1. Introduction
Hepatitis Delta Virus (HDV) is the smallest known virus that infects humans. It belongs to the Kolmoviridae family and the Deltavirus genus [
1]. It is a satellite virus that relies on the surface proteins of Hepatitis B Virus (HBsAg) for assembly, hepatocyte entry, and the release of infectious particles [
2]. HDV virions consist of HBV surface antigens and host cell lipids surrounding a ribonucleoprotein (RNP) complex formed by a single-stranded circular RNA genome of about 1700 nucleotides, along with hepatitis delta antigens (HDAg) [
2,
3,
4]. Upon hepatocyte infection, the HDV RNA genome translocates into the nucleus, where the host cell RNA polymerase initiates replication through a rolling-circle mechanism [
5,
6,
7]. This replication strategy allows the production of linear concatemers of HDV genomes and antigenomes that are self-cleaved by the HDV ribozymes, intrinsic catalytic RNA domains present in each RNA strand [
8,
9]. The antigenomic RNA encompasses a single open reading frame that encodes the short hepatitis delta antigen (S-HDAg) [
10]. Through editing by the host cell’s adenosine deaminase acting on RNA-1 (ADAR-1) at the UAG stop codon, an amber (W) site is formed. This modification results in an extended open reading frame (ORF) that gives rise to the large hepatitis delta antigen (L-HDAg) [
10]. Despite sharing most of their amino acid sequence, both the short and the large antigens differ significantly in their functions. S-HDAg is essential for HDV replication, whilst L-HDAg blocks HDV replication and is essential for viral assembly [
11,
12]. Both HDAgs contain a nuclear localization signal (NLS) located between positions 66 and 75 (EGAPPARAR), being Glu-66 (E66), and Arg-75 (R75) indispensable for the nuclear import of HDV RNP [
13,
14,
15]. The L-HDAg contains a Nuclear Export Signal (NES) and Viral Assembly Signal (VAS) domains [
16] and is responsible for the translocation of HDV RNPs from the nucleus to the cytoplasm, which is essential for viral assembly. The NES is located within the aa 198-210 of the L-HDAg sequence. More specifically, Pro-205 is the critical residue for the correct functionality of NES [
16].
The association between HBsAg and HDV RNP takes place in the ER via a lipid farnesyl-moiety present on the L-HDAg that binds to the cytoplasmic loop of the small HBV surface antigen (S-HBsAg) and is indispensable to promote viral spreading and to complete the HDV life cycle [
17,
18]. Furthermore, phosphorylation of some residues of S-HDAg plays an important role on viral replication [
19]. More specifically, phosphorylation of Ser-177 promotes HDV replication by increasing the interaction of S-HDAg with RNA Pol II [
19,
20,
21]. During the HDV replication, S-HDAg co-localizes with the host RNA pol II in the nucleoplasm, within the SC35 speckles sites that are highly active sites of transcription and RNA processing [
7,
23]. At earlier steps, S-HDAg is predominantly located in the nucleus, both in the nucleolus and the nucleoplasm. Later in the infection, when L-HDAg starts to be synthesized, the HDAgs can be found in non-SC-35 speckles sites, in the cytoplasm, and in the Golgi apparatus, where the post-translational modifications take place [
24,
25,
26]. Thus correct localization of HDAg is an important aspect of HDV viral cycle.
HDV infection is recognized as the most severe form of viral hepatitis. HDV-infected patients have a higher risk of developing cirrhosis and hepatocellular carcinoma (HCC) as well as hepatic decompensation and increased mortality in comparison with HBV mono-infected patients [
27]. Despite the severity of this disease, the underlying mechanism is still unknown and hence there is a lack of effective treatments to control HDV-induced liver damage [
28]. One of the main reasons for the scarce knowledge of the molecular mechanism/s involved in the pathology of this disease is the absence of adequate animal models that resemble the main pathological features observed in HDV patients and amenable to experimentation [
29]. Recently, we utilized adeno-associated vectors (AAVs) as delivery vehicles for HBV/HDV replication-competent viral genomes. The co-administration of recombinant AAV-HBV and AAV-HDV to WT mice resulted in the establishment of AAV-independent HDV replication and, more importantly, the animals developed a significant liver pathology characterized by transaminase elevation, lobular inflammation, cytoplasmic swelling, and sanded nuclei, that were not observed in AAV-HBV mice [
30,
31,
32]. Thanks to the easy manipulation of the system, we were able to confirm in vivo what has been previously shown in cell culture HV model e.i. that S-HDAg is essential for HDV replication and cannot be replaced by L-HDAg or host cellular proteins, and that L-HDAg is essential to produce the HDV infectious particle and inhibits its replication [
31]. Furthermore we found that the ratio L-HDAg/S-HDAg plays an important role on the magnitude of HDV-induced liver damage, the lowest it is the strongest is liver damage [
31].
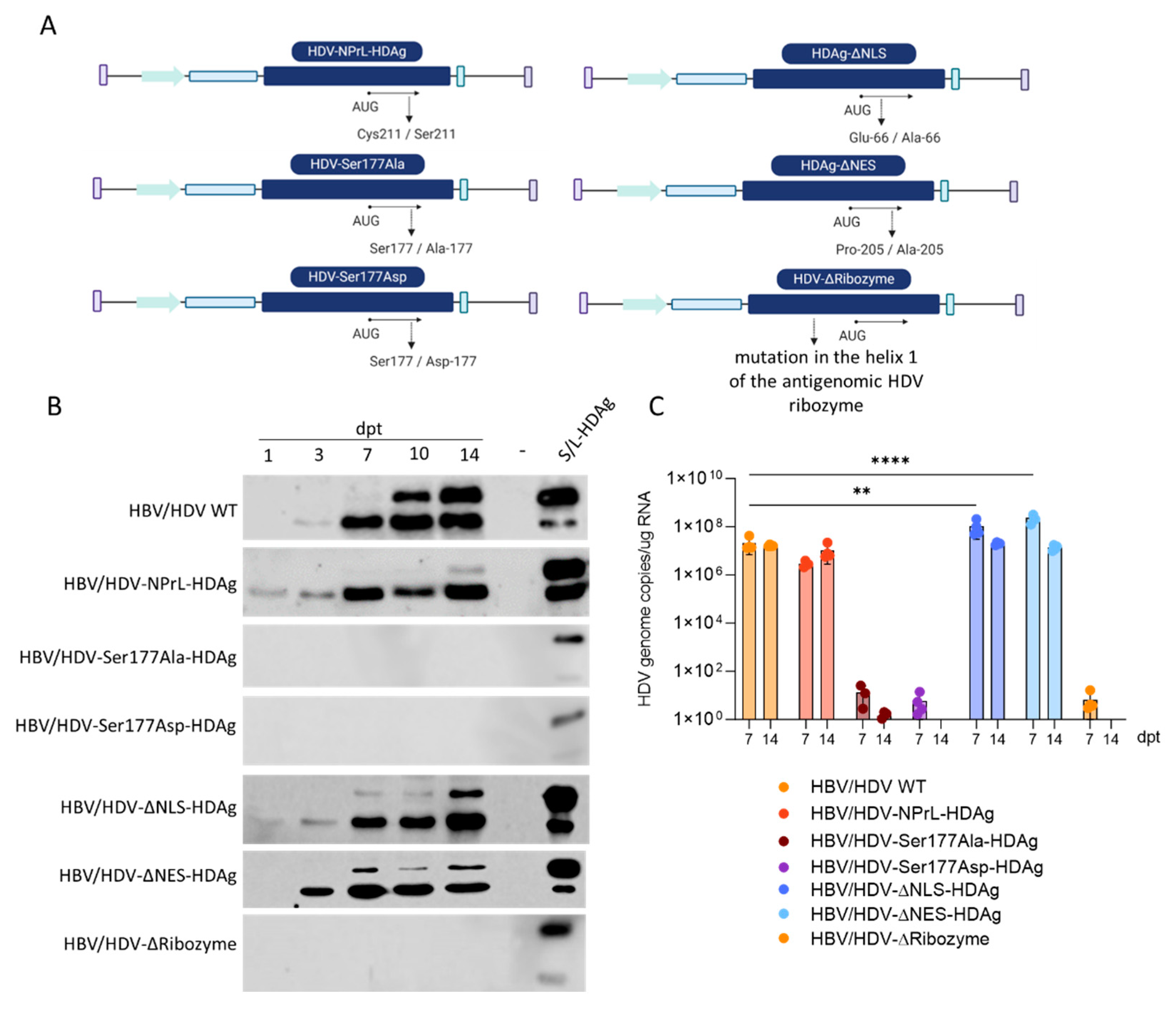
The aim of the present work was to identify the role of post-translational modifications and cellular location of HDV antigens on HDV viral cycle and in HDV-mediated liver damage. We found that Ser 177 phosphorylation and the ribozyme are essential for HDAg expression and HDV replication. Mutation in the NES and NLS signal clearly affect HDAg intracellular localization significantly affect HDV replication in vivo but not in vitro. More importantly, mutation of the isoprenylation domain prevent the formation of infectious particles and reduces the L-HDAg/S-HDAg and increase cellular toxicity both in vitro and in vivo.
2. Materials and Methods
2.1. Site-directed mutagenesis (SDM)
Site-directed mutagenesis (SDM) was handled using the TaKaRa In-Fusion Cloning Kit except for the HDV-NPrL-HDAg plasmid that was carried out using QuikChange II Site-Directed Mutagenesis. Briefly, using TaKaRa In-Fusion Cloning Kit, four oligonucleotides were designed for each HDV mutant, except for the HDV-∆HDAg mutant that required six oligonucleotides since it was necessary to introduce two mutations. Then, 10 ng of the HDV plasmid was used for the amplification of the new mutated inserts. In parallel, the plasmid was digested with the appropriate restriction enzymes. The empty vector obtained after the enzymatic digestion and the PCR products were purified and ligated for 15 minutes at 50 °C. Subsequently, Stellar competent cells were transformed by heat shock, and the obtained clones were sequenced to verify the presence of the desired mutations and the absence of unspecific mutations. When using QuikChange II Mutgenesis, two mutagenic oligonucleotide primers were designed to produce the HDV-NPrL-HDAg mutant as recommended in the QuikChange™ manual (Agilent, #200524). For the PCR amplification, 50 ng of the HDV plasmid, 125ng of primer Forward (Fw), 125ng of Primer Reverse (Rv), 1 μl of dNTP mix, 3 μl of QuikSolution and 1 μl of PfuUltra High Fidelity (HF) DNA polymerase (Agilent, #600380) were mixed at a final volume of 50 μl. And PCR amplified. Then, 10 μl of the PCR product was run in an electrophoresis gel to check the quality of the reaction. To remove the methylated DNA plasmid (in this case, the non-mutated HDV plasmid), 1 μl of the DpnI restriction enzyme was added to the reaction product and was incubated at 37 °C for 1 hour. Finally, the PCR product was electroporated on One Shot® Top10 Electrocomp™ E. coli (Thermo Fisher Scientific, #C404052). Plasmid DNA was purified using the NucleoSpin® Plasmid kit (Macherey-Nagel, #740588) and the positive colonies were identified by restriction enzyme digestion. Then, the DNA plasmids were sequenced to verify the introduction of the correct mutation and to dis-card unspecific mutations.
2.2. Cell lines
The human hepatoma cell line Huh-7 and 293T were acquired from the Glow Biologics (GBTC-099H) and ATCC (CRL-3216)., respectively. Huh-7 cells were used for the HDV plasmid transfection studies and 293-T cells for AAV vector production. Huh-7.5.1 stably expressing the human Na-taurocholate co-transporting polypeptide (hNTCP, Huh-7-hNTCP), which is essential for HBV and HDV cell entry [
33,
34] were kindly provided by Dr Urtzi Garaigorta and were employed for infectivity studies. Both cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% of L-glutamine, 1% of glucose, 100 U/ml of penicillin-streptomycin and no-essential amino acids, and incubated at 37 °C with 5% CO
2 in humidified atmosphere. In the case of Huh-7-hNTCP DMEM was supplemented with 2.5 μg/ml of blasticidine to ensure the selection of hNTCP expressing cells.
2.3. DNA Transfection
For transfection, Huh-7 cells were used at a confluence of 80-90%. The day of transfection, the culture medium was replaced by Optimem. Briefly, Lipofectamine 3000 and Optimem were mixed, vortexed and incubated 5 minutes at room temperature (RT) (reaction mix A). In parallel, plasmid DNA, the P3000 reagent and Optimem were also mixed, vortexed and incubated 5 minutes at RT (reaction mix B). Then, mix A and B were mixed, without vortex, and incubated 15 minutes at RT. After that time, the transfection mix was added drop by drop. Six hours after adding the transfection reagents, fresh DMEM 10% FBS was added on the culture plates without removing the medium. Finally, 1 day after transfection, the culture medium was removed, and the cells were washed before adding fresh DMEM 10% FBS.
2.4. Generation of AAV vectors
HEK293T cells were used as packaging cells. The constructs containing the recombinant AAV genomes were transfected together with the helper plasmid pDP8.ape (Plasmid Factory, #PF478) that provides the genes requires for the replication and encapsidation of AAV serotype 8 (a liver tropic AAV serotype in mice). The transfection was carried out using poly-ethyleneimine (PEI, Sig-ma-Aldrich, #408727). Cells were incubated for 72 hours and during this time the viral particles were assembled inside the packaging cells. Cells were collected in lysis buffer (50 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% Triton X-1000) and various freeze-thaw cycles were performed to release the viral particles. Cell debris was eliminated by centrifugation. The supernatant of the cells was collected and incubated in 8% polyethylene glycol (PEG-800, Sigma-Aldrich, #P5416) for 48-72 hours to precipitate the particles. The supernatant was centrifuged, and the pellet was resuspended in lysis buffer. Cell lysate and precipitated supernatant were mixed and treated with 0.1mg/plate of DNaseI (Roche, #10104159001) and RNaseA (Roche, #10109169001) for 1 hour at 37ºC. The viral particles were purified by ultracentrifugation in iodioxanol gradient (OPTIPREP, Atom, #415468) gradient. Iodixanol was removed and the vector was concentrated using Amicon 100K columns (MERK Millipore, #UFC910008). Viral DNA was extracted using the High Pure Viral DNA Kit (Roche, #11858874001) following the manufacturer’s instructions. The vector titer in terms of viral genomes (vg) per mL was calculated by Real Time qPCR (RT-qPCR) (#1855196, BioRad Hercules, CA, USA) using specific primers for the AAV’s inverted terminal repeat (ITR) sequences.
2.5. Animal manipulation and procedures
WT C57BL/6 mice were purchased from Harlan Laboratories (Barcelona, Spain). Six- to eight-week-old male mice were used in all experiments. Mice were kept under controlled temperature, light, and pathogen-free conditions in a BSL3 animal facility.
The animals received the vectors intravenously diluted in saline solution (Braun, #651860) in a final volume of 100 μL/mouse under inhalatory anesthesia. All mice received 5x1010 vg of the corresponding AAV-HDV wt or mutant vector together with 5x1010 vg of AAV-HBV.
The experimental design was approved by the Ethics Committee for Animal Testing of the University of Navarra (R-132-19GN).
2.6. Protein extraction from cells and liver samples
RIPA Buffer (0.75M NaCl, 5% of Tris 1M pH 8, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate diluted in water) was supplemented before each use with 1mM sodium orthovanadate, 1mM PMSF, 1mM sodium pyrophosphate and protease inhibitor cocktail. Cell pellets were resuspended and incubated for 30 minutes at 4ºC on a shaker with the RIPA lysis buffer and centrifuged at 13000 rpm for 20 minutes at 4ºC. Supernatants were collected in new tubes. Protein concentration was calculated using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, #23225).
2.7. Immunofluorescence (IF)
Cells were washed once with PBS, fixed with cold 4% formaldehyde (freshly pre-pared from methanol-free 16% formaldehyde, Thermo Scientific, #28908) for 15 minutes at RT and washed again for three times with PBS. For intranuclear staining, cells were permeabilized with 0.1% Triton X-100 in PBS for 15 minutes at RT and, after three washes with PBS, a blockade was performed with 5% BSA in PBS-0.1% tween for 30 minutes at 37ºC. Next, cells were incubated for 30 minutes at 37ºC with anti-HDV human serum provided by the BioBank of the Universidad de Navarra (CUN-28336) at dilution 1:2500. After washing three times, the secondary antibody (anti-human Alexa-Fluor 488,) was incubated for 30 minutes at 37ºC diluted 1:3000 in PBS and protected from light. Finally, coverslips were mounted in microscope slides using mounting medium with DAPI for fluorescent labelling (Vector Laboratories, #H-1200). Fluorescent samples were obtained with a confocal microscope (Zeiss LSM 880 NLO) at 40x, 60x and 100x of magnification.
2.8. Determination of HDV infectious viral particles
Huh-7-hNTCP cells seeded in BD FalconTM Culture Slides (8-well, BD Biosciences, US, #345118) or in 6- or 12-wells plate with a coverslip, were incubated 24 hours with HDV-containing cell supernatants. Supernatants were removed and cells were washed with fresh PBS. Then, cells were maintained with fresh medium that was changed every 2 days until 7 days post-infection. At this time point, cells were incubated 15 minutes at RT with 4% PFA, and HDAg expression was assessed by immunofluorescence, as described above.
2.9. Western Blot
20-30μg of extracted proteins were mixed with a SDS-PAGE loading buffer (70% Tris HCl pH 6.8, 30% glycerol, 0.35M SDS, 0.6M DTT, 0.18mM Bromophenol Blue) and boiled for 5 minutes at 95ºC. Samples were loaded on 12% SDS-polyacrylamide gels of 1.5mm and electrophoresis was performed for separating proteins. 5ul of Precision Plus Protein™ Dual Color Standard (BioRad, #161-0394), ranging from 10kDa to 250kDa, was used to determine protein size. Subsequently, the resolved proteins were transferred to a nitrocellulose membrane (Bio-Rad #162-0112) by wet electroelution at 80V for 1 hour and 15 minutes. Following, the membrane was stained with Ponceau Red dye to check the transference quality. After washing the dye with water, the membrane was blocked during 45 minutes with Tris-buffer saline (TBS)-tween 5% non-fat dry milk at RT on a shaker. Primary antibodies were added and incubated ON while shaking at 4ºC. Next, the membrane was washed 3 times with TBS-Tween 20 0.05% for 10 minutes and then, it was incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody at RT for 1h. After 3 washes of 10 minutes, the peroxidase signal was developed using SuperSignalTM West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, #34095). Odyssey CLx near-infrared Fluorescence Imaging System was used for image generation and images were analyzed with the Image Studio Lite software.
2.10. Histology and Immunohistochemistry (IHC)
Hematoxylin & Eosin (H&E): Liver sections were fixed with 4% paraformaldehyde (PFA), embedded in paraffin, sectioned (3 μm), and stained with hematoxylin and eosin. Sections were mounted and analysed by light microscopy for histologic evaluation.
Immunohistochemistry (IHC): the first steps were the same as for the H&E staining. Then, a step of antigen retrieval was performed that consisted of incubation for 30 min at 95 °C in 0,01 M Tris-1 mM EDTA pH 9. Subsequently, primary antibodies were incubated overnight at 4 °C. After rinsing in TBS-T, the sections were incubated with the corresponding secondary antibodies for 30 min at RT. Peroxidase activity was revealed using DAB+ and sections were lightly counterstained with Harris hematoxylin. Finally, slides were dehydrated in graded series of ethanol, cleared in xylene and mounted with Eukitt (Labolan, #28500, Navarra, Spain). Image acquisition was performed on an Aperio CS2 slide scanner using ScanScope Software (Leica Biosystems, Vista, CA, USA). The image analysis was performed using a plugin developed for Fiji, ImageJ (NIH, Bethesda, MD, USA). The antibodies employed were anti-HDV human serum sample (CUN-28336) and anti-F4/80 (BioLegend #123102).
2.11. RNA Extraction and RT-qPCR
Total RNA from liver samples was isolated using TRI Reagent
®® (#T9424, Sigma-Aldrich,) according to the manufacturer’s instructions. Total RNA was pre-treated with DNAse I (#AM-1907, TURBO DNA-free ™ Kit, Applied Biosystems) and reverse-transcribed into complementary DNA (cDNA) using M-MLV reverse-transcriptase (Invitrogen, #28025013). Real-time quantitative polymerase chain reactions (RT-qPCR) were performed using iQ SYBR Green Supermix (#170-8884, BioRad) in a CFX96 Real-Time Detection System (#1855196, BioRad,) and primers as specified in supplementary materials. HDV strand-specificity was analyzed as described elsewhere [
30]. GAPDH was used as a control housekeeping gene.
2.12. Statistical Analysis ç
Statistical analyses were performed using GraphPad Prism 8.0 software. The data are presented as individual values ± standard deviation. The statistical analysis performed in each experiment was specified in the legend of the figures. Significance *P<0.05, **P < 0.01, ***P < 0.001, ****P<0.0001).
4. Discussion
In this study, we comprehensively assessed in vitro and in vivo the consequences of introducing various mutations into the HDV genome, specifically targeting HDAg intracellular localization, post-translational modification, and the functionality of the HDV ribozyme. Utilizing a surrogate system developed in our laboratory involving plasmids and a liver-tropic AAV vector carrying the HDV 1.2x sequence, developed by Dr. Taylor’s group [
35], under the transcriptional control of a liver-specific promoter, we achieved transcription of the HDV anti-genome sequence. This sequence initiated HDV replication and the expression of the two HDAgs, both in vitro and, more importantly, in the livers of mice [
30].
Our in vitro investigations were conducted in the human hepatic cell line Huh-7, known for sustaining HDV replication due to the absence of a type I response [
31]. We observed that mutations in the Ser177 residue, either by replacing it with the neutral amino acid alanine or the phosphomimetic amino acid aspartic acid, resulted in the complete absence of protein expression and a substantial impact on HDV replication. Ser177 phosphorylation of S-HDAg is crucial for viral replication, as it is required for interaction with cellular RNA Pol II in the production of new genomes [
19]. Unphosphorylated S-HDAg cannot interact with the host polymerase, preventing the replication of the HDV antigenome. Furthermore, the results obtained from transfection with the mutant, where the Ser residue was replaced by Asp, underscore the importance of maintaining a delicate equilibrium between unphosphorylated and phosphorylated S-HDAg at Ser-177 in the HDV life cycle. Indeed, it has been demonstrated that only non-phosphorylated S-HDAg is incorporated into HDV particles [
22]. Our data confirm the importance of maintaining a proper balance between phosphorylated and unphosphorylated S-HDAg at Ser-177 for the correct establishment of the HDV life cycle.
Similar outcomes were observed following the transfection of the HDV mutant, where we introduced a conformational change in the ribozyme sequence of the antigenome. Drawing on previous studies involving the truncation of different regions of the HDV ribozyme, we engineered an HDV mutant with a mutation in helix 1 of the antigenomic HDV ribozyme. Transfection of this mutant into Huh-7 cells resulted in no HDAg production or HDV replication, confirming the anticipated importance of the correct folding of this sequence for establishing HDV replication [
9]. Due to the lack of protein expression and HDV replication in vitro, Ser177 and ribozyme mutants were not evaluated
in vivo.
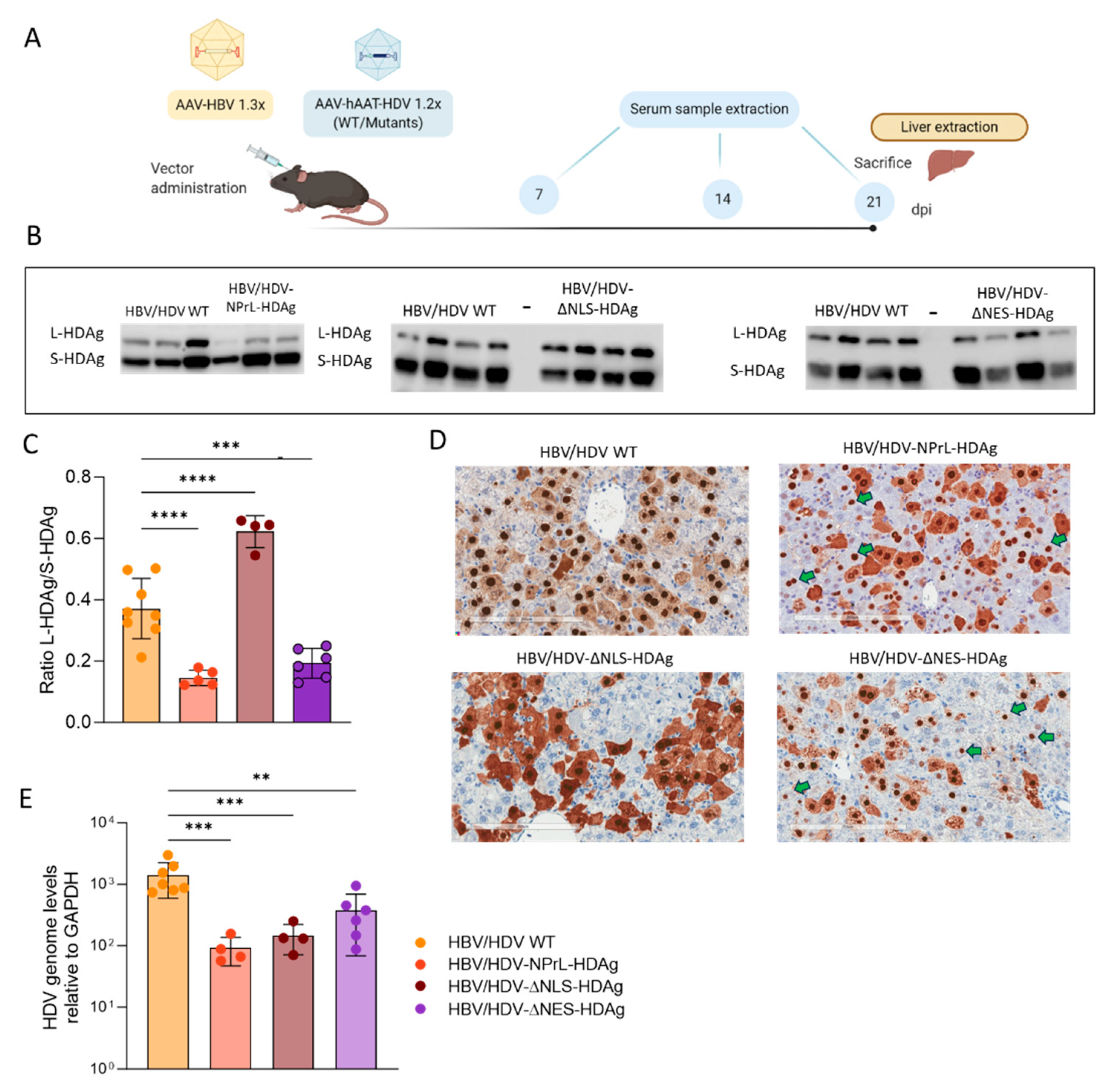
Regarding the non-prenylated mutant, we observed an altered L-HDAg/S-HDAg ratio both in vitro and in vivo through Western Blot (WB) analysis. While normal levels of S-HDAg expression were detected, the expression of non-prenylated L-HDAg was significantly lower. These data suggest that the lack of isoprenylation might affect the stability of L-HDAg, resulting in a lower L-HDAg/S-HDAg ratio compared to HDV WT. Alternatively, the absence of prenylated L-HDAg might impact HDV genome editing, reducing the expression of this antigen. However, recent findings by Verrier et al. demonstrated that treating HepaRG cells with an isoprenylation inhibitor resulted in the accumulation of edited HDV genomes, reducing the likelihood of this hypothesis [
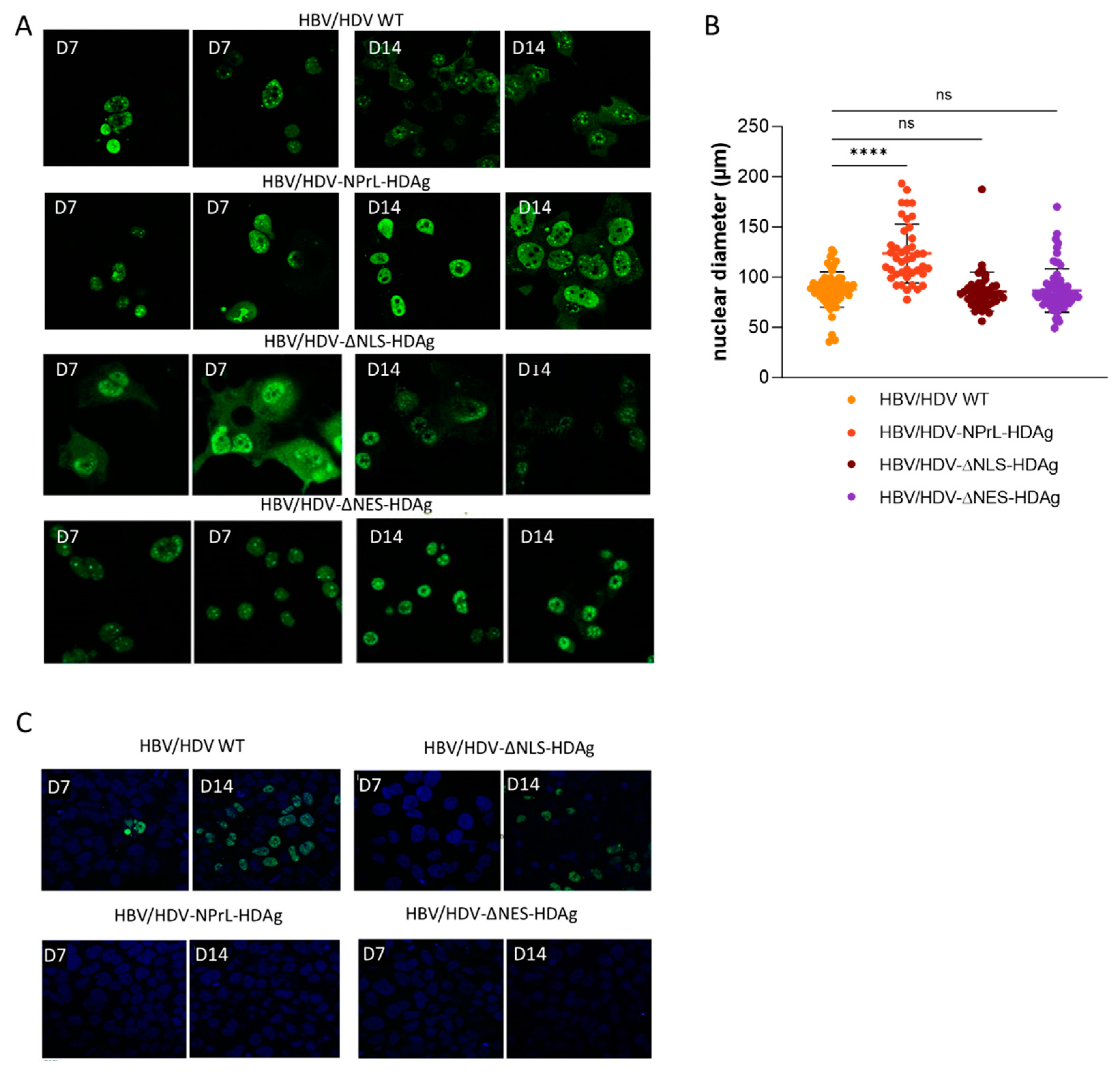
36]. Furthermore, our in vitro experiments clearly indicated that isoprenylation plays a crucial role in the cellular localization of HDAgs, leading to a predominant nuclear localization. This observation was also noted in some mouse hepatocytes. The absence of isoprenylation likely disrupts the normal translocation of HDAgs to the cytoplasm and their association with cellular membranes. Similar observations were made in HDV mutants lacking L-HDAg [
31] and in the HDV-∆NES-HDAg, highlighting the role of L-HDAg in the translocation of HDV RNPs from the nucleus to the cytoplasm.
We also found that the lack of isoprenylation leads to a delay in viral replication in vitro and a significant reduction in viral replication
in vivo. This suggests that this translational modification might play a role in the regulation of HDV replication. Interestingly, according to a previous publication, an increase in viral replication would be expected since earlier findings attributed the inhibition of viral replication to prenylated L-HDAg, however, we observed the opposite situation [
37]. One hypothesis to explain the decreased replicative capacity may be due to impaired HDAg localization and altered membrane association [
38]. Isoprenylation facilitates the association of HDV ribonucleoproteins (RNPs) with cellular membranes. Without isoprenylation, this association may be compromised, affecting the interaction of HDAgs with cellular kinases [
19,
20,
21,
22]. As expected, this mutant was not capable of producing HDV infectious particles, as this motif is essential for the interaction with HBsAg. In summary, inhibiting the isoprenylation of L-HDAg has multifaceted effects on the composition, stability, cellular localization, and membrane association of HDAgs, ultimately impacting the efficiency of HDV replication.
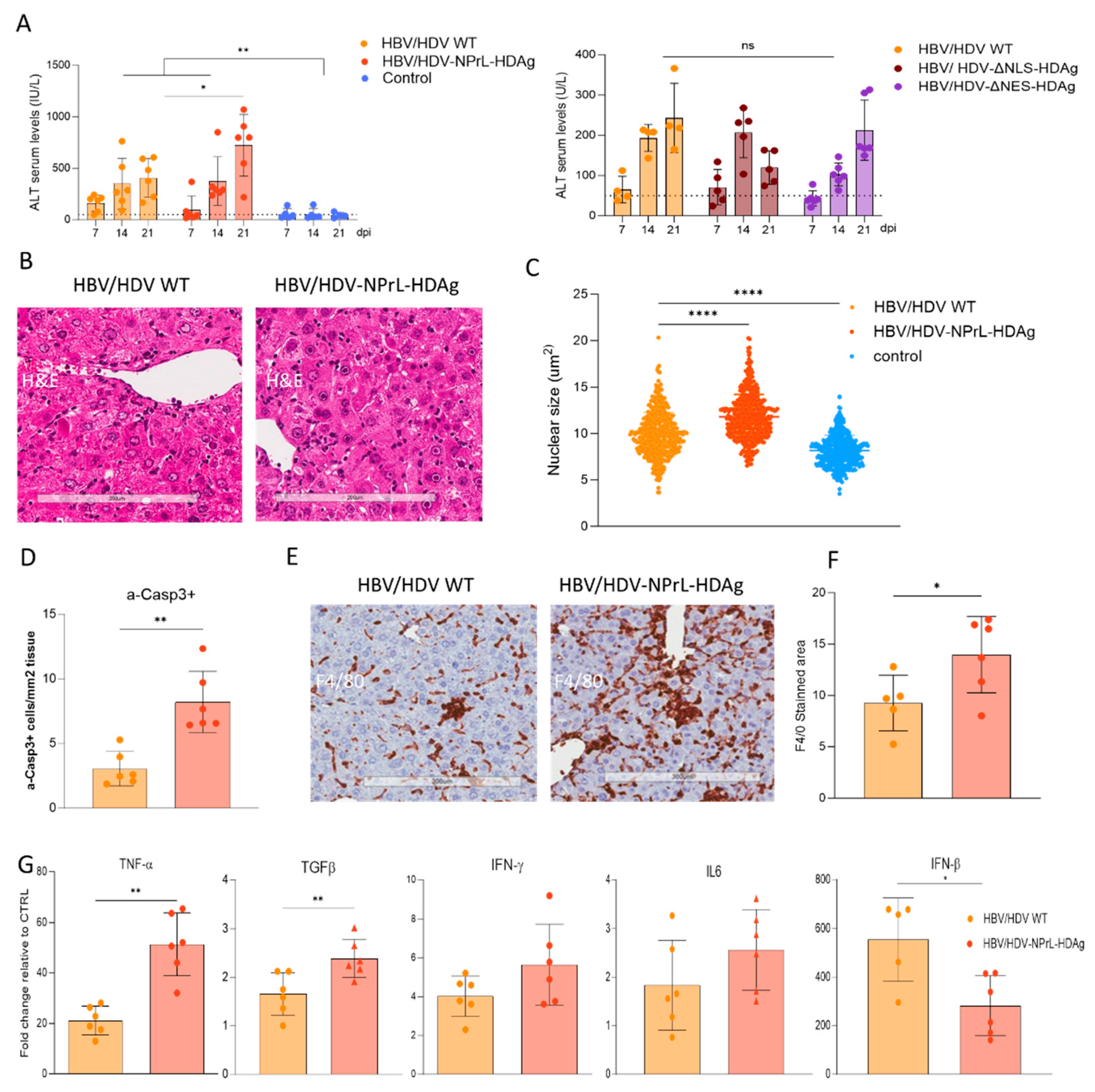
However, our more important finding associated with this mutant is its association with exacerbated liver pathology. We observed both in vitro and in vivo a higher enlargement of the nuclei compared to HDV-WT. Nuclear enlargement is indicative of cellular stress and toxicity that might be associated to alterations in cellular processes, including damage to the DNA, disruption of normal cellular functions, or induction of cellular stress responses [
39]. Even though the replicative capacity of this mutant was lower than the WT, it was able to induce stronger liver damage than HDV WT vector, as evidenced by significantly higher transaminase elevation and a higher number of apoptotic hepatocytes. This higher toxicity was accompanied by a stronger macrophage infiltrate and higher expression of proinflammatory cytokines like TNF-α or TGF-β. These results are in line with or previous that demonstrated a link between TNF-α and HDV-induced liver toxicity, as the administration of the TNF-α inhibitor etanercept resulted in the amelioration of liver injury. Interestingly, the expression of type-I IFN was lower than the one in the HDV WT, most likely an indicative of the lower replication capacity of the mutant, however, is in contradiction to observation in cell culture where treatment with isoprenylation inhibitors resulted in an increase in IFN-β production [
40].
The higher toxicity of the non-prenylated mutant could be associated with the altered L-HDAg/S-HDAg 41]; we have previously shown that an HDV mutant lacking L-HDAg expression, only expressing S-HDAg, was more toxic than the HDV WT, and that this toxicity can be reduced by L-HDAg complementation. These data points toward S-HDAg as the antigen preferentially involved in HDV cytotoxicity [
31]. However, it could not be discarded that the unprenylated L-HDAg interfered with host processes. As we know, the elimination of the isoprenylation site is detrimental for viral assembly, consequently, the HDV RNPs accumulate inside the cells. The intracellular accumulation of the HDAgs could impair biological functions and induce cell stress or hepatocyte death, as has been described for other viral proteins. One example is HCV, which induces ER stress and mitochondrial alterations through calcium signaling, leading to the production of ROS that are normally found in chronic HCV patients [
42,
43]. In the case of HBV, the HBx protein promotes the activation of the inflammasome, the production of mitochondrial ROS, and the induction of pyroptosis in the infected cell [
44]. Additional studies are needed to clarify the role of the isoprenylated and non-isoprenylated L-HDAg within the HDV-induced pathology.
Next, we determined the role of the intracellular location of HDAgs in the HDV viral cycle. For that purpose, we altered the nuclear localization signal of HDAgs by the substitution of residues Glu-66 and Arg-75 by Ala, reported as essential for the correct functionality of the NLS domain [
13,
14,
15]. The HDAgs expressed by the HDAg-ΔNLS mutant displayed a subcellular distribution pattern different from that of the HDV WT, with a predominant cytoplasmic localization of the HDAgs. However, this mutation doesn’t completely abrogate the presence of the HDAgs in the nucleus, as shown both in Huh-7 cells and mice, indicating that other sequences should be involved in the traffic of the antigens from the cytoplasm to the nucleus. The discrepancy between our results and those of Alves et al. could be due to differences in the HDV constructs since they deleted residues 66 and 75, while we substituted them with alanines [75]. Therefore, it should be considered to identify more residues implicated in the functionality of the NLS domain located between 66-75 aa. Furthermore, this mutant showed a decline in HDV replication that cannot be explained by the absence of HDAg in the nucleus but alternatively by an additional function of residue Arg-75, since it has been described to be essential for the proper recruitment of host RNA Pol by binding to chromatin-remodeling complexes [
45,
46].
Finally, we have generated a mutant in which we altered the capacity of the L-HDAg to exit the nucleus by mutating the NES signal. The NES domain is located exclusively in the L-HDAg between the residues 198-210 (ILFPADPPFSPQS). In particular, Pro-205 was demonstrated to be essential for the translocation of HDV RNP from the nuclear to the cytosol [
16]. Based on those studies, we constructed an HDV mutant with a truncated NES by replacing Pro-205 with Ala-205. Despite the loss of nuclear export, HDAg-staining was detected in the cytoplasmic compartment in both transfected Huh-7 cells and C57BL/6 mice, so some amount of protein is retained after synthesis.
Although the replication capacity of this mutant was similar to or higher than that shown by HDV-WT in vitro, it is unable to form HDV infectious particles. These results indicate that once L-HDAg is inside the nucleus, if the NES domain is not functional, the RNP particles cannot be exported to the cytoplasm, and HDV infectious particles are not formed. These findings confirm previous data reported by Lee et al., demonstrating that this residue within the NES domain is indispensable for the nuclear export of HDV RNPs mediated by L-HDAg [
16]. The trafficking of HDAgs from the nucleus to the cytoplasm is necessary for the interaction between L-HDAg and HBsAg in the ER and, consequently, is required for the production of infectious particles. Consequently, the HDVAg-ΔNES mutant was not able to form infectious progeny particles. In mice, we observed that the lack of NES altered the ratio of L-HDAg/S-HDAg and the replication capacity of HDV, which in both cases is lower than in HDV WT. The liver toxicity associated with the administration of this mutant is similar to the one of HDV-WT.