1. Introduction
Routine graft monitoring in renal transplantation relays on non-invasive biomarkers such as serum creatinine, proteinuria, and HLA antibodies. More than 30 years ago, to explore the feasibility of histological monitoring some centers started programs of protocol biopsies and observed than there were grafts with stable function depicting histological changes of rejection leading to the definition of subclinical rejection (SCR) [
1]. During the cyclosporine era, SCR was prevalent (>30%) and it was proven that its treatment better preserves renal function [
2]. However, a clinical trial addressing SCR treatment in patients on modern immunosuppression with tacrolimus/MMF yielded limited clinical benefit due to the low rate of SCR [
3]. For this reason, it has also explored whether minor histological changes are associated with graft outcome. In a study including a large set of 6-month protocol biopsies (n=957), it has been shown that interstitial inflammation (i-score >0) in otherwise normal protocol biopsies is associated with a significantly lower 15-y graft survival, comparable to SCR or interstitial fibrosis / tubular atrophy (IF/TA) with inflammation [
4].
Despite the evaluation of renal biopsies based on the Banff classification for renal transplant pathology has been refine since 1991[
5], some uncertainties persist, notably concerning the presence of borderline changes suspicious for TCMR and the incomplete phenotypes of ABMR. To further characterize underlying mechanisms leading to different histological phenotypes, analysis of the transcriptome has been incorporated [
6,
7]. It has been shown that molecular diagnostics allow detecting transcript sets strongly associated with TCMR and it has been proven useful in differentiating borderline infiltrates likely to lead to development of overt TCMR and/or graft fibrosis [
8]. Recently, we have shown that a rejection-associated gene expression score is present in 83% of protocol biopsies with SCR but only in 17% of protocol biopsies with borderline changes [
9]. Importantly, to distill information from RNA microarrays that evaluate thousands of genes, the Banff group has delineated gene-sets related to TCMR, ABMR, tissue-repair injury and other pathways implicated in graft dysfunction [
5,
10].
The presence of SCR in protocol biopsies has been associated in different cohorts with timing of biopsy, donor and recipient age, the intensity of immunosuppression and with donor-recipient HLA disparities [
1]. Our group and others have shown that a reduced exposure to tacrolimus and/or MMF is associated with a higher incidence of subclinical inflammation in protocol biopsies performed during the first year [
5,
10]. Furthermore, while certain studies have linked SCR to HLA ABDR allelic mismatch [
13], disparities at the molecular level might offer more informative insight [
14]. Notably, in liver transplant recipients reduced immunosuppression exposure and an increased number of HLA epitope mismatches between donor and recipient have been implicated in the molecular pathogenesis of subclinical liver allograft damage driven by an interferon gamma-orchestrated cellular immune response [
15].
In this current study, we employ microfluidic cards to scrutinize the transcriptome of a predefined set of genes previously described by the Banff group. Our aim is to determine whether gene expression is associated with the rejection phenotype, using normal protocol biopsies and biopsies for cause accomplishing either TCMR or ABMR Banff criteria. Subsequently, we quest these transcripts in a large set of early protocol biopsies to evaluate whether gene expression is associated with donor and recipient characteristics including the intensity of immunosuppression and donor-recipient HLA mismatch at allelic or molecular level.
2. Results
2.1. Patients and biopsies
Donor and recipient characteristics as well as transplant related variables from the 3 studied groups are shown in
Table 1. Timing of biopsy and laboratory data at the time of biopsy are detailed in
Table 2. In the rejection group II (n=12), there were a mix of cases with TCMR (n=5), active ABMR (n=5) and mixed rejection (n=2). In the group III containing only protocol biopsies (n=137), histological Banff categories were as follows: non-specific changes (n=40), subclinical TCMR (n=5), subclinical ABMR (n=3), borderline changes (n=16), interstitial fibrosis and tubular atrophy (IF/TA) without interstitial inflammation (n=59) and IF/TA with interstitial inflammation (IF/TA+i) (n=14). These results agreed with the prevalence of the different histological phenotypes in the whole cohort of early protocol biopsies (n=397) obtained at our center (40.1%, 3.1%, 2.7%, 11.5%, 34.3% and 8.3%, respectively).
To evaluate whether there was an association between subclinical inflammation and donor/recipient characteristics, transplant-related variables, or immunosuppression we compare patients with (i>0) and without interstitial infiltrates (i=0). Among the evaluated variables, subclinical inflammation was associated with prolonged cold ischemia time (p=0.040) and lower tacrolimus trough levels (TAC-C0) at the time of biopsy (p=0.002) (
Table 3 and
Table 4).
Donor and recipient demographics, HLA mismatches at allelic or molecular level, the presence of delayed graft function, timing of biopsy and renal function were not different between groups (
Table 3). At the time of biopsy, 3 out of 4 patients with DSA displayed subclinical ABMR. Multivariate logistic regression analysis showed that TAC-C0 (odds ratio [OR]: 0.76; 95% confidence interval [CI]: 0.63-0.92; p-value=0.004) was associated with i-score > 0 while cold ischemia time was in the verge of significance (OR: 1.06; 95% CI:.0.99-1.13; p-value=0.077).
2.2. Transcriptome analysis by microfluidics
The gene expression in the 3 groups of biopsies was firstly analyzed by principal component analysis (PCA) and it can be observed that biopsies from group I (normal) and biopsies from group 2 (TCMR/ABMR) cluster in different areas of the plot while the largest sample of protocol biopsies (group III) clusters in between (
Figure 1). The most relevant genes in PCA were ADAMDEC1, CCL5, CLEC4C, CXCL13 and CXCL9 for component 1; and COL1A1, NPHS1, NPHS2, SLC22A2 and SLC4A1 for component 2. As expected, the gene expression comparison between group I and group II (adjusted p value <0.01 and fold change >3) yielded as many as 111 genes differentially expressed (
Supplementary Table 1).
2.3. Transcriptome analysis and clinical variables
We evaluated whether there was an association between TAC-C0 at the time of biopsy and the 111 genes associated with rejection (
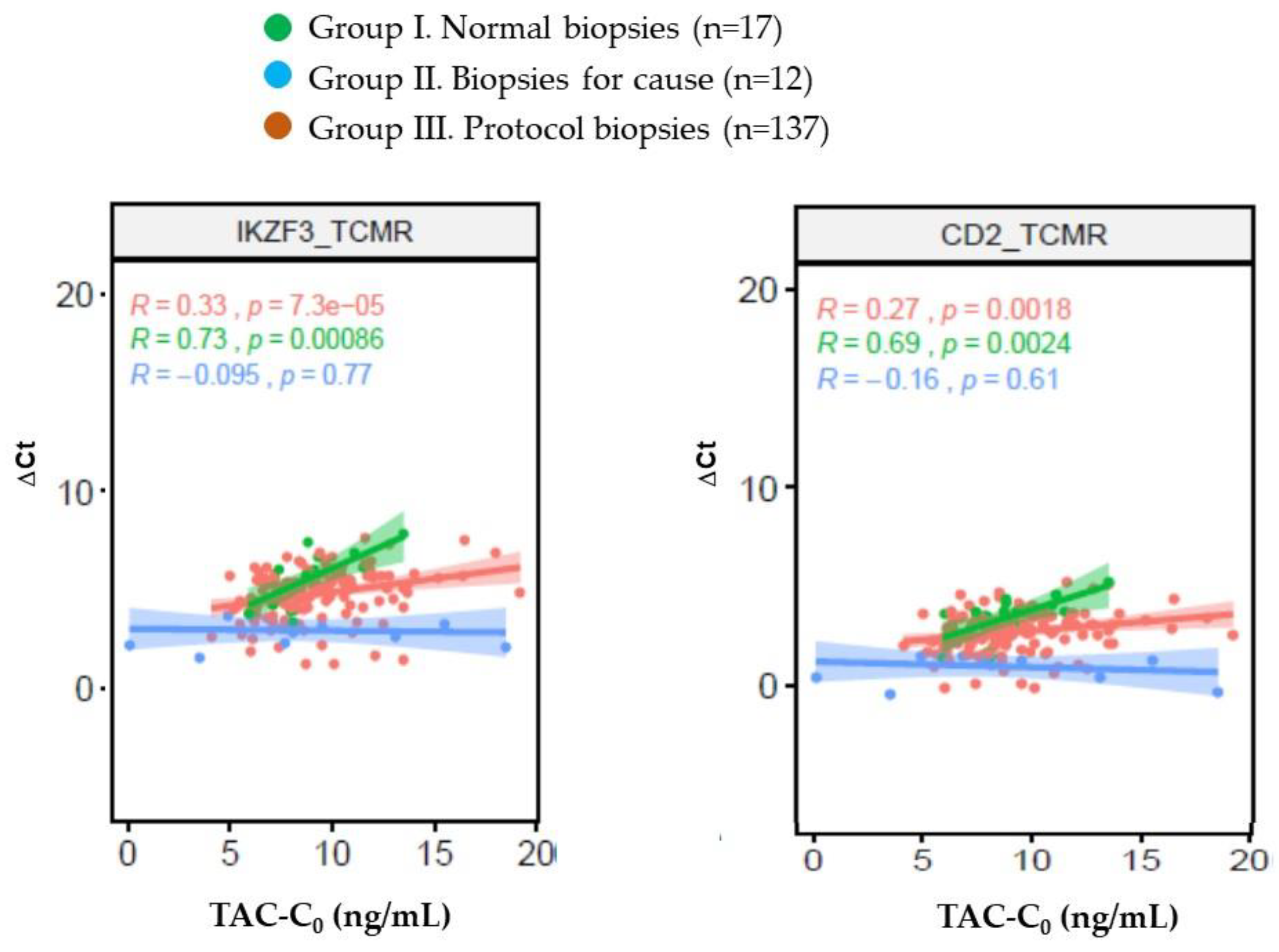
Supplementary Table 2). In group I (strictly normal protocol biopsies) there was a close correlation between TAC-C0 at the time of biopsy and expression of IKZF3 and CD2 genes (
Figure 2). Conversely, in group II (biopsies for cause with TCMR/ABMR) there was no correlation between TAC-C0 at the time of biopsy and the expression of any gene. Finally, 19 genes mainly related with TCMR (12 out of 19) correlated with TAC-C0 in the group III.
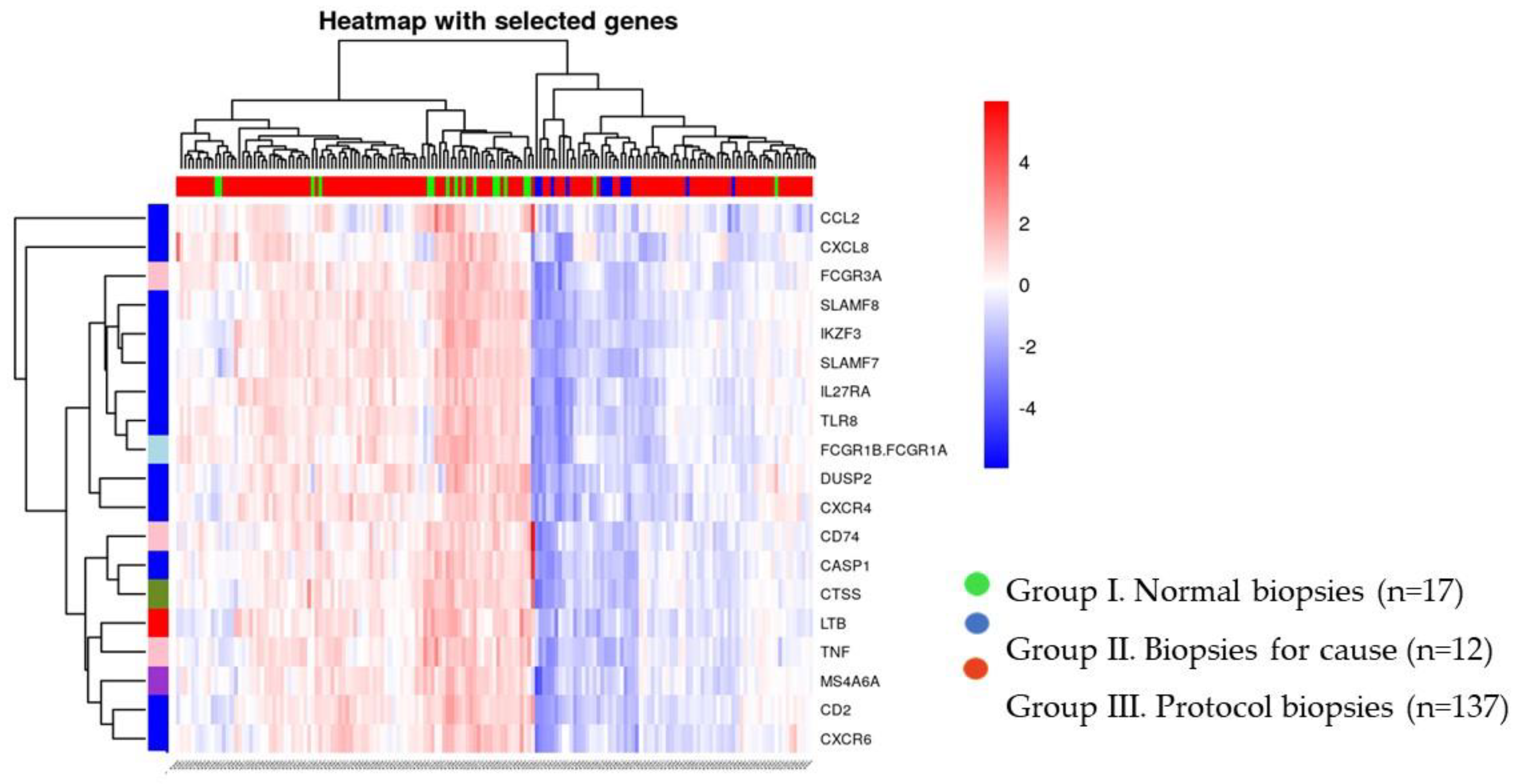
Unsupervised cluster analysis allowed defining 2 clusters of biopsies, one containing all but one normal protocol biopsies and the other containing all biopsies with rejection (
Figure 3).
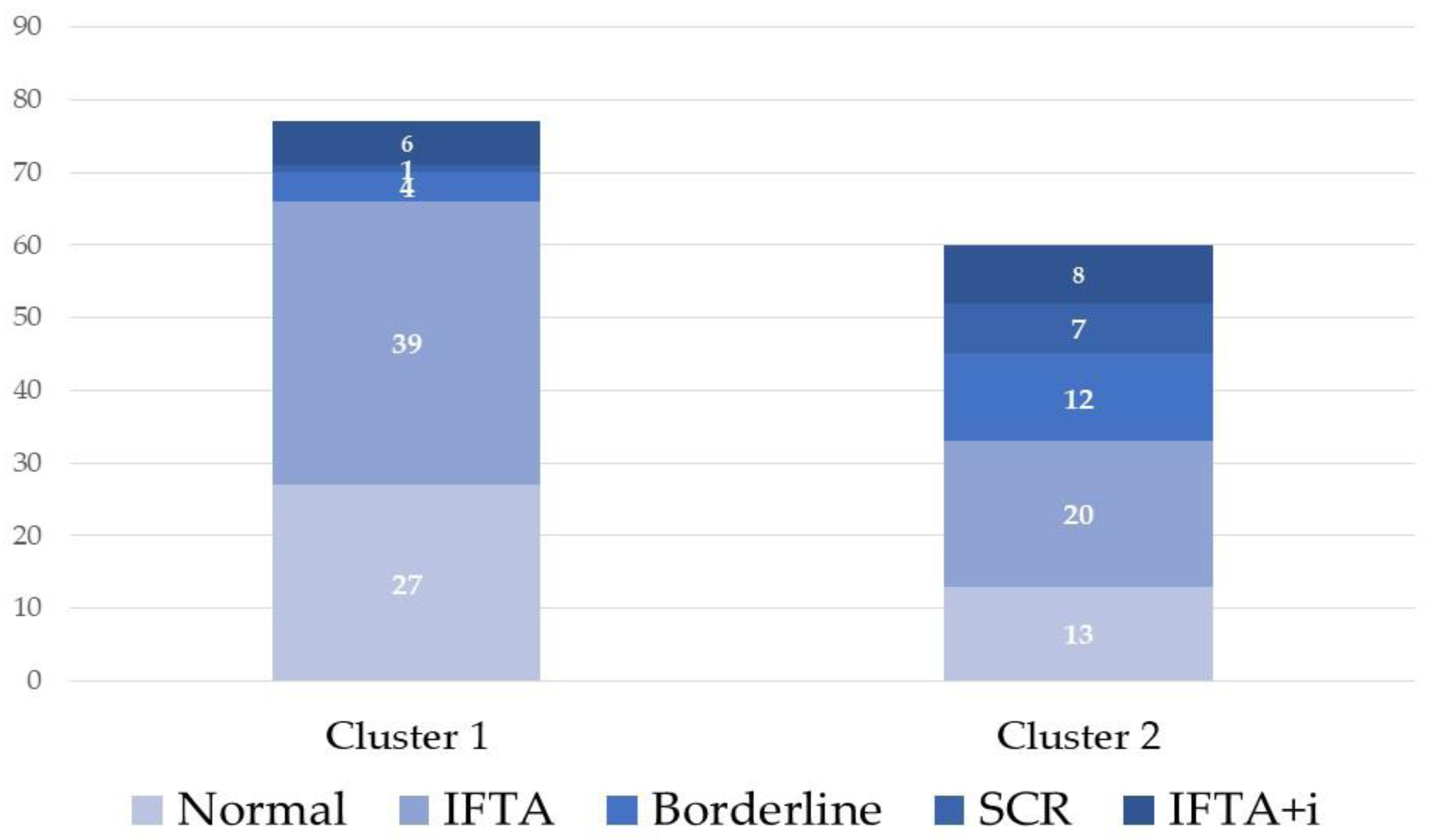
Protocol biopsies from group III distributed in a similar proportion in both clusters (77 in cluster 1 and 60 in cluster 2). We compared both clusters of biopsies from group III and observed that older donors and lower tacrolimus through levels at the time of biopsy grouped in cluster 2 (
Table 5).
Noticeably, subclinical rejection including borderline lesions (19 out of 24 cases) and biopsies with IF/TA+i (8 out of 14 cases) also tended to be grouped in cluster 2 (
Figure 4). Logistic regression analysis showed that only TAC-C0 at the time of biopsy (OR: 0.83, 95% CI: 0.72-0.96, p-value=0.0117) was associated with cluster 2.
2.4. Renal outcome and protocol biopsies
At a mean follow up of 70 ± 30 months the mean decline of renal function was -0.9±3.9 mL/min/1.73m2/year in the group of patients with a protocol biopsy (group III). While the mean annual decline of renal function did not differ significantly between patients with (i>0) or without inflammation (-1.1 ± 2.9 vs. -0.8 ± 4.3 mL/min/1.73m2/year,
Table 4), it was significantly higher in patients from cluster 2 than in patients from cluster 1 (-1.9 ± 4.1 vs. -0.2 ± 3.7 mL/min/1.73m2/year; p=0.0135;
Table 5). Moreover, this difference was independent from the presence of inflammation in the protocol biopsy (cluster 1 with i=0 (n=66) -0.20 ± 3.9; cluster 1 with i>0 (n=11) +0.19 ± 1.99; p=0.691; and cluster 2 with i=0 (n=33) -2.03 ± 4.8; cluster 2 with i>0 (n=27) -1.71 ± 3.23; p=0.763).
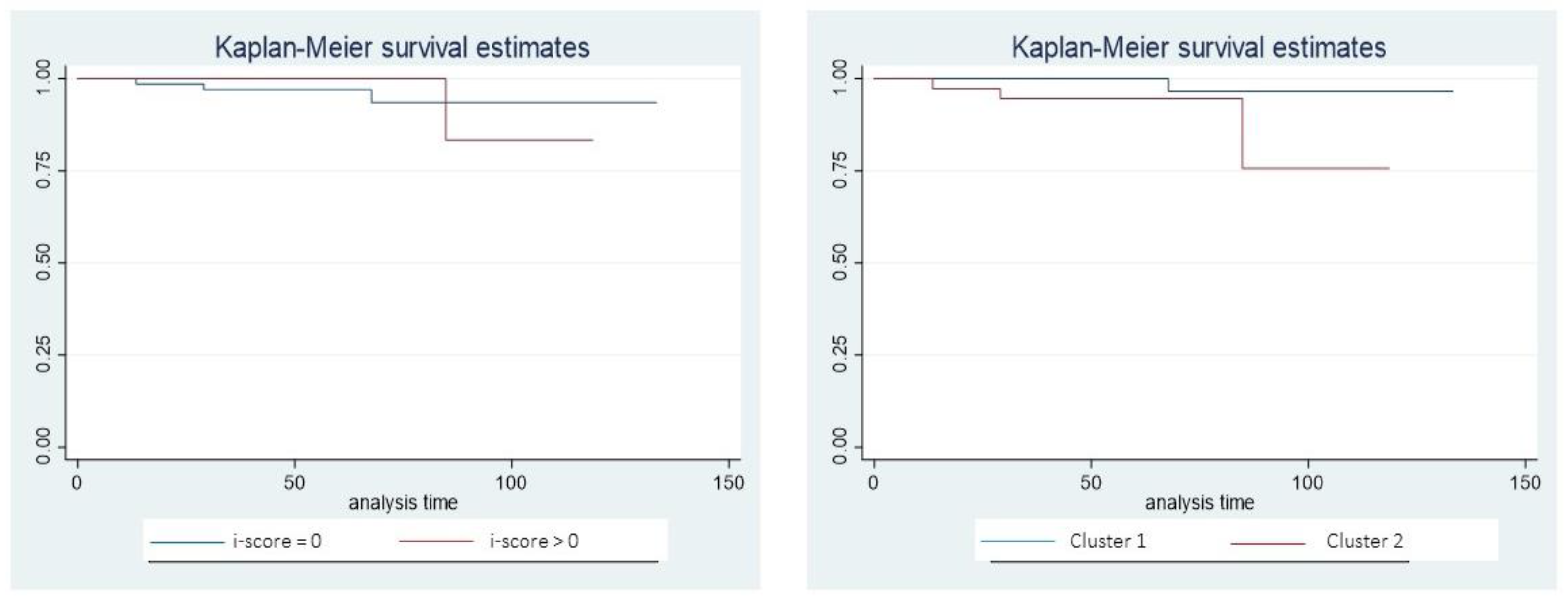
The total number of patients developing de novo DSA during follow up in our cohort was low (n=4; 2.9%). Kaplan-Meier analysis showed that patients with and without inflammation have a non-different rate of development of de novo DSA (p=0.876), while this rate tended to be higher in patients from cluster 2 (p=0.073) (
Figure 5).
3. Discussion
We conducted a prospective study on a set of 4-month protocol biopsies to evaluate whether validated rejection-associated transcripts are associated with tacrolimus exposure at the time of biopsy. The main findings of our study are that we confirm the discrimination capacity between normal and rejection biopsies of a large set of these genes and that the expression of 19 rejection-associated transcripts in early protocol biopsies are associated with tacrolimus exposure at the time of biopsy. Cluster analysis using this set of 19 genes identified a pool of patients with a higher proportion of inflammatory phenotypes including TCMR, borderline lesions and IFTA with inflammation. Interestingly, patients from this cluster received a lower exposure to tacrolimus and displayed a faster decline of renal function during follow up. The low rate of de novo DSA development in our cohort (2.9%) limits further analysis on its association with subclinical inflammation. Thus, our results suggest that a more intense immunosuppression during the early months after transplantation favors a better control of the inflammatory response without deleterious effects on renal function on the mid-term.
Tacrolimus is the mainstay of immunosuppressive regimens for kidney transplantation since it prevents T cell activation and proliferation. Although tacrolimus reduces the acute rejection rate and improves short-term outcomes after kidney transplantation, it is associated with both acute and chronic nephrotoxicity and triggers serious side effects. Despite monitoring of tacrolimus exposure relies in clinical practice on trough levels determination, there is no agreement on the target levels during the first year in renal transplant recipients. While the largest clinical trial supported minimization of tacrolimus exposure [
16], one randomized clinical trial has shown that in case of steroid discontinuation and MMF reduction, maintaining TAC-C0 >7 ng/mL after the fourth month reduces the risk of acute rejection and appearance of de novo DSAs without increasing renal toxicity [
11]. In a similar way, in low immunological risk renal transplants treated with TAC, reduced MMF and low dose steroids, TAC-C0 levels are associated with subclinical inflammation in patients monitored by protocol biopsies [
12]. Additionally, it has been described that the effect of tacrolimus trough levels was modulated by the recipient’s baseline alloimmune risk as defined by their class II HLA donor-recipient eplet mismatch [
14].
In the present study, we analyzed whether interstitial inflammation is associated with clinical characteristics of donor and recipients as well as with transplant related variables. In our cohort, the presence of interstitial inflammation was associated with lower TAC-C0 at the time of biopsy and with longer cold ischemia time, but it was not associated with mid-term renal function deterioration or the development of de novo DSA. Importantly, in our cohort like in others [
4,
17], few cases met criteria for subclinical borderline rejection (11.7%) or TCMR/ABMR (5.8%). Since the presence of interstitial inflammation (i>0) in otherwise normal biopsies has been associated with 15-year death-censored graft survival [
4] in a similar way than SCR, we choose this threshold for our analysis. Noteworthy, in other studies including patients treated with a steroid-free regimen the incidence of borderline rejection and TCMR were significantly more frequent (31% and 20.8%) [
18]. In this study, the authors did not find associations between TAC-C0 and subclinical inflammation, but it should be notice that at the time of the 3-month protocol biopsy TAC-C0 average was close to 10 ng/mL [
19]. In this study, SCR within the first post-transplant year is associated with a significantly greater hazard for subsequent clinical rejection and death-censored graft loss. On the contrary, other studies have shown that T-cell mediated inflammation detected in protocol biopsies mostly reflects the injury–repair response to implantation stresses and has little relationship to future events and outcomes [
20]. Acute kidney injury (AKI) after renal transplantation can also induced interstitial infiltration and tubulitis [
21] leading to a histological picture not distinguishable from TCMR. In this sense, in our cohort of protocol biopsies we observed an association between interstitial inflammation and longer cold ischemia time. Thus, the presence of interstitial inflammation is uncommon in our cohort of low immunological risk kidney transplants maintaining steroids (27.7%) and it is associated with tacrolimus exposure and cold ischemia time, suggesting that both immune and non-immune factors may contribute to subclinical inflammation in well-functioning grafts.
The disagreement between different studies on the prevalence of subclinical inflammation and its association with later clinical outcomes is partly explained by the inclusion of different populations and different maintenance immunosuppression regimens. However, there is general agreement that conventional biopsy assessment is limited due to poor interobserver reproducibility of individual lesions [
7,
22]. To overcome these limitations, it has been proposed to incorporate the molecular phenotyping. The application of microarrays to transplant biopsies has been an ongoing effort by many groups and the interpretation of molecular changes aided by the understanding of their biologic mechanisms led to group different transcripts [
6,
21,
23]. To summarize information derived from RNA microarrays which evaluates thousands of genes, in the last reports of the Banff meetings gene-sets containing few hundreds of genes related to TCMR, ABMR, tissue-repair injury and other pathways leading to graft dysfunction have been described [
4,
24]. In the present study, we evaluated by RT-PCR the panel of genes described in the Banff meeting 2017 [
4]. As expected, we confirm the discrimination capacity of a high number of these genes to differentiate normal protocol biopsies from biopsies for cause with rejection. From the derived gene set we were interested to evaluate its relationship with tacrolimus exposure at the time of biopsy. We found that 19 of these 111 genes, mainly related with TCMR, were mildly correlated with TAC-C0, suggesting that a higher tacrolimus exposure contributes to a better control of subclinical inflammation. Interestingly, in the small set of normal protocol biopsies we observed a close correlation between TAC-C0 and the expression of 2 out of these 19 genes (IKZF3 and CD2,
Figure 4) which are involved in T-cell regulation [
25], T cell activation and differentiation [
26]. Importantly, in the evaluated set of biopsies the expression of these 19 genes associated with TAC-C0 allowed to define two clusters of biopsies, one containing all but one normal protocol biopsies and the other containing all rejection biopsies. The large set of protocol biopsies was distributed in a similar proportion in both clusters. Patients with protocol biopsies grouped in cluster 2 received a lower exposure to tacrolimus, showed more frequently an inflammatory phenotype and displayed a faster decline of renal function on the mid-term. Thus, our results suggest that a more intense immunosuppression during the early months after transplantation favors a better control of the inflammatory response better preserving renal function on the mid-term.
Our effort to detect associations between gene expression, tacrolimus exposure and HLA compatibility at the allelic or molecular level did not show significant associations in the multivariate analysis. It should be remarked that HLA typing in this cohort was done according to clinical practice and thus, high resolution HLA typing was not done and the availability of HLA typing for all loci (especially DQ) was limited. However, in this cohort of well-immunosuppressed renal transplant recipients the number of patients developing de novo DSA was very low (2.9%) and despite patients from cluster 2 tended to develop more frequently de novo DSA, this association did not reach statistical significance. Additionally, the present study has other important limitations since associations between tacrolimus exposure and histological findings or gene transcripts were based on a single determination of TAC-C0 the day of biopsy and a more refined evaluation of tacrolimus pharmacokinetics (e.g., area under the time-concentration curve) or pharmacodynamics (e.g., calcineurin activity) was not done.
4. Materials and Methods
4.1. Patients.
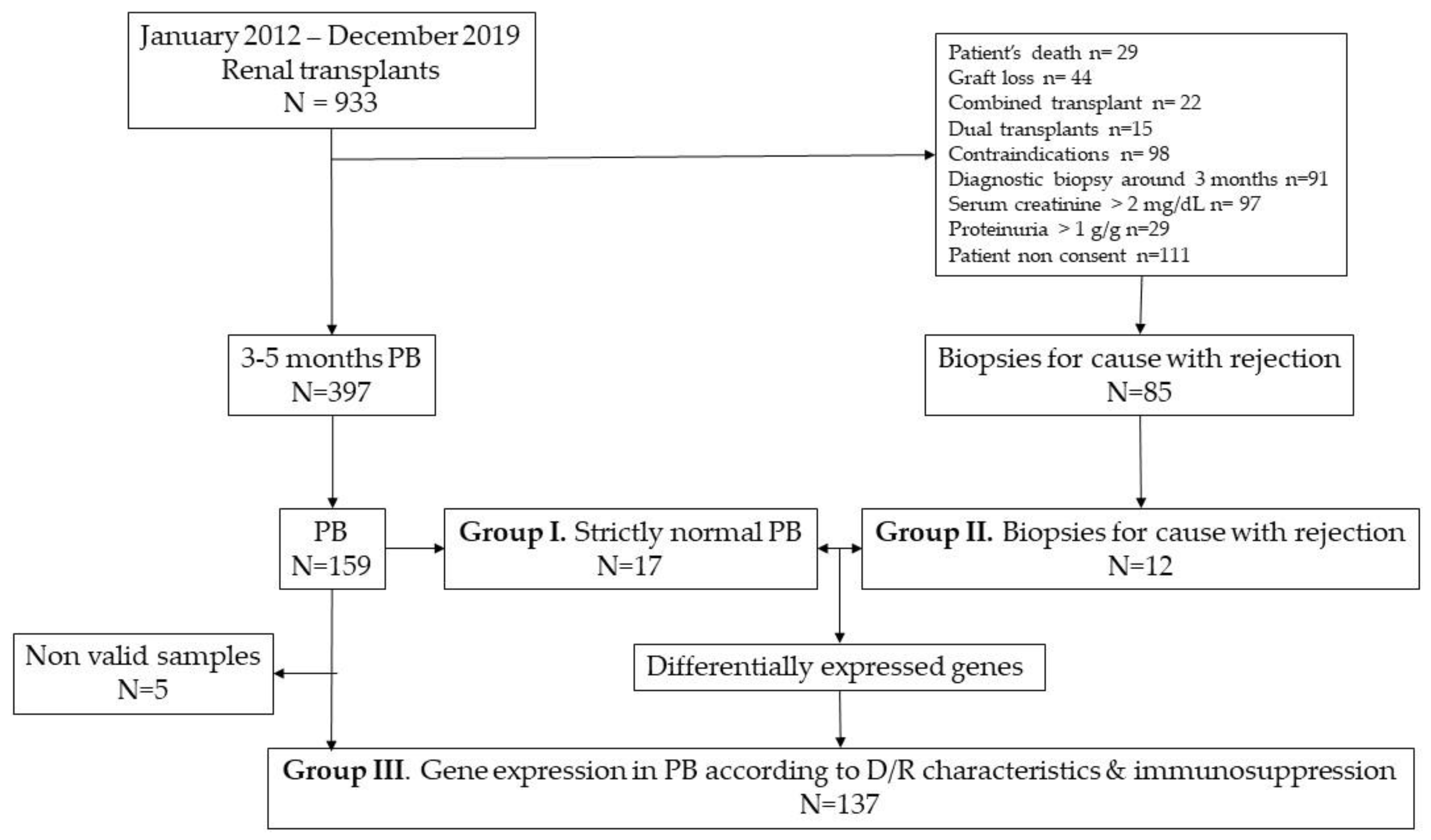
We considered renal transplants included in a prospective, observational study with an early (3-5 months) protocol biopsy performed between 2012 and 2019 as previously described [
12]. Two control groups of biopsies were selected from our biobank: strictly normal protocol biopsies (group I, n=17) and biopsies for cause with either TCMR or ABMR (group II, n=12) to generate a set of genes associated with rejection. Later, we selected a large sample of protocol biopsies (group III, n=142) to evaluate whether gene expression was associated with donor and recipient characteristics or to the intensity of immunosuppression. The flowchart of the study is shown in
Figure 6.
Demographic characteristics of donor and recipients as well as transplant-related variables were recorded. Patients were followed in the outpatient area and the decline of renal function was estimated from the linear regression of all available measurements and expressed as mL/min/1.73 m2/year to adjust for the different timing of follow up.
The present study has been approved by our Ethics Committee (Comité Etico de Investigación Clínica del Hospital Universitari Vall d’Hebron PR(AG)369/2014, approval date 1 December 2014) and all participants signed a written informed consent. The study was conducted in accordance with the Declaration of Helsinki and adhered to the Principles on the Declaration of Istanbul on Organ Trafficking and Transplant Tourism.
4.2. HLA typing and HLA antibodies
Recipient’s and donor’s HLA typing was performed by DNA-based low-resolution typing with sequence-specific primers (SSP). For class I (A and B loci) and for class II (DR loci) results were available for all donor/recipient pairs. HLA C typing was available in 116 cases from donors and 47 cases for recipients. HLA DQ loci typing was available in 40 donors and 26 recipients.
The HLA Matchmaker program (Rene Duquesnoy, 2016, University of Pittsburgh Medical Center, Pittsburgh, PA HLA-ABC Eplet Matching Version 3.1 and DRDQDP Eplet Matching Program V3.1 from
http://www.epitopes.net/downloads.html) was used to calculate eplet scores. Donor and recipient typing were converted to high resolution using a local frequency table typed by sequence-based typing. Total numbers of incompatible eplets and antibody-verified eplets were calculated.
PIRCHE-II scores were calculated using version 3.3 from
https://www.pirche.org. The NetMHCIIpan 3.0 algorithm predicted nonameric-binding cores of donor mismatched HLA-derived peptides that could bind to recipient HLA-DRB1. For cases with only low-resolution HLA typing, PIRCHE-II generates a potential high-resolution HLA typing and PIRCHE-II was calculated for each of these potential typing for both donor and recipient. These values were weighted by haplotypes frequencies in the general population as validated in a previous study [
27].
Anti HLA antibodies at the day of transplant, biopsy and during follow up were determined using single-antigen class-I and class-II flow beads-assay kit (LIFECODES, division of Immucor, Stanford, CA). Beads with a normalized MFI > 500 were considered positive if (MFI/MFI lowest bead) > 5.
4.3. Immunosuppression
Induction and maintenance immunosuppression with tacrolimus, MMF and steroids were done as previously described (12). Target TAC-C0 levels during the initial 3 months were 8-12 ng/mL and 6-10 ng/mL thereafter. Exposure to tacrolimus was evaluated using concentration dose ratio (C/D), coefficient of variation of TAC-C0 until the day of biopsy and TAC-C0 at the time of biopsy as previously described [
28].
4.4. Biopsies
Ultrasound-guided renal biopsies were performed with a 16G automated needle, and 3 cores of tissue were obtained: one was processed for optical microscopy; one was embedded in OCT for immunofluorescence and the other one was stored in RNA later. Histological lesions were evaluated according to the last Banff criteria [
5] and the definition of borderline changes were foci of tubulitis (t1-t3) with mild interstitial inflammation (i1) or mild tubulitis (t1) with moderate-severe interstitial inflammation (i2-3). C4d was stained with indirect immunofluorescence with a monoclonal antibody (Quidel, San Diego, CA, USA) and deposition in peritubular capillaries was graded according to the Banff criteria. The third core was stored with Ambion® RNAlater® Tissue Collection at −80 ◦C.
4.5. Analysis Using Fluidigm Microfluidics Dynamic Arrays
Total RNA extraction, assessment of RNA quality and cDNA synthesis were done as previously described (9). The aim of the study was the quantitative analysis of 318 genes (308 target genes and 10 housekeeping genes: ECD, EIF1, FUBP3, GGNBP2, GNB1, RPN1, RPN2, SERBP1, UBC, UBE2D3) in the biopsies. The 308 target genes were selected from a list of identified, non-repeated prime gene list reported in the Banff 2017 meeting (10). For this purpose, we used the Biomark HD Nanofluidic quantitative PCR (qPCR) system (Fluidigm) combined with GE 96.96 Dynamic Arrays IFCs. For sequence detection, predesigned Primetime qPCR primer assays or custom primer assays were used for amplification and detection using the EvaGreen fluorochrome. The assays have been divided into four IDT assay plates and all housekeeping genes have been included in all plates. Samples were treated with Exonuclease I (Exo I) (Thermo Scientific EN0582) to remove unincorporated primers. QIAGEN® Multiplex PCR Kit Cat N.206143 was used for the specific target amplification. According to the manufacturer instructions 13 genes were eliminated due to potential amplification of genomic DNA and 12 genes were also not considered due to lack of expression in more than half of the samples. The analysis of the expression of the cDNA was done with Biomark HD Nanofluidic qPCR system (Fluidigm) combined with 96.96 Dynamic Arrays IFCs by employing the Master Mix Sso FastTM Eva Green® Supermix with Low ROX (Bio-Rad Laboratories). The Ct (Cycle Threshold) data and the Quality Call of the amplification curve was done by the Fluidigm Real Time PCR Analysis Software version 4.1.3. Samples with a Ct value higher than 27 (n=5) were eliminated since they are not reliable according to Fluidigm, owner of the technology (
https://www.fluidigm.com ). All procedures were done at the genomics and proteomics service of the Universidad del País Vasco Science Park (Centro de Biotecnología María Goyri).
4.6. Statistics
Results are expressed as raw numbers for categorical variables, as the mean ± standard deviation for continuous normally distributed variables and median (interquartile range) for non-normally distributed variables. To compare unpaired data Fisher exact test, Mann-Whitney U test, Student’s t-test, Kruskal-Wallis and analysis of variance were applied according to distribution of variables. Logistic regression analysis was employed for multivariate analysis. Kaplan-Meier analysis was employed for survival analysis with log-rank test for comparisons between groups. All p-values were two-tailed and a p-value < 0.05 was considered significant.
4.7. Bioinformatic analysis
Bioinformatic analysis was performed at the Statistics and Bioinformatics Unit (UEB) of the Vall d’Hebron Institute of Research (VHIR, Barcelona, Spain). The analyses were carried out with the statistical program "R"(R version 3.6.3 (2020-02-29), Copyright (C) 2021 The R Foundation for Statistical Computing,
https://www.R-project.org/). A comprehensive quality control process was applied to assess the suitability of all samples for inclusion in the study. The calculation of relative quantification (RQ = 2−∆∆Ct) was done according to Livak’s method. A principal component analysis (PCA) was performed to describe how the samples are grouped according to the Ct values obtained. Because the variability between genes used as normalizers was low, all were used as housekeeping genes. The geometric mean of the Ct values of the housekeeping genes was obtained as described by Vandesompele et al [
29]. In the process of normalization, the Ct values of each gene were subtracted from the geometric mean value of the two housekeeping genes selected to obtain the ∆Ct values. Later it was used to make comparisons. Spearman’s correlation between the expression of each of the genes and tacrolimus levels was done to select significant genes following criteria of fold change (FC) and statistical significance (FC > 3 and p-value < 0.01).
5. Conclusions
In summary, we evaluate a cohort of patients with an early protocol biopsy and observed that a lower tacrolimus through level et the time of biopsy was associated with interstitial inflammation and a higher expression of rejection-associated transcripts in stable grafts. Cluster analysis allowed to detect a group of patients who had lower tacrolimus through levels at the time of biopsy, showed an inflammatory phenotype and displayed a faster decline of renal function on the mid-term. Thus, our results suggest that a more intense immunosuppression during the early months after transplantation favors a better control of the inflammatory response without deleterious effects on renal function on the mid-term.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
The writing of the paper was done by BC, IBT, OB and FM. The research design was done by DS and FM. The performance of the research was done by BC, IBT, AG, TJ, MM, JMZ, JS, MP, DS, OB and FM. Data analysis was done by BC, IBT and FM. All authors reviewed and approved the last version of the submitted manuscript.
Funding
The authors received grants from Red de Investigación Renal (REDinREN RD16/0009/0030), Fondo de Investigación Sanitaria del Instituto de Salud Carlos III (PI 18/01704, PI 18/01832), the Spanish Society of Transplantation and a Diaverum Spain restricted grant. Betty Chamoun has been supported by a VHIR (Vall Hebron Institute Research) grant. Jose M. Zuñiga has been supported by a Catalan Society of Transplantation grant.
Conflicts of Interest
D.S. and F.M. received grant support and personal fees from Astellas Pharma Spain and Chiesi Spain.
Abbreviations:
|
ABMR: antibody-mediated rejection. |
|
DSA: donor specific HLA antibodies. |
|
FC: fold change. |
|
FDR: false discovery rate. |
|
eGFR: estimated glomerular filtration rate by CKD-EPI formula. |
|
IF/TA: Interstitial fibrosis and tubular atrophy. |
|
IF/TA+i: Interstitial fibrosis and tubular atrophy with interstitial infiltrates. |
|
SCR: subclinical rejection. |
|
TAC-C0: tacrolimus trough levels. |
|
TCMR: T-cell mediated rejection. |
References
- Serón D, Moreso F. Protocol biopsies in renal transplantation: prognostic value of structural monitoring. Kidney Int. 2007 Sep;72(6):690–7. [CrossRef]
- Rush D, Nickerson P, Gough J, McKenna R, Grimm P, Cheang M, et al. Beneficial effects of treatment of early subclinical rejection: a randomized study. J Am Soc Nephrol. 1998 Nov;9(11):2129–34. [CrossRef]
- Rush D, Arlen D, Boucher A, Busque S, Cockfield SM, Girardin C, et al. Lack of benefit of early protocol biopsies in renal transplant patients receiving TAC and MMF: a randomized study. Am J Transplant. 2007 Nov;7(11):2538–45. [CrossRef]
- Ortiz F, Gelpi R, Helanterä I, Melilli E, Honkanen E, Bestard O, et al. Decreased Kidney Graft Survival in Low Immunological Risk Patients Showing Inflammation in Normal Protocol Biopsies. PLoS One. 2016;11(8):e0159717. [CrossRef]
- Loupy A, Haas M, Roufosse C, Naesens M, Adam B, Afrouzian M, et al. The Banff 2019 Kidney Meeting Report (I): Updates on and clarification of criteria for T cell- and antibody-mediated rejection. Am J Transplant. 2020 Sep;20(9):2318–31. [CrossRef]
- Halloran PF, Venner JM, Madill-Thomsen KS, Einecke G, Parkes MD, Hidalgo LG, et al. Review: The transcripts associated with organ allograft rejection. American Journal of Transplantation. 2018;18(4):785–95. [CrossRef]
- Moreso F, Sellarès J, Soler MJ, Serón D. Transcriptome Analysis in Renal Transplant Biopsies Not Fulfilling Rejection Criteria. Int J Mol Sci. 2020 Mar 24;21(6):2245. [CrossRef]
- de Freitas DG, Sellarés J, Mengel M, Chang J, Hidalgo LG, Famulski KS, et al. The nature of biopsies with ‘borderline rejection’ and prospects for eliminating this category. Am J Transplant. 2012 Jan;12(1):191–201. [CrossRef]
- Chamoun B, Caraben A, Torres IB, Sellares J, Jiménez R, Toapanta N, et al. A Rejection Gene Expression Score in Indication and Surveillance Biopsies Is Associated with Graft Outcome. Int J Mol Sci. 2020 Nov 3;21(21):E8237. [CrossRef]
- Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell–mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. American Journal of Transplantation. 2018;18(2):293–307. [CrossRef]
- Gatault P, Kamar N, Büchler M, Colosio C, Bertrand D, Durrbach A, et al. Reduction of Extended-Release Tacrolimus Dose in Low-Immunological-Risk Kidney Transplant Recipients Increases Risk of Rejection and Appearance of Donor-Specific Antibodies: A Randomized Study. Am J Transplant. 2017 May;17(5):1370–9. [CrossRef]
- Torres IB, Reisaeter AV, Moreso F, Âsberg A, Vidal M, Garcia-Carro C, et al. Tacrolimus and mycophenolate regimen and subclinical tubulo-interstitial inflammation in low immunological risk renal transplants. Transpl Int. 2017 Nov;30(11):1119–31. [CrossRef]
- Hernández D, Vázquez T, Alonso-Titos J, León M, Caballero A, Cobo MA, et al. Impact of HLA Mismatching on Early Subclinical Inflammation in Low-Immunological-Risk Kidney Transplant Recipients. J Clin Med. 2021 Apr 29;10(9):1934. [CrossRef]
- Wiebe C, Rush DN, Nevins TE, Birk PE, Blydt-Hansen T, Gibson IW, et al. Class II Eplet Mismatch Modulates Tacrolimus Trough Levels Required to Prevent Donor-Specific Antibody Development. J Am Soc Nephrol. 2017 Nov;28(11):3353–62. [CrossRef]
- Vionnet J, Miquel R, Abraldes JG, Wall J, Kodela E, Lozano JJ, et al. Non-invasive alloimmune risk stratification of long-term liver transplant recipients. J Hepatol. 2021 Dec;75(6):1409–19. [CrossRef]
- Ekberg H, Tedesco-Silva H, Demirbas A, Vítko Š, Nashan B, Gürkan A, et al. Reduced Exposure to Calcineurin Inhibitors in Renal Transplantation. New England Journal of Medicine. 2007 Dec 20;357(25):2562–75. [CrossRef]
- Torres IB, Reisaeter AV, Moreso F, Âsberg A, Vidal M, Garcia-Carro C, et al. Tacrolimus and mycophenolate regimen and subclinical tubulo-interstitial inflammation in low immunological risk renal transplants. Transpl Int. 2017 Nov;30(11):1119–31. [CrossRef]
- Mehta RB, Melgarejo I, Viswanathan V, Zhang X, Pittappilly M, Randhawa P, et al. Long-term immunological outcomes of early subclinical inflammation on surveillance kidney allograft biopsies. Kidney International. 2022 Dec;102(6):1371–81. [CrossRef]
- Mehta RB, Tandukar S, Jorgensen D, Randhawa P, Sood P, Puttarajappa C, et al. Early subclinical tubulitis and interstitial inflammation in kidney transplantation have adverse clinical implications. Kidney International. 2020 Aug;98(2):436–47. [CrossRef]
- Mengel M, Chang J, Kayser D, Gwinner W, Schwarz A, Einecke G, et al. The molecular phenotype of 6-week protocol biopsies from human renal allografts: reflections of prior injury but not future course. Am J Transplant. 2011 Apr;11(4):708–18. [CrossRef]
- Olsen S, Hansen ES, Jepsen FL. The prevalence of focal tubulo-interstitial lesions in various renal diseases. Acta Pathol Microbiol Scand A. 1981 Mar;89(2):137–45. [CrossRef]
- Furness PN, Taub N, Convergence of European Renal Transplant Pathology Assessment Procedures (CERTPAP) Project. International variation in the interpretation of renal transplant biopsies: report of the CERTPAP Project. Kidney Int. 2001 Nov;60(5):1998–2012. [CrossRef]
- Halloran PF, Reeve JP, Pereira AB, Hidalgo LG, Famulski KS. Antibody-mediated rejection, T cell–mediated rejection, and the injury-repair response: new insights from the Genome Canada studies of kidney transplant biopsies. Kidney International. 2014;85(2):258–64. [CrossRef]
- Mengel M, Loupy A, Haas M, Roufosse C, Naesens M, Akalin E, et al. Banff 2019 Meeting Report: Molecular diagnostics in solid organ transplantation-Consensus for the Banff Human Organ Transplant (B-HOT) gene panel and open source multicenter validation. Am J Transplant. 2020 Sep;20(9):2305–17. [CrossRef]
- Dooley BJ, Verma A, Ding R, Yang H, Muthukumar T, Lubetzky M, et al. Urinary Cell Transcriptome Profiling and Identification of ITM2A, SLAMF6, and IKZF3 as Biomarkers of Acute Rejection in Human Kidney Allografts. Transplant Direct. 2020 Jul 22;6(8):e588. [CrossRef]
- Ford ML. T Cell Cosignaling Molecules in Transplantation. Immunity. 2016 May 17;44(5):1020–33. [CrossRef]
- Geneugelijk K, Wissing J, Koppenaal D, Niemann M, Spierings E. Computational Approaches to Facilitate Epitope-Based HLA Matching in Solid Organ Transplantation. J Immunol Res. 2017;2017:9130879. [CrossRef]
- Chamoun B, Torres IB, Gabaldón A, Sellarés J, Perelló M, Castellá E, et al. Progression of Interstitial Fibrosis and Tubular Atrophy in Low Immunological Risk Renal Transplants Monitored by Sequential Surveillance Biopsies: The Influence of TAC Exposure and Metabolism. J Clin Med. 2021 Jan 4;10(1):141. [CrossRef]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002 Jun 18;3(7):RESEARCH0034. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).