Submitted:
18 January 2024
Posted:
18 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
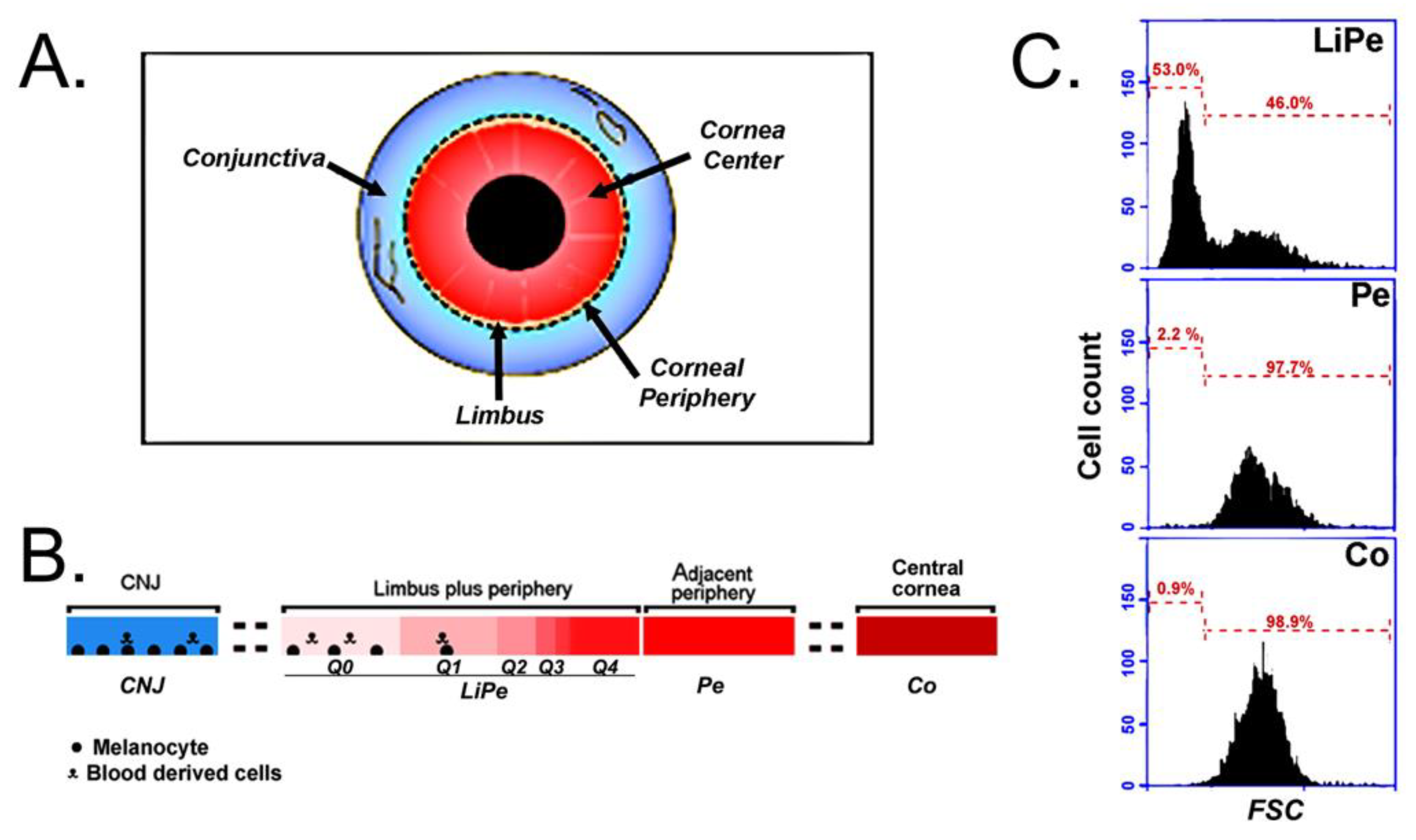
2.1. Tissue Procurement
2.2. Cell Preparation
2.3. Single Cell RNA-Seq Reading
3. Results
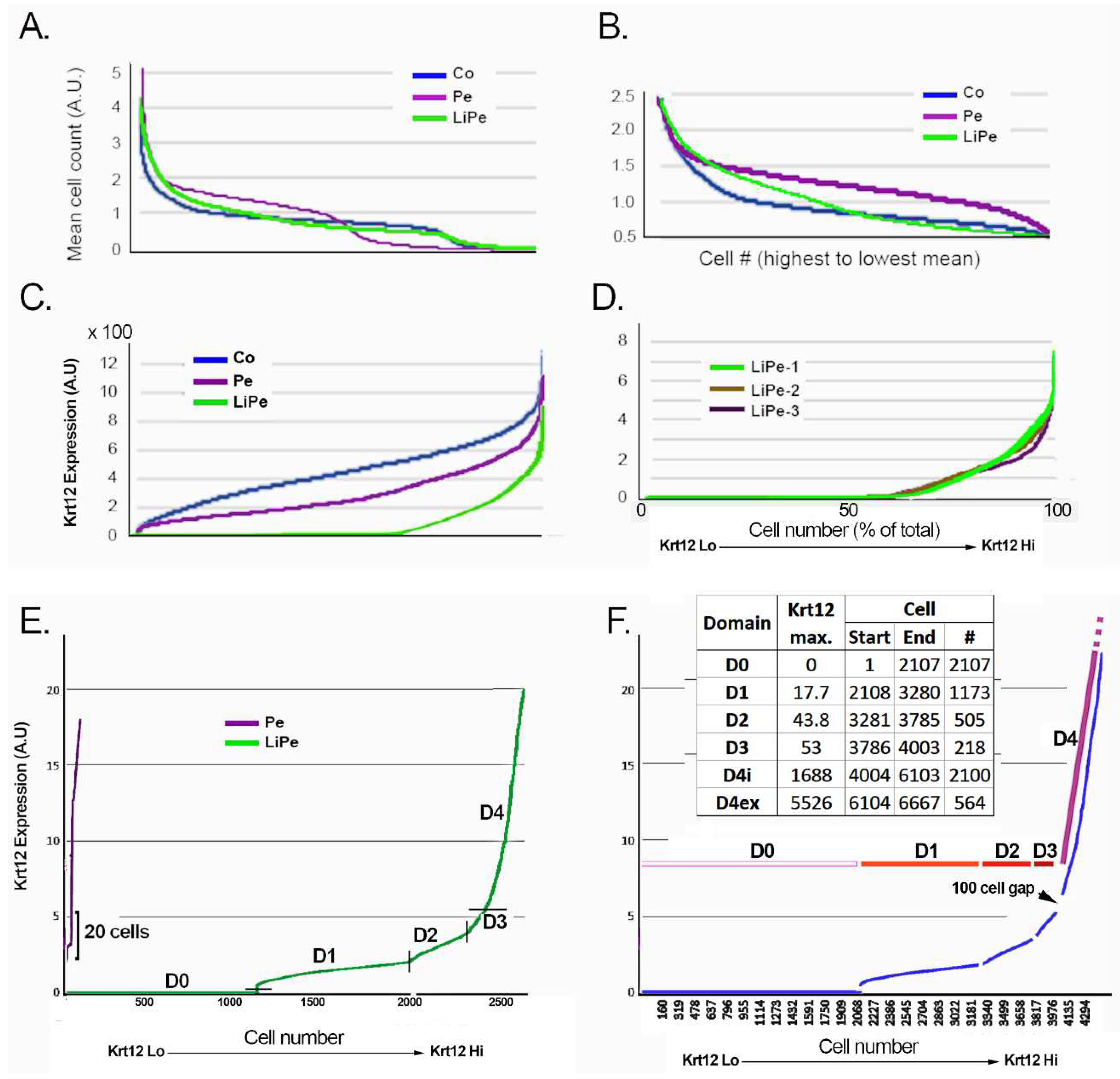
3.1. Data Pre-Processing
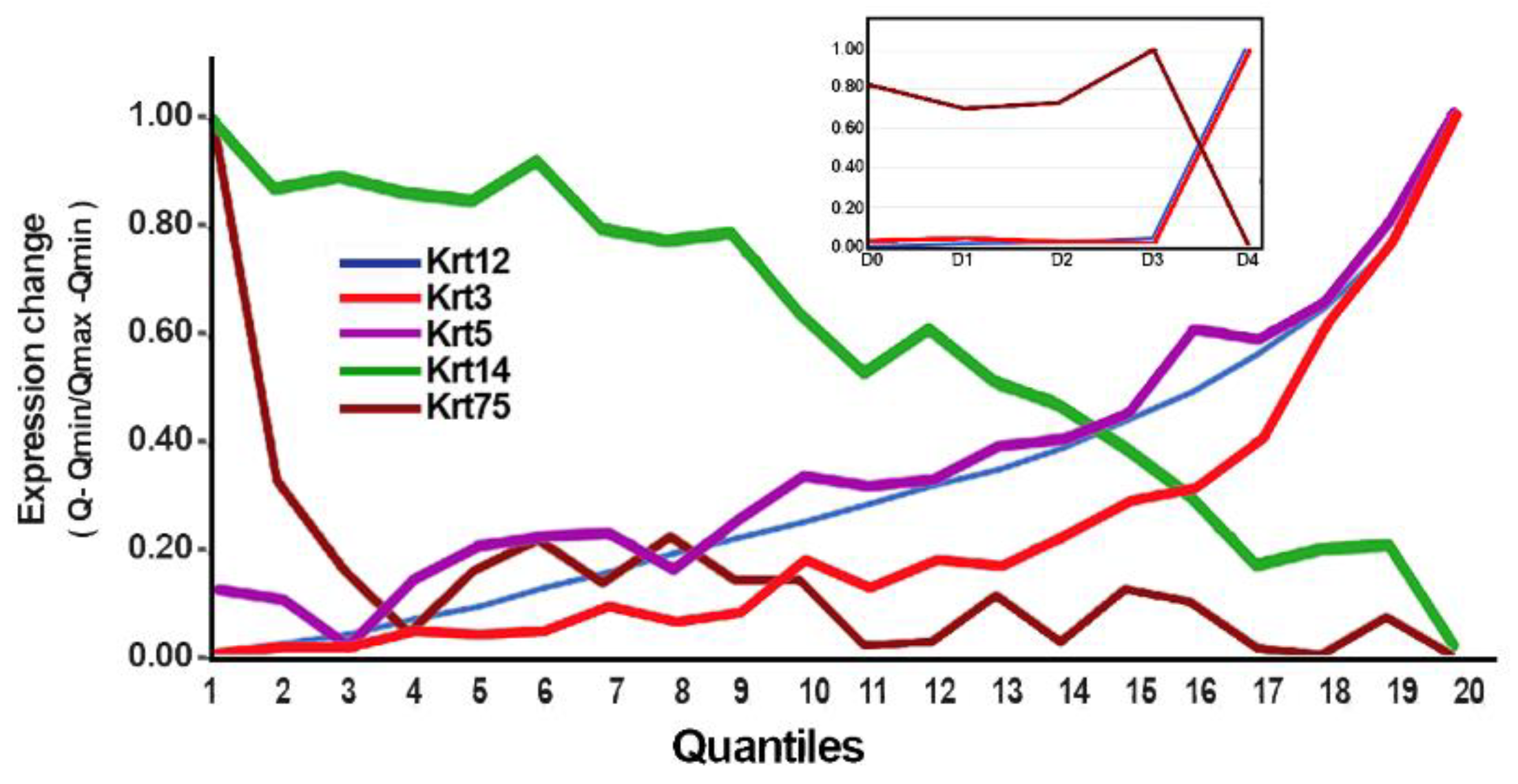
3.2. Domain Identification
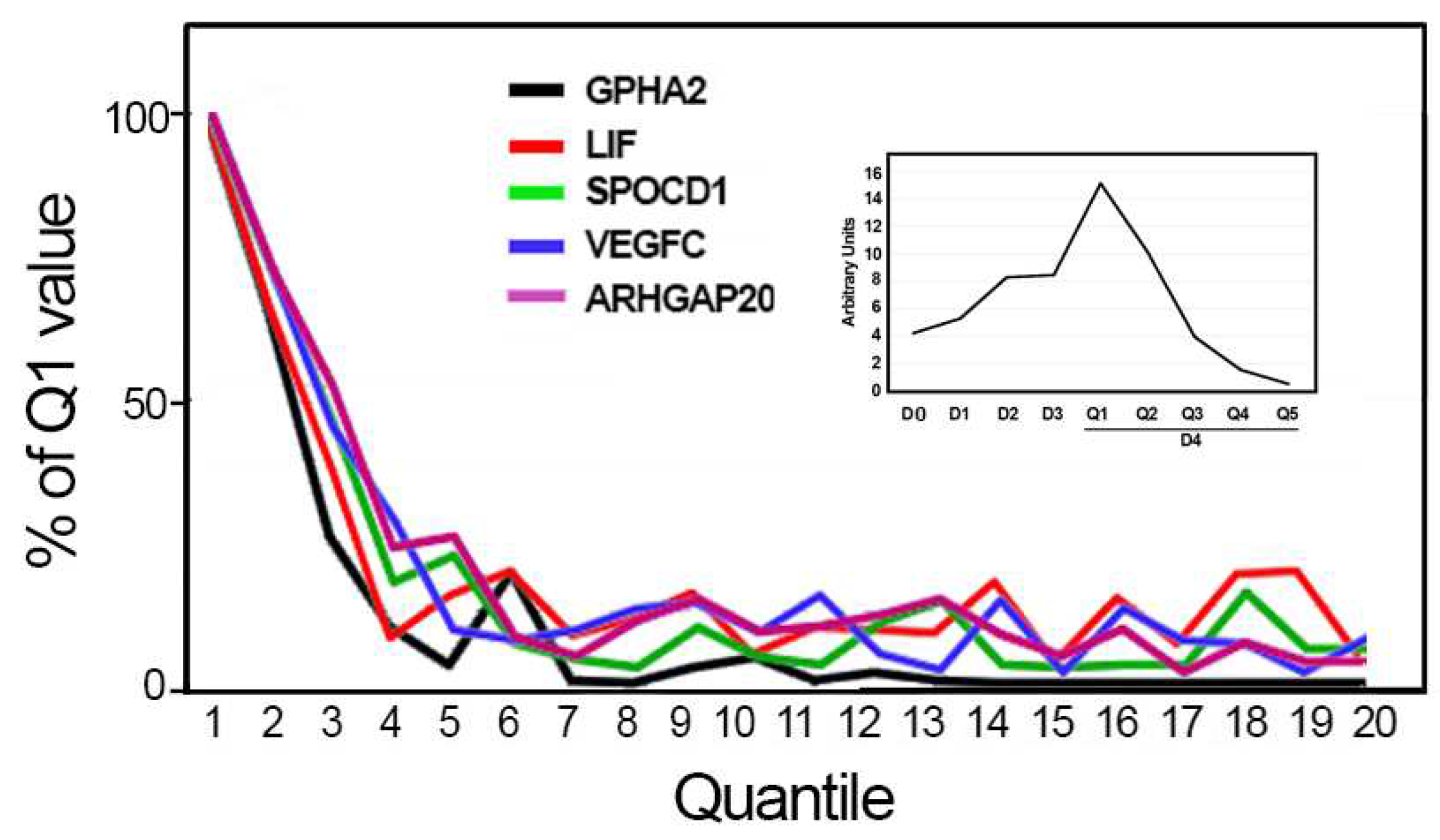
3.3. Gene Expression Differential Analysis
3.4. Gene Ontology Analysis
3.5. Gene Expression Correlation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Conflict of Interest
References
- Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM. Cell. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell 1989, 57, 201–209. [CrossRef]
- Wolosin JM, Xiong X, Schütte M, Stegman Z, Tieng A. Stem cells and differentiation stages in the limbo-corneal epithelium. Prog Retin Eye Res. 2000, 19, 223–255. [Google Scholar] [CrossRef] [PubMed]
- Moreno IY, Parsaie A, Gesteira TF, Coulson-Thomas VJ. Characterization of the Limbal Epithelial Stem Cell Niche. Invest Ophthalmol Vis Sci. 2023, 64, 48. [Google Scholar] [CrossRef] [PubMed]
- Mol R, Franke W W, Schiller D L, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell 1982, 31, 11–24. [CrossRef]
- Takahiro Nakamura 1, Ken-Ichi Endo, Leanne J Cooper, Nigel J Fullwood, Noriko Tanifuji, Masakatsu Tsuzuki, Noriko Koizumi, Tsutomu natomi, Yoichiro Sano, Shigeru Kinoshita. The successful culture and autologous transplantation of rabbit oral mucosal epithelial cells on amniotic membrane. Invest Ophthalmol Vis Sci. 2003, 44, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Schermer A, Galvin S, Sun TT. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986, 103, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Tseng, S.C. Concept and application of limbal stem cells. Eye (Lond). 1989, 3 Pt 2, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Turner HC, Budak MT, Akinci MA, Wolosin JM. Comparative analysis of human conjunctival and corneal epithelial gene expression with oligonucleotide microarrays. Invest Ophthalmol Vis Sci. 2007, 48, 2050–2061. [Google Scholar] [CrossRef] [PubMed]
- Zhou M, Li X-m, Lavker, RM Transcriptional profiling of enriched populations of stem cells versus transient amplifying cells. A comparison of limbal and corneal epithelial basal cells. J Biol Chem. 2006, 281, 19600–19609. [Google Scholar]
- Akinci MA, Turner H, Taveras M, Wolosin JM Differential gene expression in the pig limbal side population: implications for stem cell cycling, replication, and survival. Invest Ophthalmol Vis Sci. 2009, 50, 5630–5638. [CrossRef] [PubMed]
- Kaplan N, Wang J, Wray B, Patel P, Yang W, Peng H, Lavker RM. Single-Cell RNA Transcriptome Helps Define the Limbal/Corneal Epithelial Stem/Early Transit Amplifying Cells and How Autophagy Affects This Population. Invest Ophthalmol Vis Sci. 2019, 60, 3570–3583. [Google Scholar] [CrossRef] [PubMed]
- Li DQ, Kim S, Li JM, Gao Q, Choi J, Bian F, Hu J, Zhang Y, Li J, Lu R, Li Y, Pflugfelder SC, Miao H, Chen R. Single-cell transcriptomics identifies limbal stem cell population and cell types mapping its differentiation trajectory in limbal basal epithelium of human cornea. Ocul Surf. 2021, 20, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Ligocki AJ, Fury W, Gutierrez C, Adler C, Yang T, Ni M, Bai Y, Wei Y, Lehmann GL, Romano C. Molecular characteristics and spatial distribution of adult human corneal cell subtypes. Sci Rep. 2021, 11, 16323. [Google Scholar] [CrossRef] [PubMed]
- Collin J, Queen R, Zerti D, Bojic S, Dorgau B, Moyse N, Molina MM, Yang C, Dey S, Reynolds G, Hussain R, Coxhead JM, Lisgo S, Henderson D, Joseph A, Rooney P, Ghosh S, Clarke L, Connon C, Haniffa M, Figueiredo F, Armstrong L, Lako M. A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul Surf. 2021, 21, 279–298. [Google Scholar] [CrossRef] [PubMed]
- Romano, AC; Espana, EM; Yoo, SH; Budak, MT; Wolosin JM; Tseng SCG. Different Cell Sizes in Human Limbal and Central Corneal Basal Epithelia Measured by Confocal Microscopy and Flow Cytometry. Investigative Ophthalmology & Visual Science December 2003, 44, 5125–5129. [CrossRef]
- Turner HC, Budak MT, Akinci MA, Wolosin JM Comparative analysis of human conjunctival and corneal epithelial gene expression with oligonucleotide microarrays. Invest Ophthalmol Vis Sci. 2007, 48, 2050–2061. [CrossRef] [PubMed]
- Mauri F, Schepkens C, Lapouge G, Drogat B, Song Y, Pastushenko I, Rorive S, Blondeau J, Golstein S, Bareche Y, Miglianico M, Nkusi E, Rozzi M, Moers V, Brisebarre A, Raphaël M, Dubois C, Allard J, Durdu B, Ribeiro F, Sotiriou C, Salmon I, Vakili J, Blanpain C. NR2F2 controls malignant squamous cell carcinoma state by promoting stemness and invasion and repressing differentiation. Nat Cancer. 2021, 2, 1152–1169. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H, Du, M, Li, Y, Zhou, T, Lei, J, Liang, H, Zhong, Z, Al-Lamki, RS, Jiang, M, Yang J. ID proteins promote the survival and primed-to-naive transition of human embryonic stem cells through TCF3-mediated transcription Cell Death Dis. 2022, 13, 549.
- Tomlinson, RL, Lukowiak, AA, Terns, RN, Terns, MP Telomerase RNA Accumulates in Cajal Bodies in Human Cancer. Cells Mol Biol Cell 2004, 15, 81–90. [CrossRef] [PubMed]
- Bizarro J, Bhardwaj A, Smith S, Meier UT. Nopp140-mediated concentration of telomerase in Cajal bodies and regulates telomere length. Mol Biol Cell. 2019, 30, 3136–3150. [Google Scholar] [CrossRef] [PubMed]

| Limbal-Periphery-1 | Limbal-Periphery-2 | LimbalPeriphery-3 | LiPe Aver. | Periphery-1 | Central Cornea-1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| # | % | # | % | # | % | % | # | % | # | % | |
| Total cells | 5843 | 100 | 2234 | 100 | 1981 | 100 | 100.00 | 3731 | 100 | 7194 | 100 |
| Melanocytes | 585 | 10.01 | 143 | 6.4 | 40 | 2 | 6.14 | 25 | 0.67 | 25 | 0.35 |
| Langerhans | 3 | 0.05 | 0 | 0 | 1 | 0.05 | 0.03 | 0 | 0 | 2 | 0.03 |
| Blood cells | 3 | 0.05 | 2 | 0.09 | 0 | 0 | 0.05 | 0 | 0 | 0 | 0 |
| CNJE (AQP5) | 9 | 0.15 | 9 | 0.4 | 2 | 1 | 0.52 | 0 | 0 | 0 | 0 |
| Non Epithelial | 600 | 10.3 | 154 | 6.9 | 14 | 0.7 | 5.97 | 25 | 0.7 | 27 | 0.4 |
| Epithelial | 5243 | 89.7 | 2080 | 93.1 | 1924 | 97.1 | 93.30 | 3706 | 99.3 | 7167 | 99.6 |
| 0.5-2.5 limit* | 3640 | 69.4 | 1746 | 83.09 | 1281 | 66.5 | 72.32 | 1982 | 50.98 | 5060 | 71.39 |
| Krt12= 0* | 1118 | 30.2 | 543.00 | 31.3 | 446.00 | 34.6 | 32.00 | 0 | 0 | 0 | 0 |
| DEG | D0-D1 | D1-D23 | D0-D123 | D123-Di4.1 | Di (4.1-4.2) | Krt12 lo/hi | Ratio | D1/D123 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| down | up | down | up | down | up | down | up | down | up | down | up | down | up | ||
| All | 617 | 409 | 0 | 16 | 885 | 526 | 2383 | 1622 | 1107 | 1159 | 1563 | 3339 | Aver | 1.00 | 1.00 |
| R > 1.25x | 278 | 0 | 306 | 1876 | 762 | St Dev | 0.04 | 0.07 | |||||||
| 1/R> 1.25x >>1.25x | 145 | 8 | 172 | 1240 | 766 | p= | 0.62 | 0.99 | |||||||
| Gene | BHp | R | Gene | BHp= | R | Gene | BHp | R | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GRASP | 8.55E-07 | 1.76 | 21 | HSPA2 | 3.84E-07 | 1.55 | 41 | TSPAN13 | 3.13E-06 | 1.47 |
| 2 | NR2F2 | 3.27E-06 | 1.75 | 22 | USP31 | 1.40E-06 | 1.55 | 42 | PHLPP1 | 1.66E-05 | 1.47 |
| 3 | ARC | 9.04E-04 | 1.71 | 23 | SLC2A4RG | 1.32E-04 | 1.54 | 43 | FZD7 | 2.62E-04 | 1.47 |
| 4 | IER5L | 3.08E-04 | 1.7 | 24 | ABHD17B | 1.47E-04 | 1.54 | 44 | CFD | 9.02E-04 | 1.47 |
| 5 | ANKRD37 | 4.70E-04 | 1.67 | 25 | AC027031.2 | 9.36E-04 | 1.54 | 45 | EMG1 | 2.97E-05 | 1.46 |
| 6 | OSGIN1 | 1.05E-06 | 1.66 | 26 | STMN1 | 5.29E-09 | 1.53 | 46 | EID2 | 3.20E-04 | 1.46 |
| 7 | NPPC | 6.20E-05 | 1.66 | 27 | THAP11 | 3.84E-04 | 1.53 | 47 | AL035071.1 | 1.30E-03 | 1.46 |
| 8 | SOCS1 | 1.11E-05 | 1.65 | 28 | EIF1AY | 4.13E-05 | 1.51 | 48 | GYG1 | 2.56E-03 | 1.46 |
| 9 | POU3F1 | 3.88E-08 | 1.64 | 29 | FOS | 1.32E-04 | 1.51 | 49 | TSLP | 4.22E-03 | 1.46 |
| 10 | NCKAP5L | 3.86E-07 | 1.63 | 30 | CEBPA | 1.14E-03 | 1.51 | 50 | CEP290 | 5.15E-03 | 1.46 |
| 11 | TUSC1 | 7.90E-05 | 1.62 | 31 | CBX4 | 3.07E-06 | 1.5 | 51 | MYLIP | 7.66E-07 | 1.45 |
| 12 | cyp26 | 2.07E-03 | 1.61 | 32 | TXNDC11 | 3.09E-04 | 1.5 | 52 | PER1 | 8.34E-06 | 1.45 |
| 13 | TNFSF9 | 2.70E-05 | 1.6 | 33 | BDKRB1 | 4.57E-03 | 1.5 | 53 | EGLN1 | 8.49E-04 | 1.45 |
| 14 | TPM2 | 5.71E-04 | 1.6 | 34 | JUND | 9.95E-16 | 1.49 | 54 | NFIB | 4.14E-05 | 1.44 |
| 15 | SCARA3 | 1.28E-03 | 1.6 | 35 | HMGB2 | 4.86E-07 | 1.49 | 55 | RHOB | 2.80E-04 | 1.44 |
| 16 | TMEM263 | 1.03E-08 | 1.59 | 36 | POMZP3 | 2.93E-05 | 1.49 | 56 | ALKBH5 | 8.78E-04 | 1.44 |
| 17 | ID3 | 1.85E-03 | 1.59 | 37 | TMEM138 | 8.32E-05 | 1.49 | 57 | VEGFC | 2.85E-03 | 1.44 |
| 18 | ARL4A | 1.38E-07 | 1.57 | 38 | MEIS2 | 8.26E-05 | 1.48 | 58 | CXCL8 | 3.42E-03 | 1.44 |
| 19 | GEM | 3.59E-04 | 1.57 | 39 | HINT3 | 4.79E-04 | 1.48 | 59 | EGOT | 3.90E-03 | 1.44 |
| 20 | H2AFX | 2.50E-11 | 1.56 | 40 | IRX2 | 2.64E-10 | 1.47 | 60 | ID4 | 7.11E-07 | 1.43 |
| Gene | BHp | 1/R | Gene | BHp | 1/R | Gene | BHp | 1/R | |||
| 1 | SPRR2A | 1.06E-05 | 2.50 | 21 | AC010503.4 | 3.38E-04 | 1.54 | 41 | FAM83A | 5.23E-04 | 1.43 |
| 2 | LY6D | 8.83E-04 | 2.44 | 22 | ABAT | 2.43E-03 | 1.54 | 42 | GALNT7 | 9.78E-03 | 1.43 |
| 3 | SCGB2A1 | 1.37E-04 | 2.22 | 23 | ABCA12 | 5.66E-03 | 1.54 | 43 | RHOD | 4.30E-08 | 1.41 |
| 4 | UPK1B | 3.01E-05 | 2.00 | 24 | TRIP10 | 1.00E-06 | 1.52 | 44 | S100A14 | 2.14E-06 | 1.41 |
| 5 | MGST1 | 4.08E-04 | 1.89 | 25 | POLR2J3 | 4.33E-05 | 1.52 | 45 | SULT2B1 | 2.98E-06 | 1.41 |
| 6 | SH3KBP1 | 3.06E-04 | 1.85 | 26 | GPX2 | 1.28E-03 | 1.52 | 46 | PTPRE | 1.30E-05 | 1.41 |
| 7 | KRT13 | 2.35E-05 | 1.82 | 27 | CLEC7A | 4.47E-03 | 1.52 | 47 | ALDH3A1 | 9.06E-05 | 1.41 |
| 8 | CLCA2 | 5.82E-05 | 1.75 | 28 | CRTAC1 | 2.99E-06 | 1.49 | 48 | ZNF836 | 9.10E-05 | 1.41 |
| 9 | MFSD4A | 2.53E-03 | 1.75 | 29 | TJP2 | 5.08E-06 | 1.49 | 49 | AHNAK2 | 1.01E-04 | 1.41 |
| 10 | CDKN2A | 4.09E-03 | 1.75 | 30 | PLAT | 3.42E-03 | 1.49 | 50 | RAB11FIP1 | 4.20E-04 | 1.41 |
| 11 | CXCL17 | 7.30E-03 | 1.72 | 31 | CD24 | 6.01E-03 | 1.49 | 51 | CEACAM1 | 1.34E-03 | 1.41 |
| 12 | KRT6A | 6.04E-11 | 1.69 | 32 | APOBEC3A | 3.23E-07 | 1.47 | 52 | ZNF407 | 2.93E-03 | 1.41 |
| 13 | DSG1 | 2.74E-10 | 1.67 | 33 | LINC02178 | 6.87E-03 | 1.47 | 53 | MYO5B | 3.01E-09 | 1.39 |
| 14 | NME2 | 7.30E-09 | 1.61 | 34 | TMEM238 | 3.18E-05 | 1.45 | 54 | NQO1 | 4.15E-07 | 1.39 |
| 15 | LINC02154 | 4.51E-04 | 1.61 | 35 | TRPV3 | 1.26E-03 | 1.45 | 55 | ADIRF | 7.09E-07 | 1.39 |
| 16 | S100A4 | 3.14E-08 | 1.59 | 36 | EMP1 | 2.67E-15 | 1.43 | 56 | TNFRSF10A | 1.62E-06 | 1.39 |
| 17 | SLPI | 8.93E-04 | 1.59 | 37 | CST3 | 2.75E-09 | 1.43 | 57 | IL1RN | 2.42E-06 | 1.39 |
| 18 | LYPD3 | 5.15E-09 | 1.56 | 38 | SDCBP2 | 7.61E-08 | 1.43 | 58 | FRMD8 | 1.45E-04 | 1.39 |
| 19 | CDKN2B | 3.04E-06 | 1.54 | 39 | GRHL1 | 2.69E-05 | 1.43 | 59 | AC016831.1 | 1.14E-03 | 1.39 |
| 20 | PTGES | 9.10E-05 | 1.54 | 40 | TMPRSS4 | 3.91E-04 | 1.43 | 60 | PTPN21 | 1.90E-03 | 1.39 |
| Gene | BHp | R | Gene | Bhp | R | Gene | BHp | R | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | DOCK10 | 3.45E-18 | 6.86 | 21 | IL1RL2 | 1.56E-19 | 4.06 | 41 | TBX3 | 5.32E-10 | 3.41 |
| 2 | TAGLN | 2.26E-06 | 6.23 | 22 | TPM2 | 7.72E-13 | 3.97 | 42 | WFDC2 | 3.56E-29 | 3.40 |
| 3 | PAPPA | 9.99E-16 | 5.85 | 23 | IL23A | 1.86E-18 | 3.94 | 43 | C6orf141 | 1.05E-54 | 3.40 |
| 4 | GBP2 | 4.70E-13 | 5.50 | 24 | FGF13 | 5.11E-27 | 3.90 | 44 | S100A2 | 6.0E-137 | 3.39 |
| 5 | KCNH8 | 2.76E-16 | 5.47 | 25 | PTGER4 | 6.07E-32 | 3.90 | 45 | TAC1 | 2.49E-06 | 3.38 |
| 6 | C2CD4A | 1.45E-06 | 5.32 | 26 | AC145124 | 1.75E-29 | 3.83 | 46 | TMEM158 | 1.11E-49 | 3.34 |
| 7 | LPAR6 | 3.46E-06 | 5.11 | 27 | TNC | 2.07E-20 | 3.81 | 47 | SERPINE2 | 1.29E-12 | 3.32 |
| 8 | GSDME | 1.20E-24 | 4.99 | 28 | C12orf54 | 7.82E-17 | 3.79 | 48 | RCAN1 | 7.53E-19 | 3.32 |
| 9 | MSN | 2.0E-105 | 4.99 | 29 | ACTN1 | 3.74E-98 | 3.74 | 49 | PDE4B | 1.89E-41 | 3.29 |
| 10 | LIF | 4.37E-17 | 4.84 | 30 | FERMT2 | 5.04E-62 | 3.73 | 50 | MMP10 | 2.12E-11 | 3.27 |
| 11 | VEGFC | 1.70E-12 | 4.59 | 31 | TRAC | 2.09E-34 | 3.71 | 51 | TMEM173 | 3.73E-15 | 3.26 |
| 12 | TCEAL2 | 4.70E-13 | 4.55 | 32 | ZNF711 | 1.54E-12 | 3.66 | 52 | ARHGAP20 | 1.43E-17 | 3.26 |
| 13 | SERPINE1 | 2.81E-29 | 4.48 | 33 | CDH3 | 9.05E-92 | 3.64 | 53 | GPAT3 | 8.79E-23 | 3.24 |
| 14 | EGR4 | 2.15E-08 | 4.47 | 34 | EMP3 | 4.83E-64 | 3.56 | 54 | CALD1 | 2.05E-34 | 3.20 |
| 15 | SPOCD1 | 4.61E-25 | 4.42 | 35 | SERPING1 | 5.16E-21 | 3.56 | 55 | FXYD5 | 3.66E-32 | 3.18 |
| 16 | SLC43A3 | 1.29E-20 | 4.42 | 36 | C1R | 2.49E-22 | 3.52 | 56 | LIMA1 | 9.22E-89 | 3.17 |
| 17 | BEX1 | 1.17E-08 | 4.37 | 37 | SPINK6 | 2.92E-13 | 3.49 | 57 | VCL | 7.73E-99 | 3.14 |
| 18 | SERPINB10 | 1.45E-36 | 4.26 | 38 | NPPC | 5.06E-13 | 3.46 | 58 | DIRAS3 | 6.40E-13 | 3.14 |
| 19 | PPP1R12B | 4.14E-13 | 4.18 | 39 | TRAF1 | 3.22E-11 | 3.44 | 59 | HLA-G | 2.57E-14 | 3.12 |
| 20 | IL1R2 | 4.77E-16 | 4.13 | 40 | ADORA2B | 2.26E-19 | 3.43 | 60 | MUC1 | 2.09E-18 | 3.12 |
| Gene | Bhp | 1/R | Gene | Bhp | 1/R | Gene | Bhp | 1/R | |||
| 1 | LINC01474 | 1.92E-33 | 14.43 | 21 | GCHFR | 5.79E-49 | 4.15 | 41 | ADIRF | 2.4E-139 | 3.30 |
| 2 | ALDH3A1 | 3.70E-156 | 9.49 | 22 | FSIP1 | 6.59E-40 | 4.07 | 42 | NTRK2 | 5.89E-29 | 3.28 |
| 3 | KRTAP4-1 | 1.19E-07 | 9.20 | 23 | SAMD9 | 2.98E-25 | 3.97 | 43 | CPXM2 | 1.74E-64 | 3.28 |
| 4 | KRT3 | 1.11E-20 | 8.64 | 24 | ERICH5 | 3.26E-70 | 3.94 | 44 | WNT4 | 2.47E-47 | 3.28 |
| 5 | CRTAC1 | 1.61E-304 | 8.31 | 25 | AGR2 | 6.20E-48 | 3.88 | 45 | ABAT | 4.69E-42 | 3.23 |
| 6 | HTRA1 | 6.30E-65 | 8.08 | 26 | CAPS | 6.57E-35 | 3.83 | 46 | RAB40B | 3.61E-53 | 3.20 |
| 7 | SPOCK1 | 1.28E-37 | 7.89 | 27 | AC025164.1 | 5.76E-28 | 3.80 | 47 | NIPAL3 | 1.18E-27 | 3.19 |
| 8 | MAL | 3.88E-24 | 7.05 | 28 | EPB41L4B | 8.75E-42 | 3.71 | 48 | AMPD3 | 5.75E-33 | 3.19 |
| 9 | UPK1B | 7.79E-169 | 6.84 | 29 | ALDH1A1 | 3.1E-223 | 3.70 | 49 | SPINK1 | 3.72E-05 | 3.18 |
| 10 | MISP | 4.20E-38 | 6.64 | 30 | GSTA4 | 3.71E-46 | 3.69 | 50 | PNKD | 2.45E-67 | 3.15 |
| 11 | CLCA2 | 2.38E-117 | 5.73 | 31 | MIR193BHG | 6.06E-37 | 3.69 | 51 | GLRX | 3.67E-07 | 3.14 |
| 12 | HRK | 5.55E-40 | 5.61 | 32 | TSPAN1 | 2.00E-57 | 3.64 | 52 | LINC01705 | 6.23E-10 | 3.13 |
| 13 | TGFBI | 1.10E-217 | 5.09 | 33 | S100A4 | 8.4E-194 | 3.62 | 53 | CDKN2A | 3.06E-28 | 3.12 |
| 14 | CALML3 | 2.88E-29 | 4.96 | 34 | PPP1R3C | 6.73E-18 | 3.60 | 54 | SCGB2A1 | 3.90E-23 | 3.05 |
| 15 | SPINK7 | 5.12E-11 | 4.92 | 35 | TKT | 5.2E-299 | 3.54 | 55 | TRPV3 | 2.98E-47 | 3.05 |
| 16 | FA2H | 1.07E-78 | 4.52 | 36 | KRTAP4-6 | 3.19E-09 | 3.50 | 56 | PTGS2 | 6.68E-43 | 3.05 |
| 17 | NQO1 | 2.51E-156 | 4.50 | 37 | MFSD4A | 9.16E-33 | 3.39 | 57 | THBD | 9.75E-13 | 3.03 |
| 18 | DAPL1 | 6.94E-210 | 4.43 | 38 | LINC00707 | 1.57E-29 | 3.37 | 58 | KRT24 | 4.76E-04 | 3.02 |
| 19 | FABP4 | 2.92E-05 | 4.33 | 39 | KRTAP3-2 | 1.52E-09 | 3.37 | 59 | SCARA3 | 2.50E-27 | 3.00 |
| 20 | PIR | 1.25E-56 | 4.29 | 40 | SLAMF7 | 5.72E-21 | 3.32 | 60 | GAMT | 1.56E-52 | 2.97 |
| Gene | BHp | R | Gene | BHp | R | Gene | BHp | R | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GPHA2 | 3.54E-06 | 26.30 | 21 | SERPINE2 | 1.81E-06 | 3.96 | 41 | CXCL8 | 5.10E-04 | 2.88 |
| 2 | CLEC2D | 1.44E-03 | 11.85 | 22 | WFDC2 | 3.01E-11 | 3.62 | 42 | C2orf54 | 1.60E-04 | 2.80 |
| 3 | C2CD4A | 3.58E-03 | 7.53 | 23 | DIRAS3 | 1.01E-06 | 3.53 | 43 | DOCK10 | 9.15E-04 | 2.75 |
| 4 | CXCL1 | 8.61E-06 | 6.60 | 24 | KRT75 | 2.27E-03 | 3.53 | 44 | FGF2 | 2.63E-07 | 2.75 |
| 5 | CA2 | 1.72E-04 | 5.68 | 25 | C2CD4B | 5.64E-03 | 3.44 | 45 | TSLP | 2.61E-07 | 2.72 |
| 6 | SPOCD1 | 5.05E-08 | 5.52 | 26 | SERPINB10 | 7.67E-06 | 3.29 | 46 | SERPING1 | 1.31E-05 | 2.71 |
| 7 | TCEAL2 | 9.95E-05 | 5.44 | 27 | CH25H | 6.73E-05 | 3.28 | 47 | CCL20 | 7.46E-04 | 2.68 |
| 8 | PLTP | 4.92E-19 | 5.41 | 28 | PTGER4 | 2.97E-09 | 3.19 | 48 | TNC | 8.99E-07 | 2.61 |
| 9 | PAPPA | 1.26E-04 | 5.36 | 29 | F3 | 4.79E-07 | 3.14 | 49 | PDE4B | 2.39E-13 | 2.61 |
| 10 | TRAC | 5.54E-13 | 5.11 | 30 | NPPC | 4.46E-06 | 3.12 | 50 | S100A3 | 5.73E-14 | 2.60 |
| 11 | LUM | 5.31E-04 | 4.98 | 31 | TPM2 | 1.12E-03 | 3.09 | 51 | FXYD5 | 5.98E-11 | 2.60 |
| 12 | LIF | 6.72E-05 | 4.81 | 32 | GLIPR1 | 4.09E-05 | 3.08 | 52 | APOE | 1.83E-10 | 2.58 |
| 13 | KCNH8 | 1.32E-04 | 4.60 | 33 | MSN | 1.47E-18 | 3.06 | 53 | C1R | 1.06E-06 | 2.54 |
| 14 | SPINK6 | 1.81E-06 | 4.52 | 34 | GULP1 | 8.50E-10 | 3.05 | 54 | TBX3 | 6.71E-04 | 2.51 |
| 15 | C12orf54 | 1.59E-05 | 4.46 | 35 | INHBA | 3.29E-04 | 3.00 | 55 | NTNG2 | 2.55E-04 | 2.50 |
| 16 | ARHGAP20 | 1.84E-06 | 4.45 | 36 | TMEM158 | 9.55E-17 | 2.98 | 56 | CCDC88A | 7.76E-09 | 2.50 |
| 17 | VEGFC | 5.06E-05 | 4.29 | 37 | FGFR1 | 3.44E-14 | 2.95 | 57 | SOD2 | 4.35E-07 | 2.47 |
| 18 | GBP2 | 1.64E-04 | 4.27 | 38 | GBP1 | 4.16E-07 | 2.95 | 58 | MT-ND6 | 4.22E-12 | 2.46 |
| 19 | BEX1 | 5.22E-03 | 4.21 | 39 | S100A2 | 2.74E-38 | 2.94 | 59 | H1F0 | 2.06E-04 | 2.46 |
| 20 | SLC43A3 | 7.12E-05 | 4.09 | 40 | PXDN | 4.36E-06 | 2.91 | 60 | CALD1 | 5.24E-09 | 2.44 |
| Gene | BHp | 1/R | Gene | BHp | 1/R | Gene | BHp | 1/R | |||
| 1 | HSPA6 | 5.53E-05 | 5.03 | 21 | FA2H | 4.49E-33 | 2.07 | 41 | ANKRD37 | 3.77E-08 | 1.88 |
| 2 | KRT3 | 1.74E-16 | 4.85 | 22 | HSPA1B | 1.39E-12 | 2.06 | 42 | GJA1 | 2.78E-45 | 1.88 |
| 3 | LINC01474 | 1.08E-28 | 3.67 | 23 | SCARA3 | 2.03E-17 | 2.06 | 43 | SMIM30 | 6.04E-16 | 1.88 |
| 4 | KRTAP4-6 | 2.31E-07 | 3.34 | 24 | MRPL33 | 1.25E-79 | 2.06 | 44 | HOMER3 | 3.61E-28 | 1.88 |
| 5 | CALML3 | 6.58E-21 | 2.89 | 25 | AGR2 | 1.17E-21 | 2.05 | 45 | HLA-DRA | 2.73E-06 | 1.86 |
| 6 | SPOCK1 | 9.22E-30 | 2.88 | 26 | PIR | 7.67E-24 | 2.05 | 46 | PSAT1 | 2.01E-28 | 1.86 |
| 7 | ERICH5 | 5.26E-51 | 2.75 | 27 | MISP | 5.14E-13 | 1.99 | 47 | MUC15 | 1.44E-12 | 1.86 |
| 8 | HTRA1 | 6.11E-21 | 2.46 | 28 | SCIN | 2.62E-18 | 1.96 | 48 | RAD9A | 1.72E-11 | 1.85 |
| 9 | MAL | 1.13E-13 | 2.38 | 29 | CXCL14 | 9.58E-25 | 1.95 | 49 | PNKD | 3.24E-32 | 1.84 |
| 10 | GYG1 | 2.56E-25 | 2.31 | 30 | SNCG | 1.75E-12 | 1.95 | 50 | PDHB | 2.90E-14 | 1.84 |
| 11 | MGARP | 2.83E-55 | 2.24 | 31 | BEX3 | 1.26E-29 | 1.92 | 51 | WNT4 | 3.89E-18 | 1.83 |
| 12 | TSPAN1 | 1.35E-40 | 2.22 | 32 | DNAJB4 | 1.73E-09 | 1.91 | 52 | RRAD | 2.50E-05 | 1.83 |
| 13 | HSPA1A | 4.06E-14 | 2.19 | 33 | DGCR6L | 2.44E-20 | 1.91 | 53 | METTL5 | 1.44E-18 | 1.82 |
| 14 | ALDH3A1 | 1.22E-46 | 2.17 | 34 | SERINC2 | 1.56E-25 | 1.90 | 54 | NEDD9 | 6.98E-18 | 1.82 |
| 15 | CAPNS2 | 6.60E-17 | 2.16 | 35 | NQO1 | 8.90E-39 | 1.89 | 55 | RASSF6 | 2.20E-16 | 1.81 |
| 16 | TP53I3 | 1.14E-32 | 2.15 | 36 | DAPL1 | 6.04E-61 | 1.89 | 56 | EPS8L2 | 2.12E-15 | 1.81 |
| 17 | CLCA4 | 4.86E-13 | 2.15 | 37 | ALDH1A1 | 2.83E-95 | 1.89 | 57 | PAX6 | 1.72E-20 | 1.81 |
| 18 | GJB6 | 2.79E-56 | 2.14 | 38 | ECM1 | 2.61E-19 | 1.88 | 58 | CRTAC1 | 1.58E-71 | 1.81 |
| 19 | KIF22 | 6.36E-16 | 2.13 | 39 | FSIP1 | 1.30E-13 | 1.88 | 59 | FAM114A1 | 5.06E-17 | 1.81 |
| 20 | CAPS | 2.97E-19 | 2.10 | 40 | OTUD1 | 2.21E-07 | 1.88 | 60 | LAMTOR2 | 6.57E-29 | 1.80 |
| Gene | R | Gene | R | Gene | 1/R | Gene | 1/R | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | VEGFC | 25.37 | 21 | HSPB8 | 4.68 | 1 | ALDH3A1 | 28.92 | 21 | ECM1 | 4.50 |
| 2 | TRAC | 22.44 | 22 | SLC12A2 | 4.57 | 2 | UPK1B | 27.18 | 22 | DSG1 | 4.48 |
| 3 | TPM2 | 20.38 | 23 | SLC6A6 | 4.52 | 3 | CRTAC1 | 26.76 | 23 | CLU | 4.45 |
| 4 | NPPC | 17.20 | 24 | TSC22D1 | 4.37 | 4 | CLCA2 | 18.94 | 24 | ENO1 | 4.42 |
| 5 | CXCL8 | 10.73 | 25 | CCDC66 | 4.26 | 5 | NQO1 | 12.59 | 25 | POLR2J3 | 4.39 |
| 6 | ID4 | 8.15 | 26 | TRIM27 | 4.23 | 6 | MFSD4A | 9.10 | 26 | TRIP10 | 4.29 |
| 7 | SAPCD2 | 7.97 | 27 | LARP6 | 4.14 | 7 | ALDH1A1 | 9.00 | 27 | CTSD | 4.26 |
| 8 | cyp26 | 7.52 | 28 | MTERF3 | 3.92 | 8 | S100A4 | 8.94 | 28 | TMEM238 | 4.06 |
| 9 | ZNF22 | 7.41 | 29 | MBD3 | 3.90 | 9 | SCGB2A1 | 8.57 | 29 | FAM83A | 3.81 |
| 10 | TSLP | 7.01 | 30 | CDC42EP1 | 3.89 | 10 | TKT | 7.87 | 30 | ABCA12 | 3.78 |
| 11 | CREB5 | 6.46 | 31 | C12orf65 | 3.87 | 11 | ADIRF | 7.80 | 31 | GALNT7 | 3.65 |
| 12 | ATP1B1 | 5.92 | 32 | FOXC1 | 3.85 | 12 | ABAT | 7.13 | 32 | AC010503.4 | 3.56 |
| 13 | PLEKHO1 | 5.44 | 33 | TNFAIP8 | 3.79 | 13 | CLEC7A | 6.95 | 33 | ASPH | 3.30 |
| 14 | SPATA2L | 5.28 | 34 | COQ10A | 3.75 | 14 | PTGES | 6.41 | 34 | LMTK2 | 3.25 |
| 15 | MEIS2 | 5.23 | 35 | FAM129A | 3.74 | 15 | TRPV3 | 5.63 | 35 | GIPC1 | 3.20 |
| 16 | SOX4 | 5.08 | 36 | DDIT3 | 3.69 | 16 | TMPRSS4 | 5.42 | 36 | ARL5B | 3.11 |
| 17 | ZC2HC1A | 5.04 | 37 | DST | 3.66 | 17 | GSN | 5.25 | 37 | ANXA11 | 3.08 |
| 18 | NCOA7 | 4.97 | 38 | DDX28 | 3.60 | 18 | CST3 | 4.88 | 38 | EMP1 | 3.01 |
| 19 | PALLD | 4.95 | 39 | MYC | 3.59 | 19 | SDC1 | 4.76 | 39 | MYH14 | 2.83 |
| 20 | NUDT11 | 4.90 | 40 | Thap7 | 3.59 | 20 | GPRC5A | 4.73 | 40 | DBI | 2.80 |
| Gene | R1 | R123 | Gene | R1 | R123 | Gene | 1/R1 | 1/R123 | |||
| 1 | NR2F2 | 1.75 | 1.74 | 21 | SHARPIN | 1.41 | 1.39 | 1 | HIST1H1E | 1.30 | 1.30 |
| 2 | ID3 | 1.67 | 1.65 | 22 | PRPSAP1 | 1.40 | 1.15 | 2 | ITGA2 | 1.27 | 1.20 |
| 3 | SCARA3 | 1.60 | 1.58 | 23 | CKS1B | 1.39 | 1.35 | 3 | SLFN5 | 1.25 | 1.18 |
| 4 | GEM | 1.57 | 1.55 | 24 | RFXANK | 1.38 | 1.23 | 4 | ADAMTS9 | 1.25 | 1.31 |
| 5 | SLC2A4RG | 1.54 | 1.48 | 25 | CITED2 | 1.38 | 1.35 | 5 | KIAA1551 | 1.25 | 1.20 |
| 6 | ABHD17B | 1.54 | 1.43 | 26 | TOB1 | 1.38 | 1.30 | 6 | GJB5 | 1.23 | 1.19 |
| 7 | THAP11 | 1.53 | 1.49 | 27 | MAZ | 1.37 | 1.24 | 7 | SEMA4B | 1.23 | 1.15 |
| 8 | FOS | 1.51 | 1.36 | 28 | TENT5C | 1.37 | 1.40 | 8 | S100A16 | 1.23 | 1.20 |
| 9 | HMGB2 | 1.49 | 1.55 | 29 | GLA | 1.36 | 1.31 | 9 | KRT17 | 1.22 | 1.21 |
| 10 | JUND | 1.49 | 1.43 | 30 | KLF4 | 1.36 | 1.28 | 10 | PITPNC1 | 1.22 | 1.13 |
| 11 | TSPAN13 | 1.47 | 1.41 | 31 | BOLA3 | 1.36 | 1.23 | 11 | SLC38A1 | 1.22 | 1.15 |
| 12 | CEP290 | 1.46 | 1.48 | 32 | RPS4Y1 | 1.35 | 1.22 | 12 | PLS3 | 1.21 | 1.18 |
| 13 | GYG1 | 1.46 | 1.27 | 33 | CUEDC2 | 1.35 | 1.23 | 13 | RPS17 | 1.19 | 1.17 |
| 14 | PER1 | 1.45 | 1.43 | 34 | INAVA | 1.35 | 1.34 | 14 | AQP3 | 1.19 | 1.14 |
| 15 | EGLN1 | 1.45 | 1.32 | 35 | MRPS11 | 1.34 | 1.31 | 15 | RPL17 | 1.16 | 1.14 |
| 16 | ALKBH5 | 1.44 | 1.30 | 36 | ATN1 | 1.33 | 1.26 | 16 | WWTR1 | 1.16 | 1.12 |
| 17 | C9orf3 | 1.43 | 1.44 | 37 | MXD4 | 1.33 | 1.36 | 17 | GARS | 1.16 | 1.10 |
| 18 | FBXL15 | 1.43 | 1.39 | 38 | ATG4D | 1.33 | 1.30 | 18 | TRIM44 | 1.15 | 1.12 |
| 19 | CLEC2B | 1.41 | 1.40 | 39 | VDAC3 | 1.33 | 1.26 | 19 | ZMAT2 | 1.15 | 1.13 |
| 20 | HSPA1A | 1.41 | 1.53 | 40 | SUMO3 | 1.31 | 1.21 | 20 | ISG20 | 1.14 | 1.13 |
| Gene | BHp | R | Gene | BHp | R | Gene | BHp | R | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MEG3 | 4.0E-17 | 76.68 | 21 | TAC1 | 2.8E-18 | 11.14 | 41 | LINC01127 | 3.0E-54 | 7.60 |
| 2 | ACTN1 | 5.2E-107 | 48.81 | 22 | KCNMA1 | 3.3E-72 | 11.09 | 42 | GPNMB | 4.6E-65 | 7.56 |
| 3 | VIT | 1.5E-72 | 43.58 | 23 | S100A2 | 1.3E-65 | 11.05 | 43 | PPIF | 2.7E-189 | 7.50 |
| 4 | SPARC | 8.3E-131 | 35.52 | 24 | CDH3 | 5.7E-118 | 10.81 | 44 | SYNJ2 | 4.9E-178 | 7.31 |
| 5 | MFHAS1 | 7.1E-70 | 31.24 | 25 | NTRK2 | 2.5E-71 | 10.77 | 45 | MOXD1 | 2.1E-111 | 7.30 |
| 6 | CCL20 | 2.0E-05 | 21.54 | 26 | CRABP2 | 5.9E-103 | 10.44 | 46 | PALLD | 3.8E-168 | 7.18 |
| 7 | DRAM1 | 2.0E-45 | 18.60 | 27 | BMP2 | 5.6E-29 | 10.22 | 47 | FST | 4.3E-58 | 7.17 |
| 8 | TRAC | 3.6E-36 | 18.48 | 28 | CAVIN1 | 8.9E-191 | 10.01 | 48 | COTL1 | 3.4E-47 | 6.98 |
| 9 | LGALS1 | 7.5E-17 | 17.00 | 29 | KRT16 | 2.3E-43 | 9.78 | 49 | AC020916.1 | 3.0E-101 | 6.92 |
| 10 | KRT14 | 3.0E-264 | 16.66 | 30 | LAMB1 | 1.5E-72 | 9.74 | 50 | MEIS3 | 5.5E-85 | 6.91 |
| 11 | IL13RA2 | 2.3E-46 | 16.63 | 31 | TMEM158 | 4.2E-90 | 9.49 | 51 | OXTR | 4.1E-51 | 6.86 |
| 12 | LAMA3 | 3.4E-151 | 15.95 | 32 | NFATC1 | 1.1E-58 | 9.31 | 52 | TRIML2 | 3.7E-37 | 6.78 |
| 13 | TGM2 | 1.2E-61 | 15.80 | 33 | TNNT1 | 2.3E-40 | 9.18 | 53 | CPVL | 9.1E-163 | 6.77 |
| 14 | ANXA5 | 0.0E+00 | 15.25 | 34 | MGLL | 3.5E-37 | 8.53 | 54 | EGR3 | 7.0E-42 | 6.76 |
| 15 | CCNA1 | 2.2E-19 | 14.15 | 35 | WNT9A | 1.3E-78 | 8.38 | 55 | CDK6 | 2.3E-127 | 6.75 |
| 16 | SMOX | 4.0E-53 | 13.13 | 36 | OSBP2 | 7.1E-64 | 8.34 | 56 | ZFP42 | 2.9E-17 | 6.71 |
| 17 | FERMT2 | 1.4E-31 | 13.07 | 37 | FAM180A | 6.6E-27 | 7.96 | 57 | ETS1 | 1.1E-145 | 6.70 |
| 18 | FLNA | 1.9E-242 | 12.68 | 38 | SSUH2 | 4.7E-46 | 7.85 | 58 | PLAU | 1.9E-223 | 6.61 |
| 19 | SALL3 | 1.2E-23 | 12.30 | 39 | SLC7A11 | 4.4E-59 | 7.75 | 59 | MSANTD3 | 1.8E-52 | 6.58 |
| 20 | UGDH | 8.3E-120 | 11.79 | 40 | SLC9A2 | 3.8E-63 | 7.66 | 60 | CARD10 | 2.1E-95 | 6.58 |
| Gene | BHp | 1/R | Gene | BHp | 1/R | Gene | BHp | 1/R | |||
| 1 | CYP26A1 | 1.3E-77 | 33.40 | 21 | SLC26A2 | 1.4E-160 | 9.69 | 41 | MUC15 | 5.3E-248 | 7.88 |
| 2 | PSCA | 2.9E-35 | 25.62 | 22 | CXXC5 | 6.3E-202 | 9.69 | 42 | MUC16 | 1.4E-43 | 7.80 |
| 3 | APBB1IP | 5.4E-46 | 14.64 | 23 | RRAD | 4.9E-41 | 9.37 | 43 | SIK1 | 1.9E-55 | 7.72 |
| 4 | SMIM5 | 2.9E-105 | 14.25 | 24 | TJP3 | 2.8E-55 | 9.27 | 44 | FOS | 4.9E-189 | 7.70 |
| 5 | CA6 | 5.8E-31 | 14.06 | 25 | DHRS9 | 1.6E-15 | 9.20 | 45 | ABLIM2 | 1.8E-53 | 7.50 |
| 6 | CLIC3 | 1.3E-59 | 13.92 | 26 | HS3ST6 | 9.9E-152 | 9.09 | 46 | RAB40C | 2.6E-50 | 7.46 |
| 7 | KRTAP4-8 | 3.9E-09 | 12.87 | 27 | TMEM246 | 4.3E-46 | 8.95 | 47 | OVOL2 | 1.9E-47 | 7.31 |
| 8 | ADH7 | 0.0E+00 | 12.12 | 28 | RAET1E | 6.9E-96 | 8.94 | 48 | BEND5 | 1.3E-60 | 7.26 |
| 9 | FAM3D | 4.3E-53 | 11.53 | 29 | FBP1 | 7.1E-112 | 8.75 | 49 | EFS | 2.2E-38 | 7.19 |
| 10 | HES5 | 9.8E-118 | 11.40 | 30 | PLBD1 | 5.5E-64 | 8.68 | 50 | KRTAP4-9 | 2.4E-25 | 7.19 |
| 11 | NUDT7 | 5.3E-41 | 10.96 | 31 | ERICH5 | 0.0E+00 | 8.58 | 51 | RAB6B | 7.8E-159 | 6.96 |
| 12 | GNE | 3.8E-48 | 10.80 | 32 | GGT6 | 4.8E-73 | 8.50 | 52 | CXCL2 | 2.7E-08 | 6.92 |
| 13 | SLURP1 | 6.9E-91 | 10.70 | 33 | RELN | 2.3E-07 | 8.26 | 53 | CAPN5 | 2.6E-39 | 6.91 |
| 14 | ST6GALNAC1 | 5.0E-57 | 10.54 | 34 | CNGA1 | 8.2E-96 | 8.18 | 54 | ICMT | 4.7E-63 | 6.87 |
| 15 | ADRB1 | 1.7E-44 | 10.46 | 35 | SCGB2A1 | 2.2E-169 | 8.10 | 55 | RGS16 | 3.1E-05 | 6.83 |
| 16 | CALML5 | 9.7E-59 | 10.13 | 36 | C10orf99 | 7.0E-28 | 8.07 | 56 | IL20RA | 5.3E-103 | 6.59 |
| 17 | MIR210HG | 1.3E-70 | 9.94 | 37 | MUC21 | 4.3E-20 | 8.06 | 57 | GPX2 | 1.8E-157 | 6.58 |
| 18 | RHOU | 4.5E-252 | 9.93 | 38 | TLDC2 | 9.1E-51 | 8.05 | 58 | PLEKHH3 | 2.0E-118 | 6.51 |
| 19 | PCP4L1 | 7.8E-66 | 9.87 | 39 | BBOX1 | 1.8E-161 | 7.97 | 59 | HSPA6 | 1.5E-10 | 6.49 |
| 20 | KRTAP4-7 | 6.2E-11 | 9.85 | 40 | OTUD1 | 2.7E-99 | 7.89 | 60 | SLC39A2 | 4.8E-30 | 6.47 |
| D0 unique go terms | FE | FDR | D1 unique GO terms | FE | FDR |
|---|---|---|---|---|---|
| pos. reg. of core promoter binding | 24.65 | 7.43E-03 | reg. of plasma membrane organization | 15.59 | 5.12E-03 |
| integrated stress response signaling | 11.2 | 2.84E-05 | cytoplasmic translation | 13.34 | 8.35E-18 |
| neg. reg. of mRNA splicing, via spliceosome | 9.64 | 7.73E-03 | reg. of translation in response to stress | 12.75 | 9.43E-03 |
| reg. of transcript. from RNA pol. II promoter in stress | 8.45 | 1.69E-03 | intermediate filament cytoskeleton organization | 6.64 | 4.93E-04 |
| mitochondrial electron transport, NADH to ubiquinone | 6.43 | 6.76E-03 | ribosomal small subunit biogenesis | 5.99 | 9.17E-04 |
| pos. reg. of miRNA metabolic process | 6.37 | 1.38E-03 | cell-cell junction assembly | 5.14 | 1.35E-03 |
| proton motive force-driven mitochondrial ATP synthesis | 5.2 | 9.92E-03 | keratinocyte differentiation | 5.07 | 7.82E-04 |
| aerobic electron transport chain | 5.16 | 1.30E-03 | epithelial cell development | 5.05 | 5.62E-05 |
| neg. reg. of mRNA metabolic process | 5.01 | 8.21E-04 | actin filament organization | 5.01 | 1.94E-07 |
| mitochondrial ATP synthesis coupled electron transport | 4.88 | 1.96E-03 | pos. reg. of protein-containing complex assembly | 3.93 | 3.61E-03 |
| oxidative phosphorylation | 4.7 | 3.58E-04 | supramolecular fiber organization | 3.8 | 6.86E-09 |
| cellular response to topologically incorrect protein | 4.57 | 5.80E-03 | actin cytoskeleton organization | 3.72 | 6.68E-08 |
| reg. of mRNA splicing, via spliceosome | 4.53 | 1.82E-03 | neg. reg. of apoptotic signaling pathway | 3.68 | 3.85E-03 |
| response to unfolded protein | 4.35 | 1.41E-03 | skin development | 3.63 | 9.20E-04 |
| cellular response to leukemia inhibitory factor | 4.28 | 9.10E-03 | neg. reg. of kinase activity | 3.54 | 8.13E-03 |
| reg. of TGFb receptor signaling pathway | 3.7 | 5.40E-03 | wound healing | 3.49 | 4.16E-04 |
| aerobic respiration | 3.47 | 5.76E-03 | epithelial cell differentiation | 3.38 | 1.95E-07 |
| neg. reg. of transcription by RNA polymerase II | 3.04 | 3.36E-15 | cell junction assembly | 3.17 | 8.53E-03 |
| in utero embryonic development | 2.95 | 3.75E-05 | epidermis development | 3.01 | 6.59E-03 |
| reg. of cellular response to growth factor stimulus | 2.83 | 1.40E-03 | response to wounding | 2.95 | 8.34E-04 |
| neg. reg. of DNA-templated transcription | 2.82 | 2.53E-17 | neg. reg. of catalytic activity | 2.94 | 3.45E-05 |
| neg. reg. of RNA biosynthetic process | 2.79 | 4.85E-17 | neg. reg. of phosphorylation | 2.9 | 9.08E-03 |
| neg. reg. of nucleobase-containing metabolic process | 2.72 | 1.72E-18 | ribonucleoprotein complex biogenesis | 2.84 | 2.00E-03 |
| neg. reg. of macromolecule biosynthetic process | 2.71 | 2.03E-18 | neg. reg. of phosphorus metabolic process | 2.77 | 7.29E-03 |
| neg. reg. of cellular biosynthetic process | 2.69 | 1.74E-18 | pos. reg. of cellular component biogenesis | 2.75 | 1.47E-03 |
| reg. of hemopoiesis | 2.66 | 6.53E-04 | reg. of protein kinase activity | 2.65 | 7.31E-04 |
| neg. reg. of biosynthetic process | 2.63 | 4.61E-18 | reg. of cell migration | 2.47 | 8.07E-05 |
| neg. reg. of cellular metabolic process | 2.37 | 1.48E-18 | cytoskeleton organization | 2.38 | 6.20E-06 |
| embryonic organ development | 2.34 | 4.19E-03 | |||
| D123 unique GO terms | FE | FDR | D4i.1 unique Go terms | FE | FDR |
| pos. reg. of protein localization to Cajal body | 7.01 | 4.61E-03 | Golgi lumen acidification | 10.14 | 1.00E-03 |
| protein folding in endoplasmic reticulum | 7.01 | 4.58E-03 | desmosome organization | 8.88 | 4.91E-03 |
| maturation of LSU-rRNA | 4.9 | 3.32E-04 | synaptic vesicle lumen acidification | 7.92 | 4.81E-04 |
| maturation of SSU-rRNA | 4.78 | 1.15E-07 | cellular response to arsenic-containing substance | 7.75 | 2.20E-04 |
| maturation of 5.8S rRNA | 4.76 | 3.36E-05 | lysosomal lumen acidification | 5.76 | 2.87E-03 |
| mitochondrial RNA metabolic process | 4 | 1.46E-04 | vacuolar acidification | 4.85 | 1.19E-03 |
| mitochondrial gene expression | 3.98 | 7.60E-15 | proton motive force-driven mitochondrial ATP synthesis | 4.56 | 4.81E-06 |
| neg. reg. of stem cell differentiation | 3.98 | 8.91E-03 | neg. reg. of stress-activated protein kinase cascade | 4.06 | 9.15E-04 |
| spliceosomal snRNP assembly | 3.96 | 7.97E-04 | mitochondrial ATP synthesis coupled electron transport | 3.76 | 8.48E-06 |
| ribosome biogenesis | 3.95 | 1.61E-32 | ribonucleoside triphosphate metabolic process | 3.72 | 3.02E-09 |
| rRNA processing | 3.95 | 1.74E-22 | cellular aldehyde metabolic process | 3.68 | 8.49E-04 |
| pos. reg. of transcription by RNA polymerase I | 3.81 | 2.99E-03 | glycolytic process | 3.66 | 9.11E-03 |
| protein-RNA complex assembly | 3.74 | 3.75E-18 | NADH dehydrogenase complex assembly | 3.48 | 2.26E-03 |
| protein-RNA complex organization | 3.72 | 1.16E-18 | ERBB signaling pathway | 3.17 | 7.76E-03 |
| tRNA aminoacylation for protein translation | 3.67 | 1.61E-03 | cell-cell junction organization | 3.14 | 1.29E-07 |
| reg. of hematop. progenitor cell differentiation | 3.35 | 8.46E-03 | cellular oxidant detoxification | 2.99 | 2.09E-03 |
| reg. of RNA splicing | 3.23 | 2.86E-07 | pos. reg. of protein localization to membrane | 2.93 | 9.46E-04 |
| reg. of telomere maintenance via telomerase | 3.18 | 6.42E-09 | cellular response to toxic substance | 2.9 | 3.80E-04 |
| ncRNA processing | 3.17 | 1.24E-06 | vacuolar transport | 2.7 | 7.62E-05 |
| pos. reg. of protein localization to nucleus | 3.1 | 3.88E-27 | neg. reg. of apoptotic signaling pathway | 2.66 | 2.32E-06 |
| pos. reg. of chromosome organization | 3.04 | 7.98E-04 | pos. reg. of ubiquitin-dependent prot. catabolic process | 2.63 | 6.03E-03 |
| reg. of stem cell population maintenance | 2.74 | 3.95E-06 | cell-cell junction assembly | 2.61 | 1.81E-03 |
| endoplasm. ret.to Golgi vesicle-med. transp. | 2.7 | 6.08E-04 | organelle disassembly | 2.58 | 9.15E-03 |
| reg. of DNA-templated transcription elongation | 2.69 | 6.68E-24 | process utilizing autophagic mechanism | 2.52 | 1.54E-07 |
| peptidyl-lysine acetylation | 2.68 | 6.17E-03 | energy derivation by oxidation of organic compounds | 2.51 | 4.30E-06 |
| reg. of response to endoplasmic ret. stress | 2.65 | 3.18E-04 | pos. reg. of proteolysis involved in prot. catabolic process | 2.48 | 3.15E-03 |
| Gene | Crl | Gene | Crl | Gene | Crl | Gene | Crl | Gene | Crl | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SPINT2 | 0.99 | 21 | ERICH5 | 0.97 | 41 | CLU | 0.96 | 1 | ANXA5 | -0.98 | 21 | RPS13 | -0.95 |
| 2 | KRT5 | 0.99 | 22 | TPD52 | 0.97 | 42 | GJB6 | 0.96 | 2 | FLNA | -0.97 | 22 | RPS8 | -0.95 |
| 3 | ADIRF | 0.99 | 23 | NMRK1 | 0.97 | 43 | FXYD3 | 0.96 | 3 | CAVIN1 | -0.97 | 23 | ILF2 | -0.95 |
| 4 | COMT | 0.98 | 24 | TP53I3 | 0.97 | 44 | PLEKHH3 | 0.96 | 4 | SYNJ2 | -0.96 | 24 | DEK | -0.95 |
| 5 | SNCG | 0.98 | 25 | UPK1B | 0.97 | 45 | WLS | 0.96 | 5 | EGR3 | -0.96 | 25 | EFNB2 | -0.95 |
| 6 | ASPH | 0.98 | 26 | SUCO | 0.97 | 46 | CSRP2 | 0.96 | 6 | CRABP2 | -0.96 | 26 | CREBRF | -0.95 |
| 7 | MGARP | 0.98 | 27 | SPOCK1 | 0.97 | 47 | CD99 | 0.96 | 7 | PPIF | -0.96 | 27 | H2AFZ | -0.95 |
| 8 | LINC01474 | 0.98 | 28 | PAX6 | 0.97 | 48 | BCAP31 | 0.96 | 8 | PLAU | -0.96 | 28 | RPS16 | -0.95 |
| 9 | GJA1 | 0.98 | 29 | C4orf3 | 0.97 | 49 | SH3RF1 | 0.96 | 9 | CDK6 | -0.96 | 29 | RPS23 | -0.94 |
| 10 | ASAH1 | 0.98 | 30 | KRT3 | 0.97 | 50 | GLOD4 | 0.96 | 10 | AJAP1 | -0.96 | 30 | PABPC1 | -0.94 |
| 11 | TSTD1 | 0.98 | 31 | TCEA3 | 0.97 | 51 | EFNA1 | 0.96 | 11 | PTMS | -0.95 | 31 | RPS19 | -0.94 |
| 12 | SCIN | 0.98 | 32 | PSAT1 | 0.97 | 52 | CRYBG1 | 0.96 | 12 | PALLD | -0.95 | 32 | SUB1 | -0.94 |
| 13 | SLC20A1 | 0.98 | 33 | PEBP1 | 0.97 | 53 | SLC2A1 | 0.96 | 13 | SLC9A2 | -0.95 | 33 | MEPCE | -0.94 |
| 14 | GSN | 0.98 | 34 | ENO1 | 0.97 | 54 | RB1 | 0.96 | 14 | HIVEP1 | -0.95 | 34 | EIF5 | -0.94 |
| 15 | PDP1 | 0.97 | 35 | MFSD4A | 0.96 | 55 | PIR | 0.96 | 15 | ETS1 | -0.95 | 35 | STK17A | -0.94 |
| 16 | SDC1 | 0.97 | 36 | GSTP1 | 0.96 | 56 | PPDPF | 0.96 | 16 | OSBP2 | -0.95 | 36 | RPS12 | -0.94 |
| 17 | FABP5 | 0.97 | 37 | ATP6V1F | 0.96 | 57 | LAD1 | 0.96 | 17 | OAF | -0.95 | 37 | NPM1 | -0.94 |
| 18 | MAL | 0.97 | 38 | AGR2 | 0.96 | 58 | ALDH1A1 | 0.96 | 18 | MEIS3 | -0.95 | 38 | RPL18 | -0.94 |
| 19 | CAPS | 0.97 | 39 | CAPN1 | 0.96 | 59 | UQCR11 | 0.96 | 19 | COQ8B | -0.95 | 39 | RPS6 | -0.94 |
| 20 | MUC15 | 0.97 | 40 | ANXA7 | 0.96 | 60 | CTSD | 0.96 | 20 | RAB31 | -0.95 | 40 | CFL1 | -0.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




