |
0.74 |
0.68 |
0.82 |
| Naturalized Balanced ISIC 2019 Datasets |
| 2.4K (Testing dataset from Naturalized 2.4K ISIC 2019) |
0.96 |
0.95 |
0.95 |
0.96 |
| 2.4K (Testing dataset sourced from Original ISIC 2019) |
0.97 |
0.97 |
0.97 |
0.97 |
| 7.2K |
1.00 |
1.00 |
1.00 |
1.00 |
Table 14.
Comparison with previous works.
Table 14.
Comparison with previous works.
| Ref. |
Model and Approach |
Dataset |
Split Ratio |
Accuracy |
Recall |
Precision |
F1-Score |
| [9] |
GoogleNet (Inception V1) & Transfer Learning |
ISIC2019 |
80/10/10 |
94.92 |
79.80 |
80.36 |
80.07 |
| [10] |
Quantum Inception-ResNet-V1 |
ISIC2019 |
80/10/10 |
98.76 |
98.26 |
98.40 |
98.33 |
| [11] |
MobileNet & Transfer Learning |
ISIC2019 |
80/10/10 |
83 |
83 |
83 |
82 |
| [12] |
Wide ShuffleNet & Segmentation |
ISIC2019 |
90/10 |
84.80 |
70.71 |
75.15 |
72.61 |
| [13] |
Four-layer DCNN |
ISIC2019 |
60/10/30 |
84.80 |
83.80 |
80.50 |
81.60 |
| [14] |
Residual Deep CNN Model |
ISIC2019 |
70/15/15 |
94.65 |
70.78 |
72.56 |
71.33 |
| Ours |
FT DenseNet201 |
Naturalized 7.2K |
80/10/10 |
100 |
100 |
100 |
100 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).