1. Introduction
The rising popularity of Brain-Computer Interface (BCI) applications is closely related to the increasing accessibility of Electroencephalography (EEG) hardware (EEG headsets), which are noninvasive, portable, wireless and often with open software. While BCI software platforms do exist, there is a limited number of systems specifically tailored to help inexperienced programmers in the development of BCI applications: NeuroBlock [
1], Blockly block framework for dynamic brain-based virtual environment [
2], BrainFlow [
3], Neuromore Studio [
4], NeuroScale [
5], NeuroPype [
6], EmotivBCI Node-RED toolkit [
7]. From them, software technologies for BCI programming in the Internet of Things (IoT) are only two: NeuroScale [
5], designed to integrate with various popular EEG devices commonly used in research and clinical settings, while EmotivBCI Node-RED toolkit [
7,
8] is device specific. However, the BCI toolkit for programing EEG-based BCI applications is only for Emotiv EPOC+ and Insight devices. The benefit of using the EmotivBCI Node-RED toolkit in comparison to the NeuroScale approach lies in its simplification of the integration process. For example, while both methods aim to interface EEG headsets with robotic arms, the EmotivBCI Node-RED toolkit offers prebuilt blocks specifically designed for use with Node-RED [
9], a visual programming tool, and streamline the integration. In contrast, NeuroScale’s approach determines the user’s intent from processed signals and communicates with a robot arm using an interface or middleware. The integration method can vary, involving APIs, communication protocols or custom development, depending on the specific hardware and software components in use. Therefore, node-based visual programming can be the more appropriate solution in this context.
The paper focuses on the introduction of new nodes in the Node-RED library, configured within the OpenBCI category, for the design and integration of an EEG-based Brain-Machine Interface in the context of the Internet of Things (IoT). We created a new BCI toolkit in Node-RED for more than 20 open-source EEG boards based on
BrainFlow software library [
10] that supports and provide uniform data acquisition API for them. Additionally, BrainFlow provides a robust API for signal processing and EEG featuring, which are open-source, can be modified and utilized independently of the type of the BCI headsets. Although, the BrainFlow library assists inexperienced programmers in developing BCI applications, several obstacles we have encountered. They include various challenges related to programming skills, signal acquisition and processing, managing multiple python scripts for board interaction, real-time EEG data streaming, data sampling and sliding window techniques, decomposition of power spectral density into different frequencies, real-time data streaming, data sampling, power spectral density analysis, and integration with different devices and services.
To overcome these difficulties and enhance the development of BCI applications for integrating with IoT devices, processes or services, several contributions have been made:
A model for visual node-based programming has been designed utilizing BrainFlow library within the Node-RED platform, tailored to help inexperienced programmers in the development of BCI applications;
An openBCI Node-RED toolkit based on the proposed model has been developed, which can be applied to more than 20 EEG-based devices.
The proposed openBCI toolkit was implemented and validated through a case study, wherein a BCI application utilizing BrainFlow performance metrics was employed to control a TinkerKit Braccio robot arm via OpenBCI Cyton + Daisy boards
The rest of the paper is organized as follows:
Section 2 describes the system architecture of an EEG-based BCI for communication with devices and services in the IoT using openBCI toolkit in Node-RED;
Section 3 describes the new nodes in openBCI toolkit in Node-RED library.
Section 4 presents the flowchart illustrating the algorithm behind the ‘openBCI-data’ node. At the end we present our conclusions and future work.
2. System Architecture
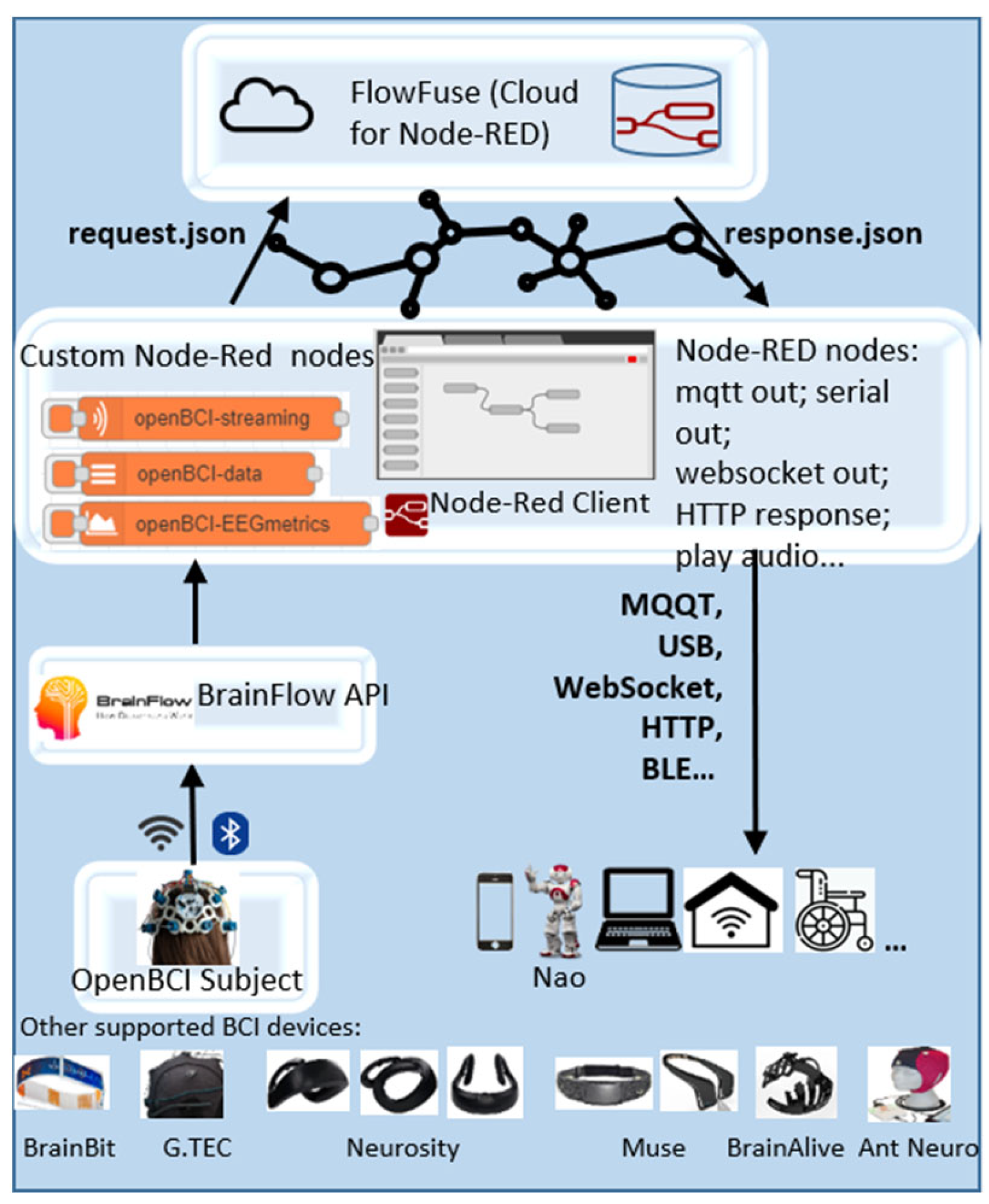
Designed and developed is a system architecture of an EEG-based BCI for communication with devices and services in the IoT using openBCI toolkit in Node-RED with practical application capabilities (
Figure 1).
To register EEG signals, a non-invasive and portable EEG device OpenBCI [
11] was utilized. However, within the established architecture, other devices supported by the BrainFlow library can also be employed. The measured EEG data is transmitted via Bluetooth to the Node-RED platform [
12], nevertheless the architecture is also allowing data transmission over Wi-Fi. Node-RED is utilized as a browser-based tool for streamlining programming flows and acts as a gateway to the IoT. It facilitates sending JSON-type requests to the FlowFuse platform [
13], employed for cloud computing. The architecture provides users the flexibility to use various EEG devices, and for this purpose, three new nodes have been designed and added to the Node-RED library (
Figure 2)—‘openBCI-streaming’ ‘openBCI-Data’ and ‘openBCI-EEGmetrics’. These are customized nodes with access to the BrainFlow API according to the selected device and processing needs. BrainFlow is a software framework for building BCI applications using conventional programming languages, supporting over twenty EEG-based BCI devices.
The new nodes contribute to transforming these BCI devices into ‘things’ only through visual programming in Node-RED, eliminating the need for any programming code. This way, the capabilities of Node-RED are expanded, offering an easy way for OpenBCI users to specify the board ID, data type, and electrodes of interest. Based on the input values entered in the node’s user settings, a JavaScript file within the node processes and transmits the parameters to the BrainFlow API. The parameter transmission is done through a child process linking the Node.js server in Node-RED to the BrainFlow API, generating the output in JSON type. The connection to the BrainFlow API has been established through a newly developed Python file with Python software code, utilizing the BrainFlow libraries. Each of the newly created nodes contains such a file in its directory, differed by the functionalities of the specific node. Node-RED also provides various output nodes such as mqtt out, serial out, http response, play audio, generic-BLE out, etc., for subsequent data transmission to IoT.
3. Overview of the openBCI toolkit within Node-RED
The developed original methods and algorithms for visual programming and integration of a BCI in a web-based streaming environment, successfully validated using OpenBCI device, BrainFlow library, Node-RED and Arduino-based robotic arm are combined into a newly created toolkit openBCI within the Node-RED platform, (
https://flows.nodered.org/collection/W7dKrufq2WWR), and applicable to more than 20 EEG-based devices.
At the beginning of the development, it was observed that when EEG devices transmit data through the COM port via a USB dongle, it is not possible to initiate more than one process to a specific port. In other words, only one session can be started on one COM port. This limitation restricts the parallel collection and processing of data of different nature and/or from different electrodes, requiring all conditions to be structured in a single process and session. This restriction contradicts the concept of visual programming to design logically separated processes into multiple micro-processes, enabling various combinations/scenarios tailored to the specific needs of the user. To overcome this limitation, an original method was developed in the Node-RED platform, based on the BrainFlow streaming board. This board can stream data to various destinations such as files, sockets, etc., directly from BrainFlow, practically acting like a consumer for data received from the main process. In the developed method, when initiating a session with the OpenBCI board, a streaming function to a socket with a valid multicast address and port is added. For each subsequent data collection and/or processing process, a session is not started with the main board but with the BrainFlow streaming board. This way, the main process to the COM port remains only one, while the user can configure multiple micro-processes through visual programming.
3.1. Node ‘openBCI-streaming’
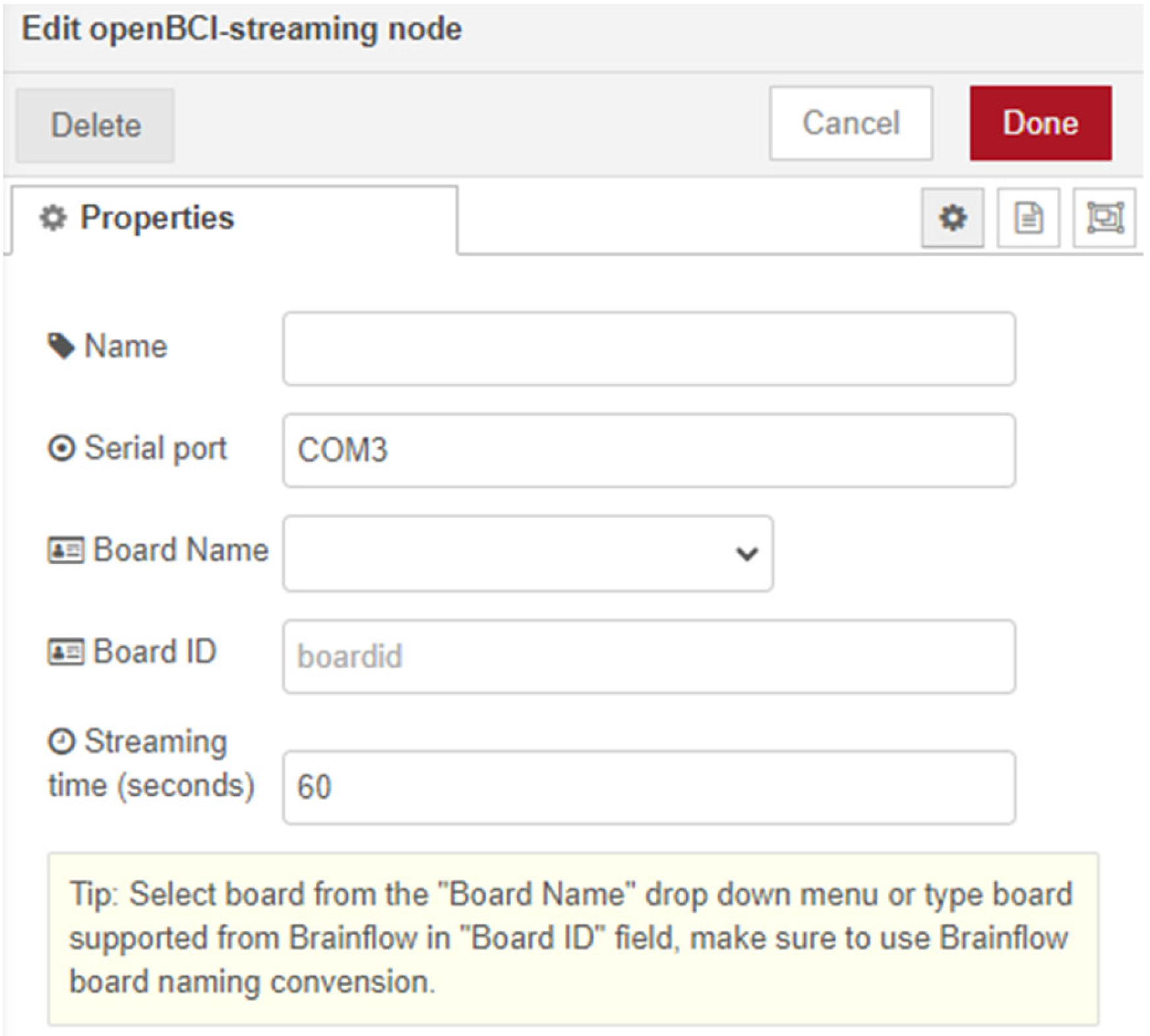
The node ‘openBCI-streaming’ initiates the main process to the OpenBCI board in Node-RED. The node is configured with a button for instant start, as well as an input for initiation via another Node-RED node. The node is defined to be located after installation in the newly created openBCI category for the purpose of the dissertation in the Node-RED palette. Upon opening the node by double-clicking, the user settings of the node are visualized (
Figure 3).
The algorithm of the node includes mandatory user input fields such as ‘Board Name’ or ‘Board ID,’ ‘Serial port,’ and ‘Streaming time (in seconds).’ The ‘Name’ field is optional, used if the user wishes to rename the node. When configuring user settings, the user needs to know the exact COM port used by their EEG device for the session to be registered. Upon opening, the node is configured to display an example value ‘COM3’ in the ‘Serial port’ field and a value of ‘60’ in the ‘Streaming time (seconds)’ field for user convenience. The ‘Board Name’ field is defined as a drop-down menu of OpenBCI boards in the BrainFlow API. When selecting an option from the menu, the ‘Board ID’ text field is deactivated via JavaScript. Similarly, JS deactivates the ‘Board Name’ field when the ‘Board ID’ field is filled. Only one of the two fields can be used to start a session, and this automation has been added to avoid user errors when initiating the node. This automation is implemented in all three new nodes and a tip has been added to the node visualization: ‘Select board from the ‘Board Name’ drop-down menu or type board supported from Brainflow in the ‘Board ID’ field’. After initiating the node, the algorithm starts a session to the ‘streaming’ Brainflow board and returns a ‘Start stream’ debug message. After the user-defined time in the ‘Streaming time (seconds)’ field elapses, the node sends a ‘Stop stream’ JSON type message to signal the end of the session.
3.2. Node ‘openBCI-data’
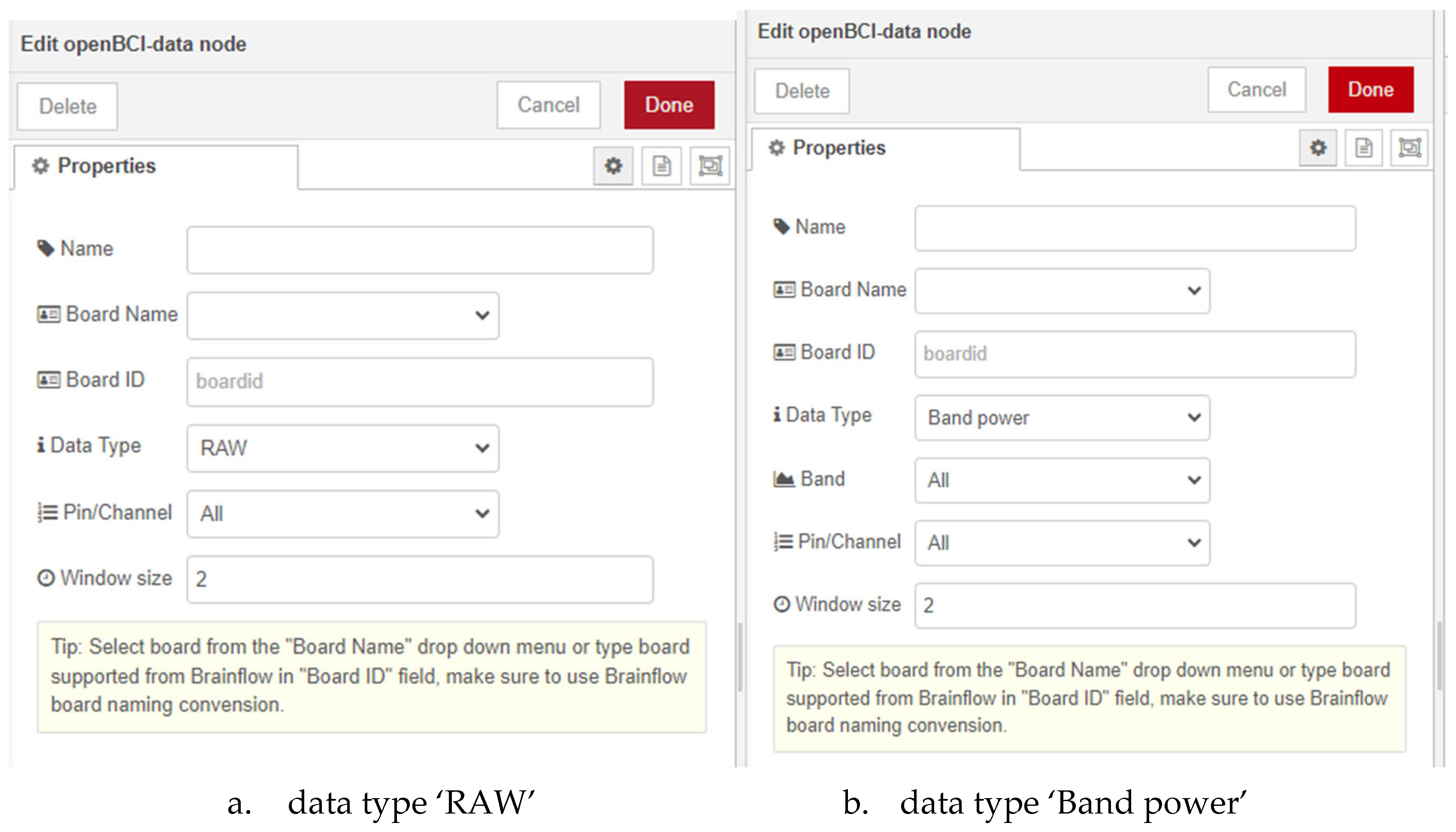
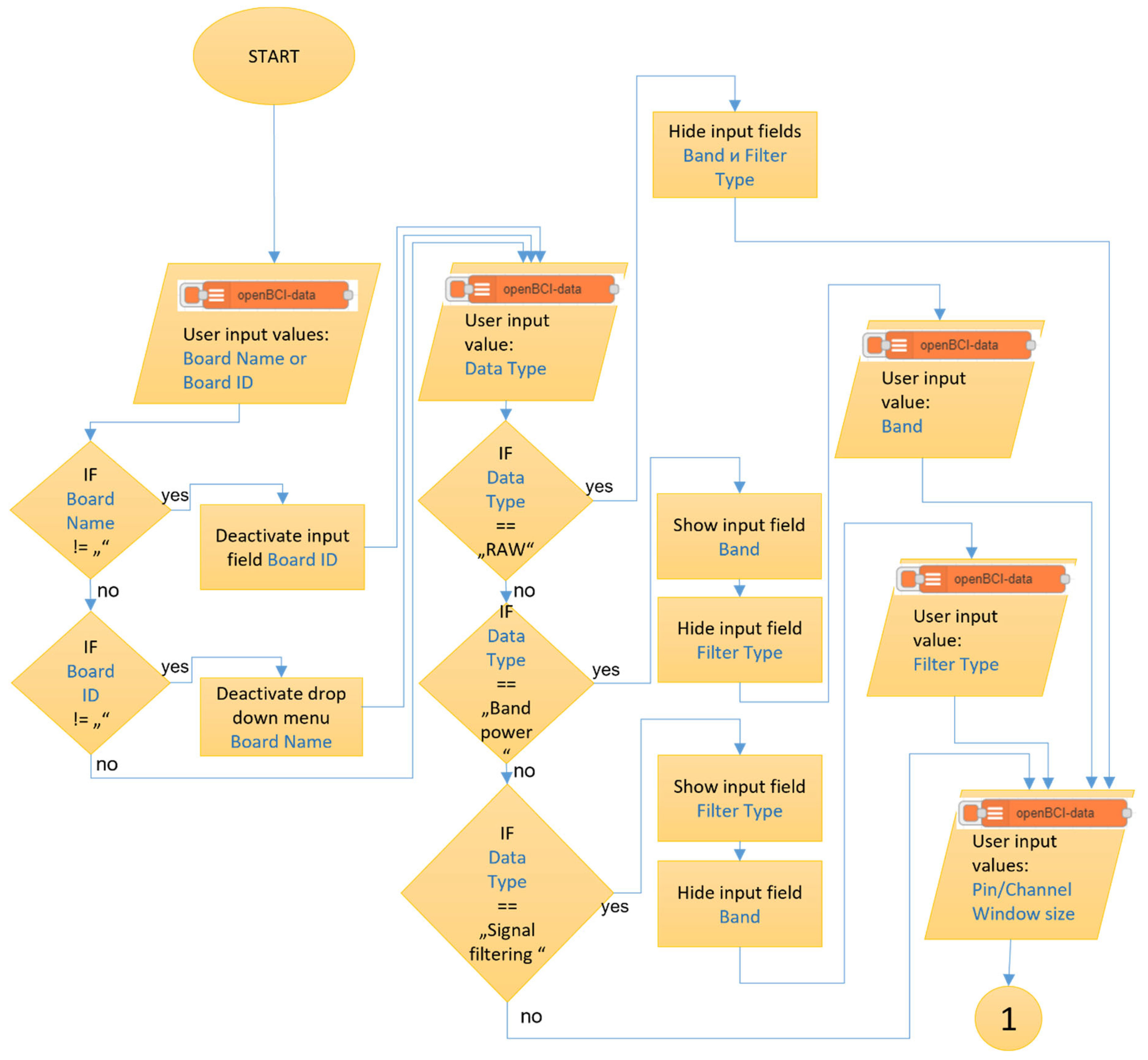
The ‘openBCI-data’ node has the most functionalities among the three newly created nodes and executes an algorithm to access raw data from the EEG device, process it through predefined filters and calculate the powers of six frequency bands of the EEG signal in real-time. This node operates only when there is an already started and correctly functioning ‘openBCI-streaming’ node since the algorithm reads the data directly from it. In the user settings of the node (
Figure 4 a) is mandatory to enter either the ‘Board Name’ or ‘Board ID’ field.
Upon initially opening the node settings, the dropdown menu ‘Data type’ is set to ‘RAW.’ This option allows the user access to the raw data obtained from the EEG device. The time window for visualizing the data is set through the ‘Window size’ field, with the value provided in seconds. When selecting the second option ‘Band power’ (
Figure 4b), an additional ‘Band’ field is displayed in the user settings menu. It includes the options ‘Alpha’, ‘Theta’, ‘Gamma’, ‘HighBeta’, ‘LowBeta’, ‘Delta’, ‘Gamma’ and ‘All’. When selecting one of the options, the algorithm calculates the power of the chosen frequency band for the selected channel of the used board and returns the result in type JSON. The last option in the ‘Data type’ dropdown menu is ‘Signal filtering’. An additional menu for choosing the filter type includes the options ‘Bandpass’, ‘Bandstop’, ‘Lowpass’, etc. Upon starting the node with a selected filter type from the menu, the node’s algorithm filters the raw data from the chosen channel based on the specified filter and returns the result in type JSON.
3.3. Node “openBCI-EEGmetrics”
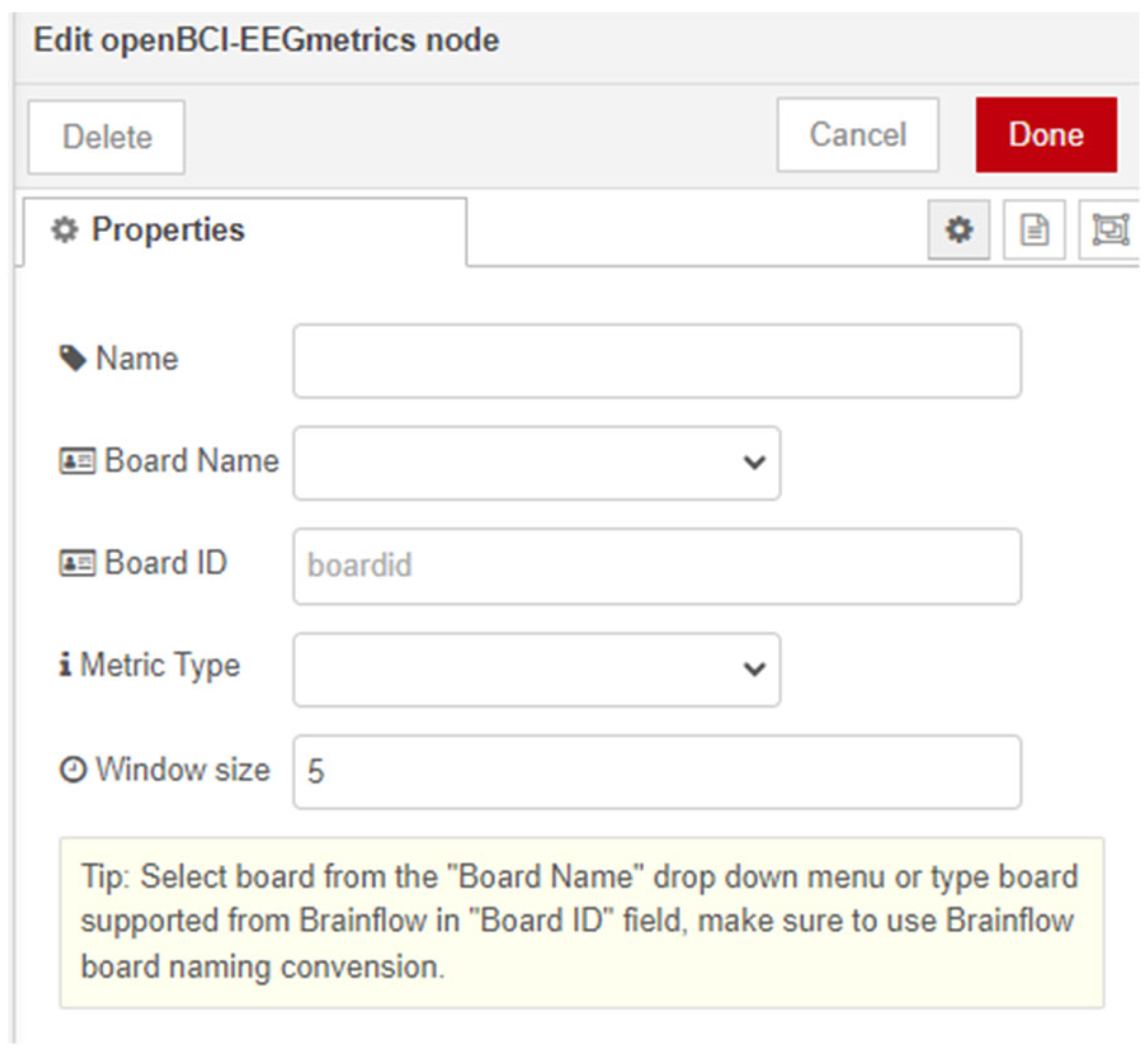
The node “openBCI-EEGmetrics” allows users to monitor the levels of focus on the object connected to the EEG device. In the node’s settings (
Figure 5), a dropdown menu labeled “Metric type” is configured with options “Relaxation” and “Concentration.”
The algorithm for calculating the metrics is integrated from the Brainflow API. For “Relaxation,” it considers FFT values associated with delta, theta, and alpha brain waves, while for “Concentration,” it examines beta and gamma brain waves. Relaxation is typically achieved through “meditation” with closed eyes, while concentration can be attained through focused attention with open eyes. The algorithm returns values in the range between 0 and 1. The Brainflow API provides the option to specify a classifier of choice (Regression, KNN, SVM or LDA) for this algorithm. However, according to OpenBCI’s recommendation, the newly created node is configured to always use Regression as the classifier. All channels of the used EEG device are employed for the calculations, and the user can set the time window (in seconds) for computing the average power of the frequency band in the “Window size” field in the user settings.
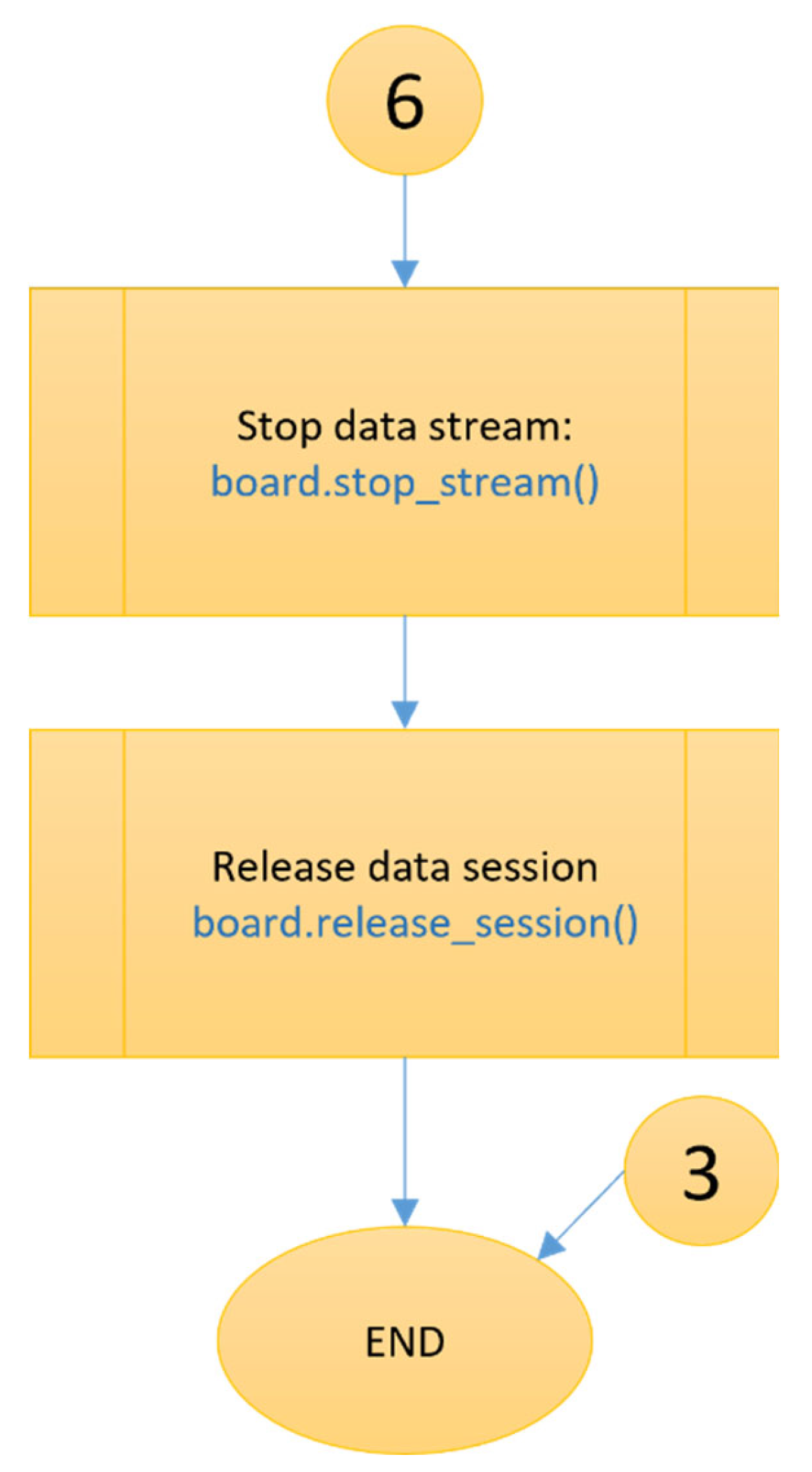
4. The algorithms’ flowchart
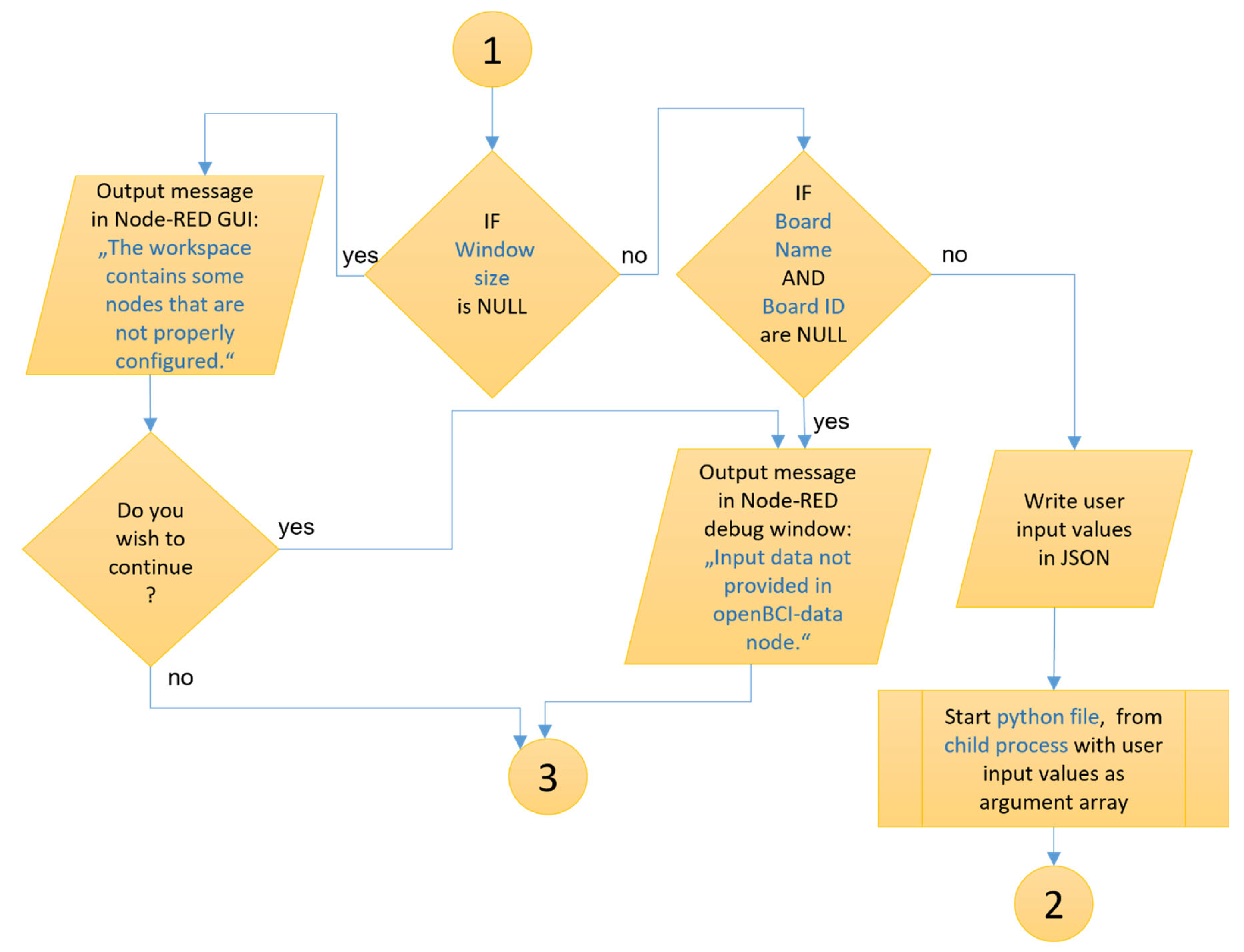
A detailed flowchart has been developed for the new “openBCI-data” node. It illustrates the working principle of the developed original methods and algorithms in the overall operational process of the new node. The node itself combines various software technologies and languages such as HTML, JavaScript, Python, Express and others, and the flowchart illustrates the logical connections between them. The developed diagram is specific to the “openBCI-data” node, but its fundamental principles are applicable to the other two newly created nodes—“openBCI-streaming” and “openBCI-EEGmetrics”. The front-end of the “openBCI-data” node (
Figure 6), responsible for visualizing the node, is built using HTML and JavaScript.
Figure 6 illustrates the logical principles in a file with the extension .html in the node’s directory.
The back-end of the “openBCI-data” node (
Figure 7) is entirely constructed from JavaScript code.
Figure 6 illustrates the logical principles in the two .js files in the node’s directory.
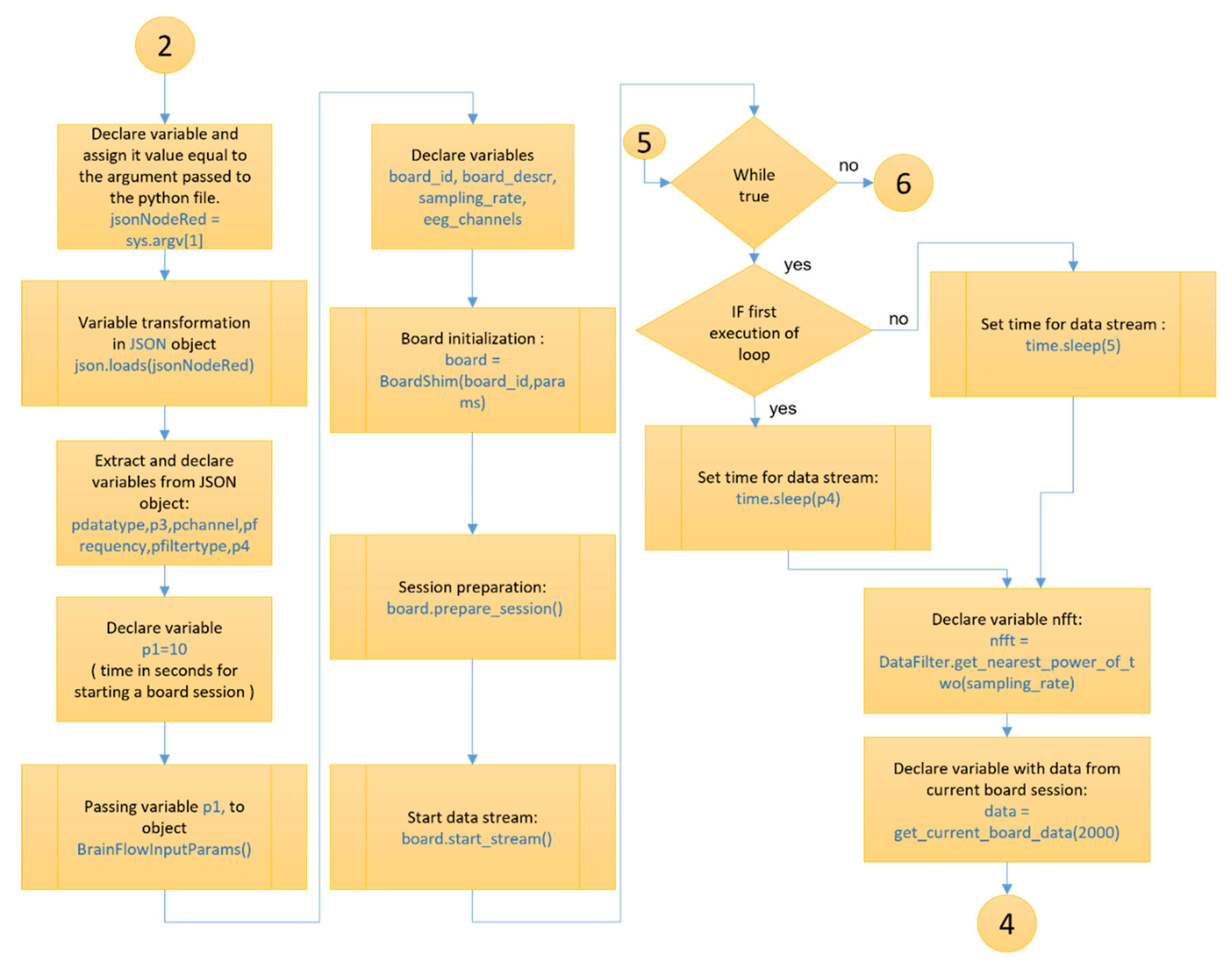
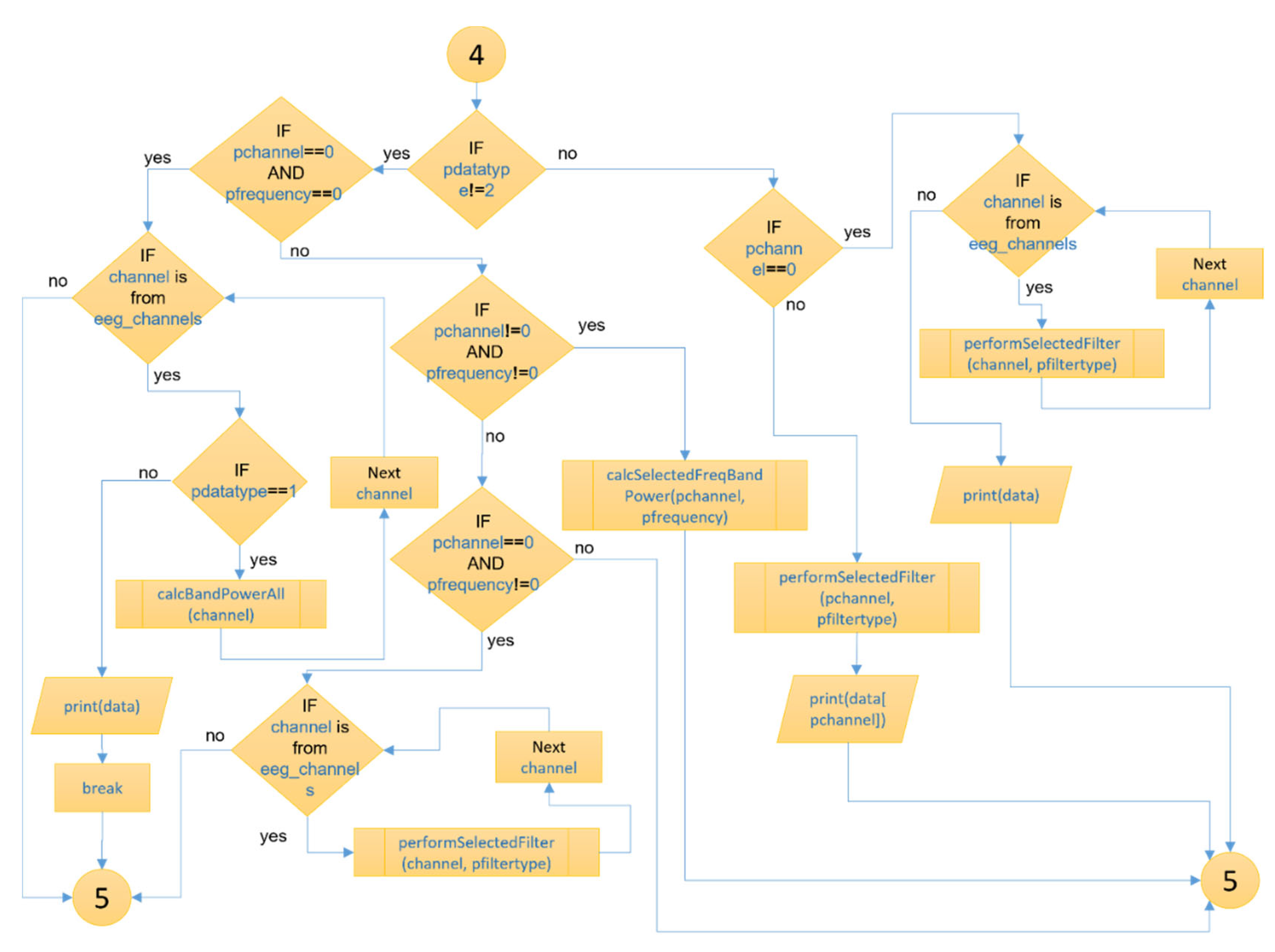
Figure 8 and
Figure 9 show the section of the “openBCI-data” node for collecting and processing EEG data, executed by the .py file from the node’s directory. This is the part where the Brainflow API is integrated through the Python programming language. The end of the processes of the “openBCI-data” node is illustrated in
Figure 10
5. Specific in creating OpenBCI nodes in the Node-RED library
To create a customized node, a directory containing three types of files—package.json, js, and html have to be initially created in the file system. The package.json file, a standard file used by Node.js modules to describe their content, is used to package all files in the directory as an npm module.
5.1 HTML File (.html)
The front-end of the node is defined in the HTML file, executed in the Node-RED editor within a web browser. It contains three separate parts, each wrapped in its own HTML <script> tag. The first tag defines the main node, registered in the platform editor, where properties such as palette category, editable defaults, and the icon to be used are specified. It is within a regular JavaScript script tag with type=“text/javascript”. This tag ensures the node is registered in the editor using the RED.nodes.registerType function. The second tag represents an editing template, defining the content of the node’s editing dialog window. Automatic show/hide functionalities for parts of the node’s content are created within this tag. The tag is defined as a text/html script with a specified data-template-name based on the node type. The node type is used throughout the editor to identify the node and must correspond to the value used in the RED.nodes.registerType function call in the respective .js file. The third tag is a help text tag displayed in the Node-RED platform’s information sidebar.
5.2. JavaScript File (.js)
The back-end of the node is defined in the JavaScript file, executed during Node-RED’s runtime. The file, with a .js extension, determines the behavior of the node. Nodes are defined by a constructor function used to create new instances of the node. This function is registered upon the platform’s startup, allowing it to be invoked when nodes of the corresponding type are placed in the flow. The function is passed an object containing properties set in the flow editor. The constructor function should first invoke RED.nodes.createNode to initialize features shared by all nodes. Subsequently, the node-specific code is executed. Communication between the front-end and back-end of each node is achieved by creating an HTTP endpoint within the node’s runtime, and HTTP calls to this endpoint are made from the individual nodes’ editors. The HTTP endpoint is created using the Express routing application API in Node-RED’s runtime. All Express routing methods are available through the RED.httpAdmin API, however only the post method is being used in the developed software. Additionally, the newly created nodes utilize Express’ authentication middleware through the RED.auth.needsPermission API, setting specific write permissions for the node type’s endpoint.
5.3. Child_Process Module
A connection from the Node-RED Node.js server to the Brainflow API was established. This connection was facilitated by a second .js file using the child_process module to execute a newly developed Python file. The Node.js child_process module allows access to operating system functionalities by executing any system command within a child process. It provides control over the arguments passed to the OS command and allows the use of the command’s output. There are four different ways to create a child process in Node.js: spawn(), fork(), exec(), and execFile(). The spawn function was used in the newly created nodes, as it starts a command in a new process and enables passing all arguments to that command. The command executed in the nodes for this dissertation is the Python file from the node’s directory. Arguments to the command executed by the spawn function are passed as the second argument to the function. The result of the spawn function’s execution is a ChildProcess instance that implements the EventEmitter API. This means that event handlers can be directly registered on this child object. Events that can be registered with the ChildProcess instance are exit, close, disconnect, error, and message. In the newly created nodes, event handlers for exit and error are registered. Each child process also receives the three standard stdio streams, which can be accessed using child.stdin, child.stdout, and child.stderr. When these streams are closed, the child process using them emits the close event. In the newly created nodes, the most crucial readable streams stdout and stderr are used by listening for the data event, which will contain the command’s output or any error during its execution. The command’s output is passed to the node’s output in JSON type, while any execution errors are visualized in the Node-RED platform’s debug console.
5.4. Publishing to npm
The developed nodes have been published to npm using the npm publish command and are available for use by the community. User installation commands are individual for each specific node and are as follows:
npm install node-red-contrib-openbci
npm install node-red-contrib-openbci-EEGmetrics
npm install node-red-contrib-openbci-streaming
6. Conclusions
The proposed user-friendly approach for BCI application development will enable a larger circle of users to develop BCI applications without requiring technical expertise. The new nodes in the openBCI toolkit contribute to transforming BrainFlow supported BCI devices into ‘things’ only through visual programming in Node-RED, eliminating the need for any programming code. The proposed toolkit has been validated through pilot tests, demonstrating its effectiveness in controlling a robotic arm using the OpenBCI headset. Future validation will involve testing the toolkit in different Node-RED applications through additional research experiments, supplemented by detailed results assessment, including graphical representations.
References
- Crawford, C.S.; Gilbert, J.E. NeuroBlock: A block-based programming approach to neurofeedback application development. In Proceedings of the 2017 IEEE Symposium on Visual Languages and Human-Centric Computing (VL/HCC), Raleigh, NC, USA; 2017; pp. 303–307. [Google Scholar] [CrossRef]
- Crawford, C.S.; Andujar, M.; Jackson, F.; Applyrs, I.; Gilbert, J.E. Using a visual programing language to interact with visualizations of electroencephalogram signals. In Proceedings of the ASEE-SE Annual Meeting; 2016. [Google Scholar]
- BrainFlow. Retrieved [January 2024]. Available online: https://brainflow.org/.
- Neuromore. Retrieved [January 2024]. Available online: https://www.neuromore.com/.
- Neuroscale. Retrieved [January 2024]. Available online: https://neuroscale.intheon.io/.
- Neuropype. Retrieved [January 2024]. Available online: https://www.neuropype.io/.
- EmotivBCI Node-RED toolkit. Retrieved [January 2024]. Available online: https://emotiv.gitbook.io/emotivbci-node-red-toolbox/.
- Rușanu, O.A. The Development of Brain-Computer Interface Applications Controlled by the Emotiv Insight Portable Headset Based on Analyzing the EEG Signals Using NODE-Red and Python Programming Software Tools. In Open Science in Engineering; REV 2023. Lecture Notes in Networks and Systems; Auer, M.E., Langmann, R., Tsiatsos, T., Eds.; Springer: Cham, 2023; Volume 763. [Google Scholar] [CrossRef]
- Torres, D.; Dias, J.P.; Restivo, A.; Ferreira, H.S. Real-time Feedback in Node-RED for IoT Development: An Empirical Study. In Proceedings of the 2020 IEEE/ACM 24th International Symposium on Distributed Simulation and Real Time Applications (DS-RT), Prague, Czech Republic; 2020; pp. 1–8. [Google Scholar] [CrossRef]
- BrainFlow software library. Retrieved [January 2024]. Available online: https://github.com/brainflow-dev/brainflow.
- OpenBCI. Retrieved [January 2024]. Available online: https://openbci.com/.
- Node-RED. Retrieved [January 2024]. Available online: https://nodered.org/.
- FlowFuse. Retrieved [January 2024]. Available online: https://flowfuse.com/.
- openBCI toolkit within the Node-RED platform. Retrieved [January 2024]. Available online: https://flows.nodered.org/collection/W7dKrufq2WWR.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).