1. Introduction
Escherichia coli (E. coli), a Gram-negative bacterium residing in the lower intestines of warm-blooded animals, is a multifaceted microorganism with both beneficial and pathogenic strains. While certain strains contribute to digestive processes, pathogenic variants can lead to infections, transmitted through contaminated food, water, or person-to-person contact. Clinical manifestations range from gastrointestinal symptoms to severe complications, posing public health concerns. Additionally, E. coli serves as a vital research model and is extensively employed in biotechnology for various applications [
1,
2,
3]. This communication centers on the application of molecular docking techniques to explore multiple targets within Escherichia coli (E. coli). The study delves into the investigation of various natural compounds, assessing their binding energies with specific targets. Through computational analyses [
4,
5], the study aims to elucidate potential interactions between these natural compounds and key proteins within E. coli.
2. Material and Methods
-Beta-lactamase was taken by Protein Data Bank (PDB Code: 3g34); Grid box Coordinates of binding Center X ( 28,556), Y( 93,9135), Z(-5,7545).
-MccE protein was taken by Protein Data Bank (PDB Code: 3r9f); Grid box Coordinates of binding Center X ( 32,50962), Y( 16,8891), Z(-19,3926).
-Penicillin G acylase was taken by Protein Data Bank (PDB Code:1gm7) Grid box Coordinates of binding Center X ( 15,0177), Y( -1,6784), Z(4,3186).
-DNA polymerase II was taken by Protein Data Bank (PDB Code: 3maq ); Grid box Coordinates of binding Center X ( 14,5451), Y( 10,7065 ), Z(12,0351).
3. Results and Discussion
Escherichia coli (E. coli) is a bacterium commonly found in the intestinal flora of humans and animals. While most strains of E. coli are harmless and contribute to the intestinal microbiota, some pathogenic strains can cause severe illnesses [
1,
2,
3,
4,
5]. Enterohemorrhagic E. coli (EHEC), such as the well-known serotype O157:H7, can lead to foodborne infections characterized by symptoms like hemorrhagic diarrhea, abdominal cramps, vomiting, and fever[
2]. Infections often result from the consumption of contaminated food or water or direct contact with infected animals[
1,
2,
3,
4,
5].
E. coli has been extensively used in biomedical research and pharmaceutical production due to its ability to produce recombinant proteins. Its well-studied genetics make it a model organism in molecular biology. Preventive measures, including proper hygiene practices, thorough cooking of food, and consumption of safe water, are crucial to reducing the risk of E. coli infections. Understanding the various strains and their impact on human health is essential for developing effective prevention and treatment strategies.
For the first time, this work was explored the antimicrobial potential of Hypericin with several proteins of E.coli, through molecular docking studies [7.10]. The findings reveal promising binding interactions between Hypericin and essential protein targets associated with bacterial survival.
This investigation focuses on the specific examination of four targets within Escherichia coli (E. coli): Beta-lactamase, MccE protein, Penicillin G acylase, and DNA polymerase II.
Molecular docking techniques were employed to scrutinize the binding interactions of various natural compounds with these targets. Notably, Hypericin demonstrated outstanding binding energy scores, approximately -10.5 kcal/mol, within the Ligand Binding Site of the investigated proteins.
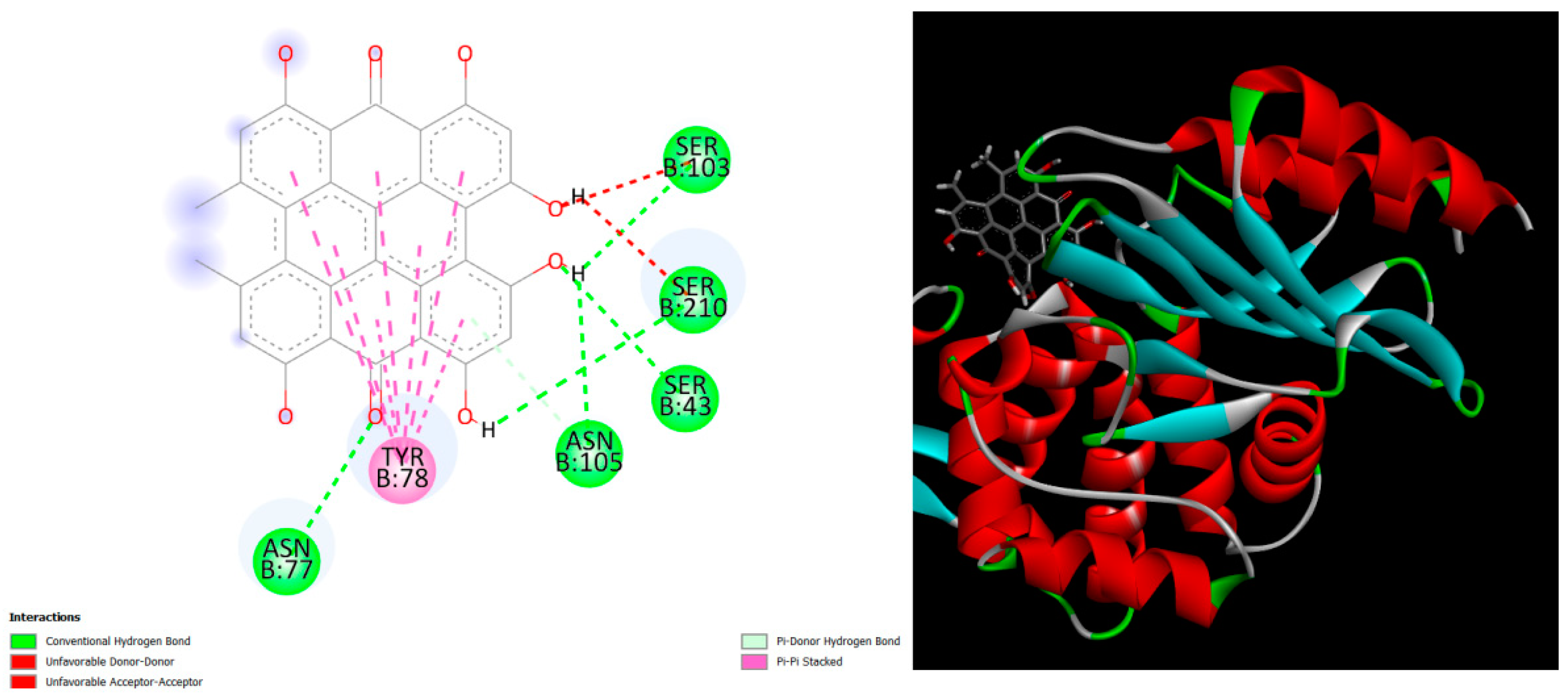
Figure 1.
displays the docking outcomes of Beta-lactamase in conjunction with Hypericin -9.6 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Hypericin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Hypericin.
Figure 1.
displays the docking outcomes of Beta-lactamase in conjunction with Hypericin -9.6 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Hypericin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Hypericin.
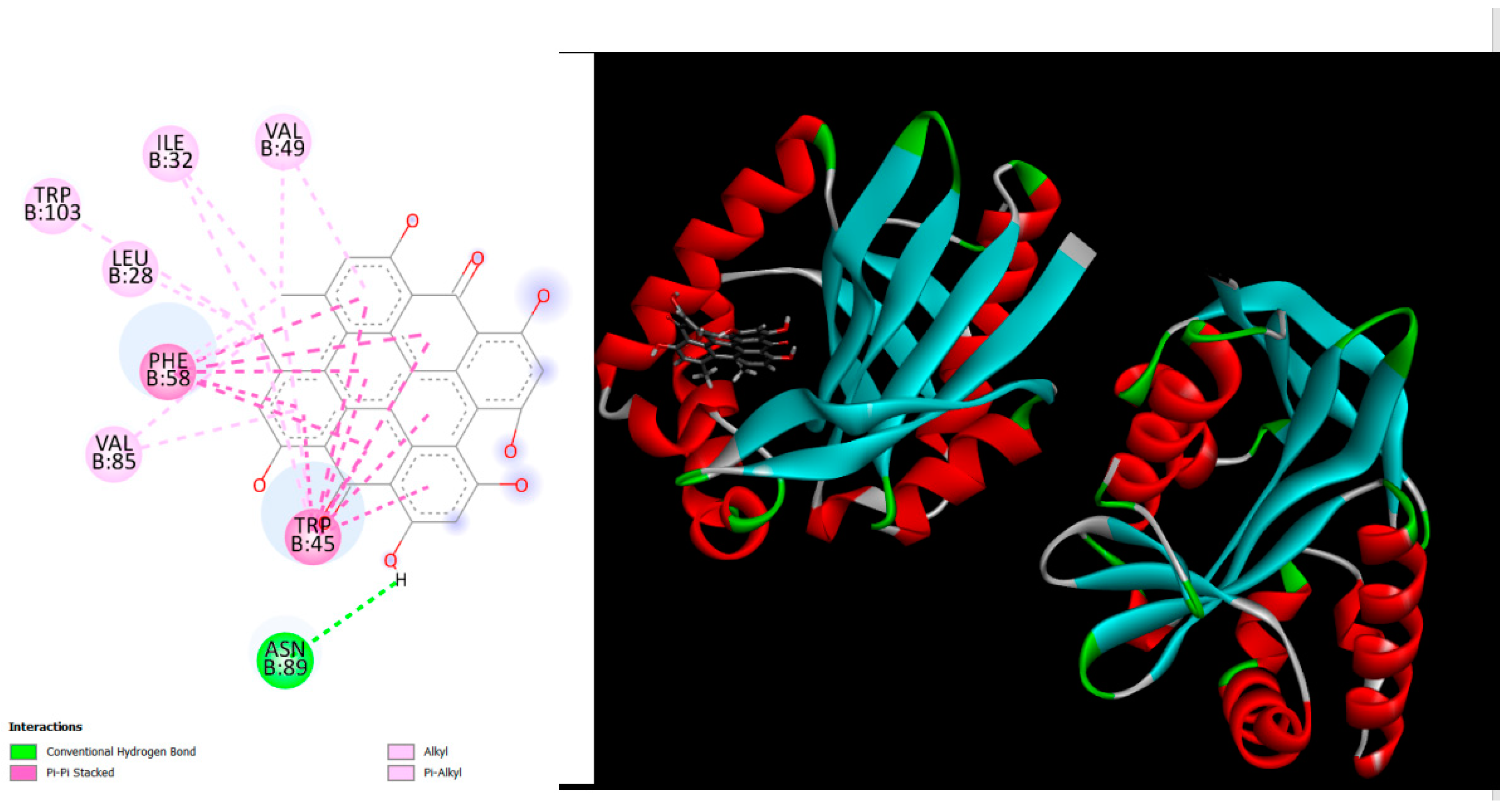
Figure 2.
displays the docking outcomes of MccE protein in conjunction with Hypericin -12.7 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Hypericin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Hypericin.
Figure 2.
displays the docking outcomes of MccE protein in conjunction with Hypericin -12.7 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Hypericin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Hypericin.
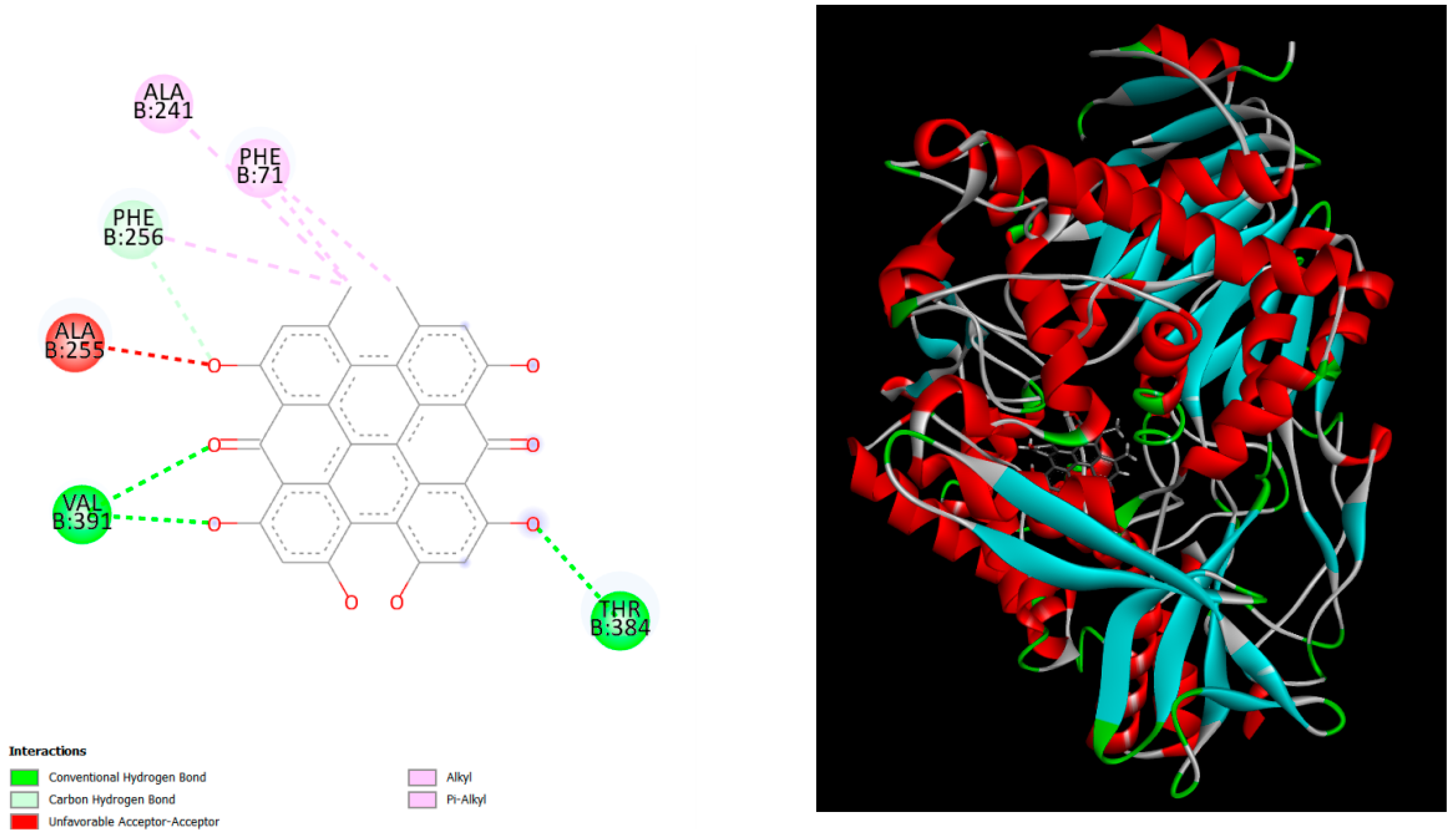
Figure 3.
displays the docking outcomes of Penicillin G acylase in conjunction with Hypericin -9.8 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Hypericin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Hypericin.
Figure 3.
displays the docking outcomes of Penicillin G acylase in conjunction with Hypericin -9.8 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Hypericin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Hypericin.
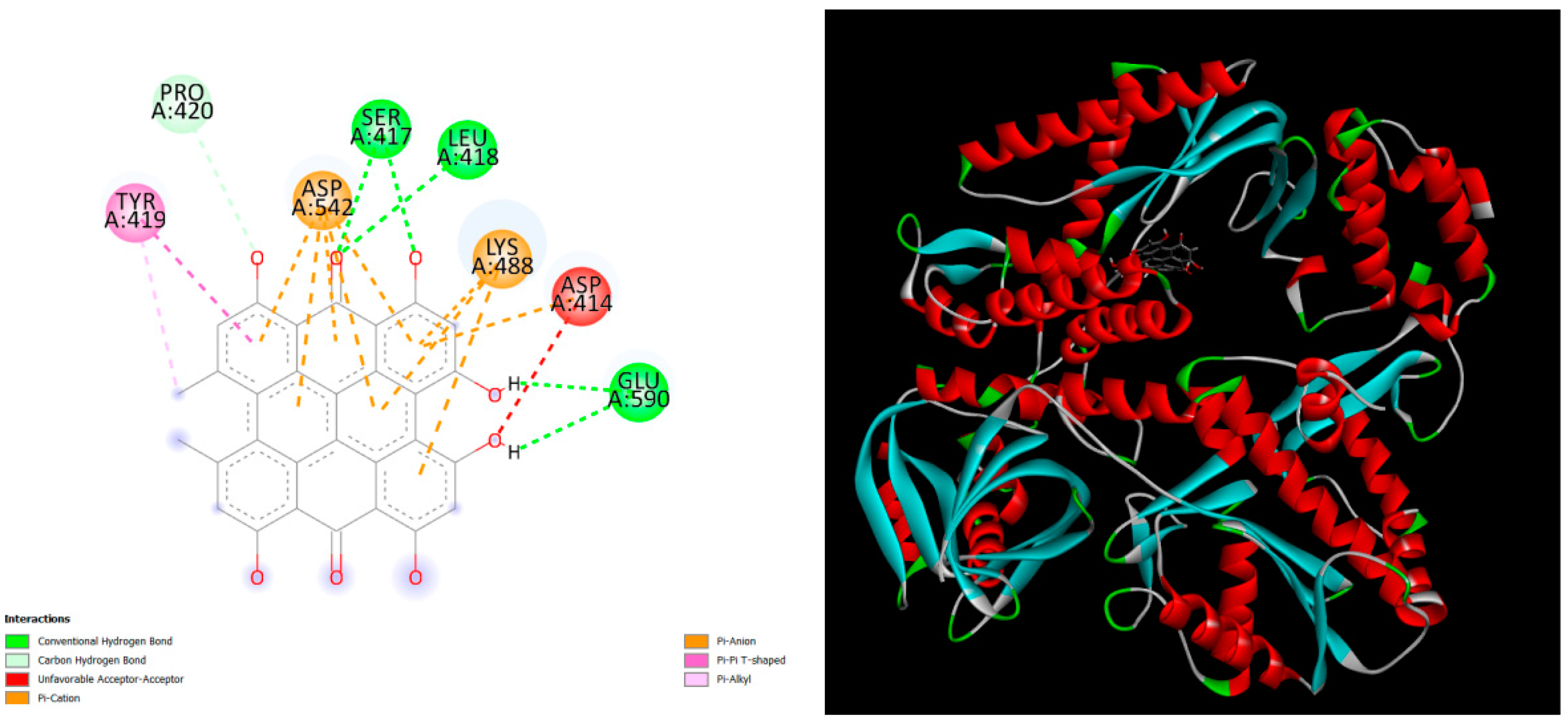
Figure 4.
displays the docking outcomes of DNA polymerase II in conjunction with Hypericin -9.7 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Hypericin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Hypericin.
Figure 4.
displays the docking outcomes of DNA polymerase II in conjunction with Hypericin -9.7 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina through the Mcule Database. On the left side, 2D diagrams illustrate the residue interactions between the protein and Hypericin. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Hypericin.
4. Conclusion
In conclusion, this study has utilized molecular docking techniques to probe the interactions between various natural compounds, with a particular emphasis on Hypericin, and key targets within Escherichia coli (E. coli). The investigation focused on four significant proteins: Beta-lactamase, MccE protein, Penicillin G acylase, and DNA polymerase II. The notable finding of Hypericin exhibiting exceptional binding energy scores of approximately -10.5 kcal/mol within the Ligand Binding Site of these proteins suggests its potential as an effective modulator of E. coli targets.
While these computational results provide promising insights, further experimental validation is imperative to confirm Hypericin's actual efficacy and safety.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pokharel, P., Dhakal, S., & Dozois, C. M. The diversity of escherichia coli pathotypes and vaccination strategies against this versatile bacterial pathogen. Microorganisms 2023, 11, 344. [CrossRef]
- Huerta-Saquero, A., Chapartegui-González, I., Bowser, S., Khakhum, N., Stockton, J. L., & Torres, A. G. P22-Based Nanovaccines against Enterohemorrhagic Escherichia coli. Microbiology Spectrum 2023, 11, e04734-22. [CrossRef]
- Cobo-Simón, M., Hart, R., & Ochman, H. Escherichia coli: What is and Which are? Molecular Biology and Evolution 2023, 40, msac273. [CrossRef]
- Nataro, J. P., & Kaper, J. B. Diarrheagenic escherichia coli. Clinical microbiology reviews 1998, 11, 142–201. [CrossRef]
- Gomes, T. A., Elias, W. P., Scaletsky, I. C., Guth, B. E., Rodrigues, J. F., Piazza, R. M., ... & Martinez, M. B. Diarrheagenic escherichia coli. brazilian journal of microbiology 2016, 47, 3–30. [CrossRef]
- Allocati, N., Masulli, M., Alexeyev, M. F., & Di Ilio, C. Escherichia coli in Europe: an overview. International journal of environmental research and public health 2013, 10, 6235–6254. [CrossRef]
- Odhar, H. A., Rayshan, A. M., Ahjel, S. W., Hashim, A. A., & Albeer, A. A. M. A. Molecular docking enabled updated screening of the matrix protein VP40 from Ebola virus with millions of compounds in the MCULE database for potential inhibitors. Bioinformation 2019, 15, 627. [CrossRef]
- Trott, O., & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry 2010, 31, 455–461. [CrossRef]
- Callil-Soares, P. H., Biasi, L. C. K., & Pessoa Filho, P. D. A. Effect of preprocessing and simulation parameters on the performance of molecular docking studies. Journal of Molecular Modeling 2023, 29, 251. [CrossRef]
- Ding, J., Tang, S., Mei, Z., Wang, L., Huang, Q., Hu, H., ... & Wu, J. Vina-GPU 2.0: Further Accelerating AutoDock Vina and Its Derivatives with Graphics Processing Units. Journal of Chemical Information and Modeling 2023, 63, 1982–1998. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).