Submitted:
24 January 2024
Posted:
26 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Data compilation and processing
2.2. Explainable Tree-based Models
2.2.1. Model evaluation
2.2.2. Assessment statistics
3. Results
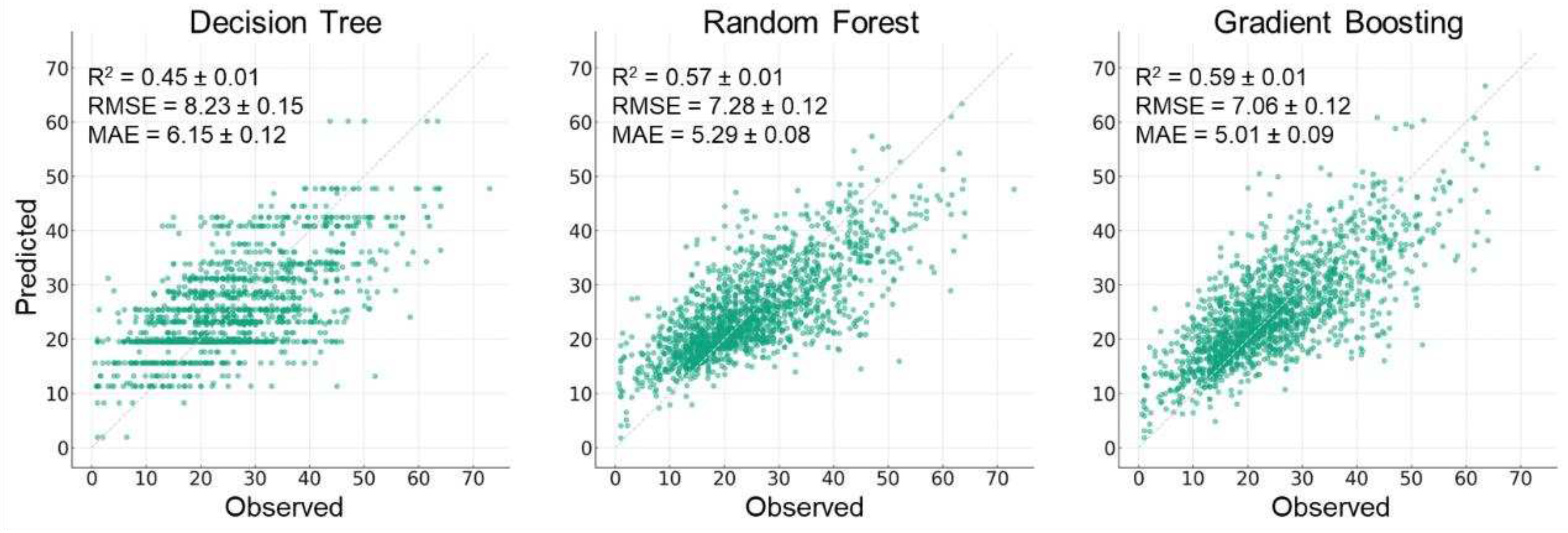
3.1. Comparison of Selected Tree-based Models
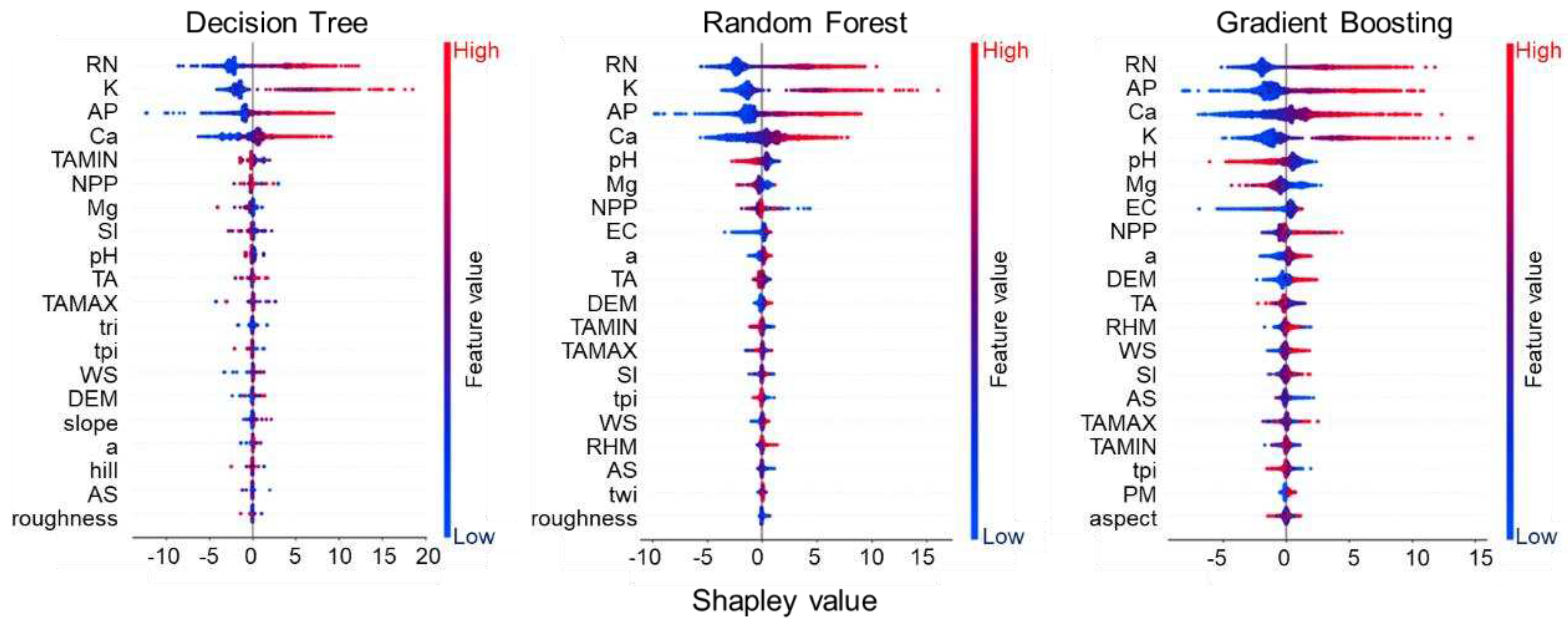
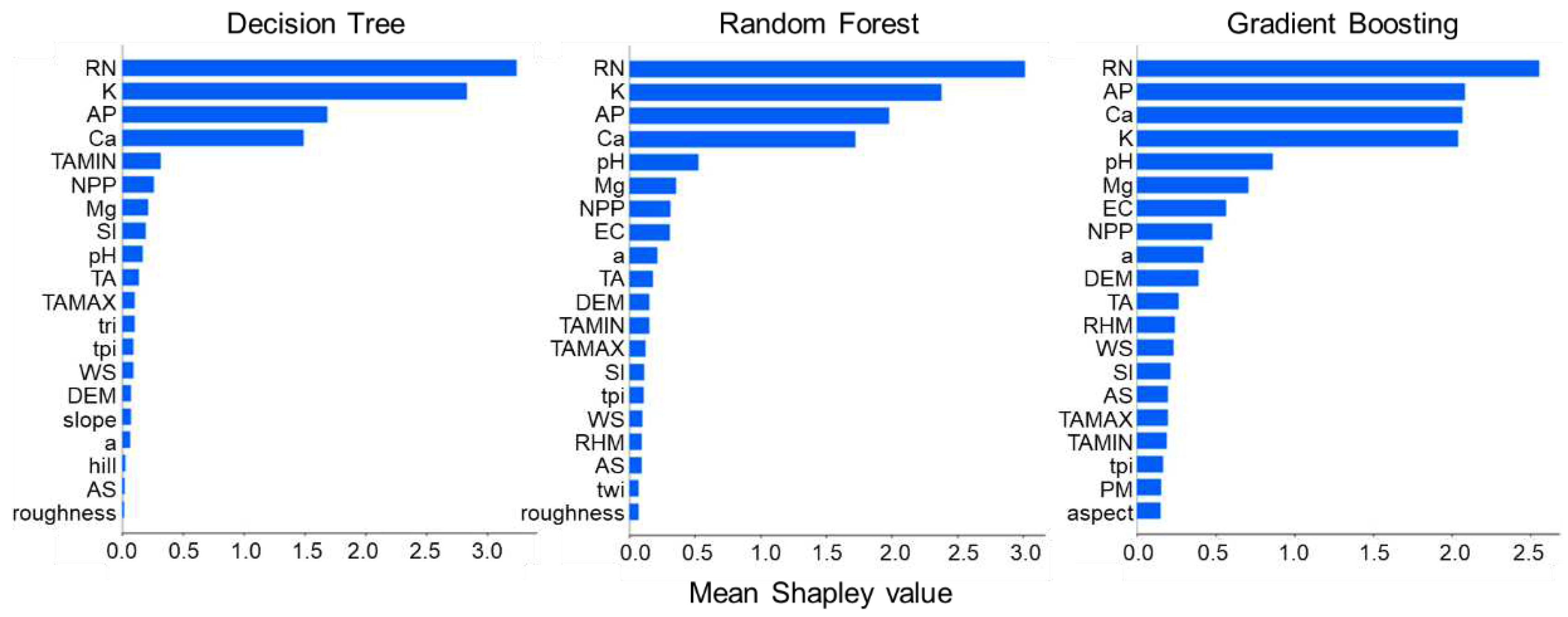
3.2. Feature Overall Importance Analysis
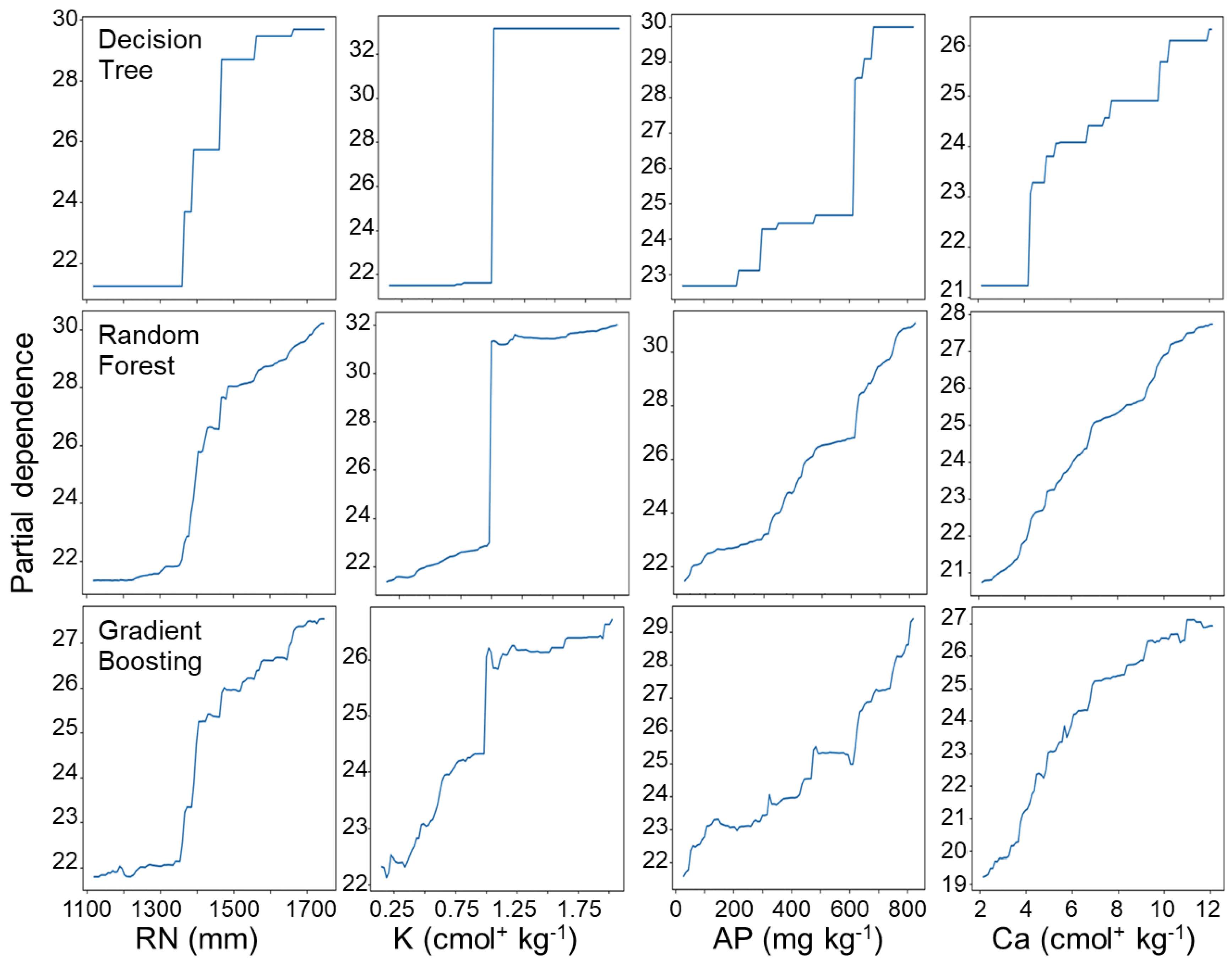
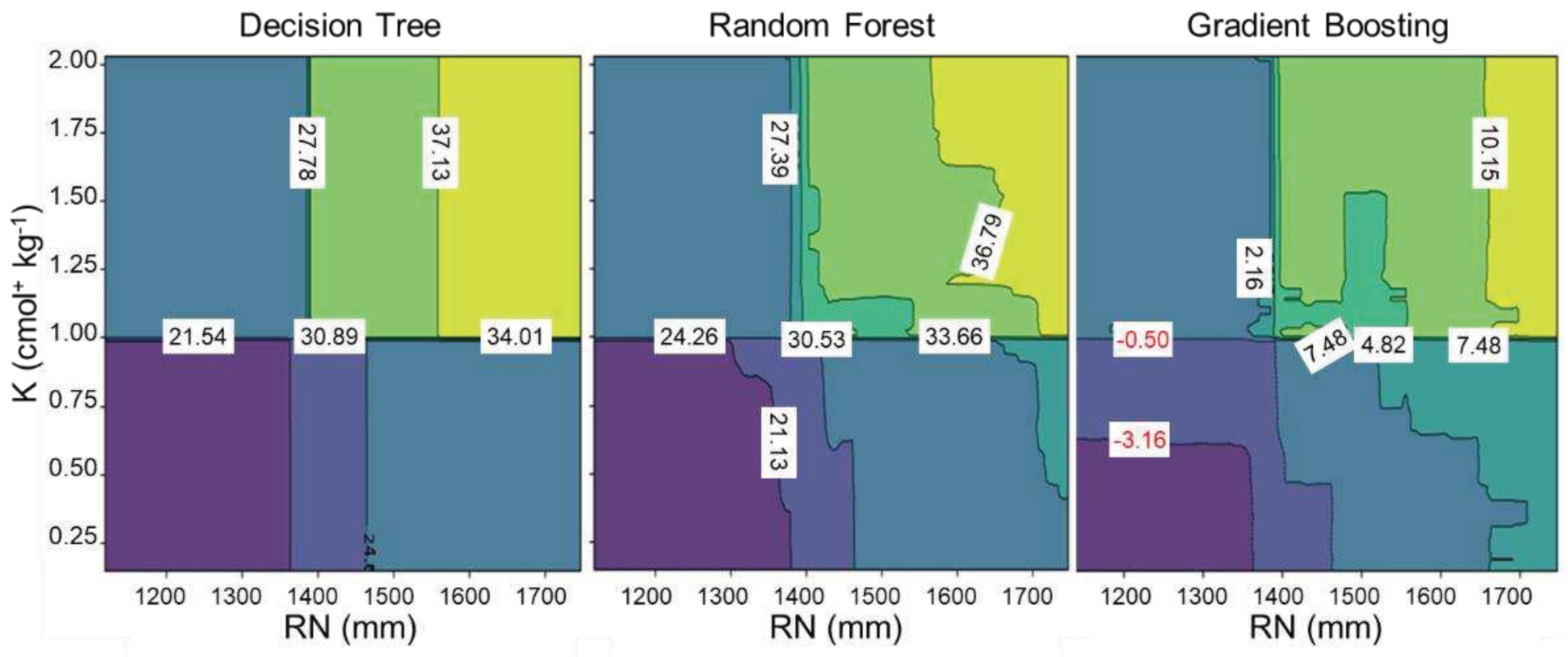
3.2. Feature Partial Dependence Analysis
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bodria, F.; Giannotti, F.; Guidotti, R.; Naretto, F.; Pedreschi, D.; Rinzivillo, S. Benchmarking and survey of explanation methods for black box models. Data Mining and Knowledge Discovery 2021, 37, 1719–1778. [Google Scholar] [CrossRef]
- Ryo, M. Explainable artificial intelligence and interpretable machine learning for agricultural data analysis. Artificial Intelligence in Agriculture 2022, 6, 257–265. [Google Scholar] [CrossRef]
- Miller, T. Explanation in Artificial Intelligence: Insights from the Social Sciences. Artif. Intell. 2017, 267, 1–38. [Google Scholar] [CrossRef]
- Belle, V.; Papantonis, I. Principles and practice of explainable machine learning. Frontiers in Big Data 2021, 4. [Google Scholar] [CrossRef] [PubMed]
- Saeed, W.; Omlin, C. Explainable AI (XAI): A systematic meta-survey of current challenges and future opportunities. Knowledge-Based Systems 2023, 263, 110273. [Google Scholar] [CrossRef]
- Wang, M.; Fu, W.J.; He, X.N.; Hao, S.J.; Wu, X.D. A survey on large-scale machine learning. Ieee T Knowl Data En 2022, 34, 2574–2594. [Google Scholar] [CrossRef]
- Visser, O.; Sippel, S.R.; Thiemann, L. Imprecision farming? Examining the (in)accuracy and risks of digital agriculture. Journal of Rural Studies 2021, 86, 623–632. [Google Scholar] [CrossRef]
- Dundon, S.J. Agricultural ethics and multifunctionality are unavoidable. Plant Physiol 2003, 133, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nature Machine Intelligence 2019, 1, 206–215. [Google Scholar] [CrossRef]
- Finger, R. Digital innovations for sustainable and resilient agricultural systems. Eur Rev Agric Econ 2023, 50, 1277–1309. [Google Scholar] [CrossRef]
- Hoang, N.T.; Taherzadeh, O.; Ohashi, H.; Yonekura, Y.; Nishijima, S.; Yamabe, M.; Matsui, T.; Matsuda, H.; Moran, D.; Kanemoto, K. Mapping potential conflicts between global agriculture and terrestrial conservation. Proceedings of the National Academy of Sciences 2023, 120, e2208376120. [Google Scholar] [CrossRef] [PubMed]
- Chouldechova, A. Fair prediction with disparate impact: A study of bias in recidivism prediction instruments. Big data 2016, 5 2, 153–163. [Google Scholar] [CrossRef]
- Schuett, J. Risk management in the Artificial Intelligence Act. European Journal of Risk Regulation 2023, 1–19. [Google Scholar] [CrossRef]
- Thomson Reuters. LAWnB IP Exclusive Report: 2023 Domestic and International AI Regulatory and Policy Trends; 10/24/2023 2023; pp. 5-8.
- Breiman, L. Statistical modeling: the two cultures. Statistical Science 2001, 16, 199–231. [Google Scholar] [CrossRef]
- Guidotti, R.; Monreale, A.; Turini, F.; Pedreschi, D.; Giannotti, F. A survey of methods for explaining black box models. ACM Computing Surveys 2018, 51, 1–42. [Google Scholar] [CrossRef]
- Adadi, A.; Berrada, M. Peeking inside the black-box: a survey on explainable artificial intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Barredo Arrieta, A.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Information Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Theissler, A.; Spinnato, F.; Schlegel, U.; Guidotti, R. Explainable AI for time series classification: A review, taxonomy and research directions. IEEE Access 2022, 10, 100700–100724. [Google Scholar] [CrossRef]
- Yuan, H.; Yu, H.; Gui, S.; Ji, S. Explainability in graph neural networks: A taxonomic survey. IEEE Transactions on Pattern Analysis and Machine Intelligence 2020, 45, 5782–5799. [Google Scholar] [CrossRef]
- Miller, T. Explanation in artificial intelligence: Insights from the social sciences. Artificial Intelligence 2019, 267, 1–38. [Google Scholar] [CrossRef]
- Pichler, M.; Hartig, F. Machine learning and deep learning—A review for ecologists. Methods in Ecology and Evolution 2023, 14, 994–1016. [Google Scholar] [CrossRef]
- Rudin, C.; Chen, C.; Chen, Z.; Huang, H.; Semenova, L.; Zhong, C. Interpretable machine learning: Fundamental principles and 10 grand challenges. Statistics Surveys 2022, 16, 1–85. [Google Scholar] [CrossRef]
- Antle, J.M.; Basso, B.; Conant, R.T.; Godfray, H.C.J.; Jones, J.W.; Herrero, M.; Howitt, R.E.; Keating, B.A.; Munoz-Carpena, R.; Rosenzweig, C.; et al. Towards a new generation of agricultural system data, models and knowledge products: Design and improvement. Agric Syst 2017, 155, 255–268. [Google Scholar] [CrossRef]
- Smith, P.; Davies, C.A.; Ogle, S.; Zanchi, G.; Bellarby, J.; Bird, N.; Boddey, R.M.; McNamara, N.P.; Powlson, D.; Cowie, A.; et al. Towards an integrated global framework to assess the impacts of land use and management change on soil carbon: current capability and future vision. Global Change Biol 2012, 18, 2089–2101. [Google Scholar] [CrossRef]
- Hu, T.; Zhang, X.; Bohrer, G.; Liu, Y.; Zhou, Y.; Martin, J.; Li, Y.; Zhao, K. Crop yield prediction via explainable AI and interpretable machine learning: Dangers of black box models for evaluating climate change impacts on crop yield. Agricultural and Forest Meteorology 2023, 336, 109458. [Google Scholar] [CrossRef]
- Paustian, K.; Lehmann, J.; Ogle, S.; Reay, D.; Robertson, G.P.; Smith, P. Climate-smart soils. Nature 2016, 532, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Lal, R. Challenges and opportunities in soil organic matter research. Eur J Soil Sci 2009, 60, 158–169. [Google Scholar] [CrossRef]
- Conway, G.R. The properties of agroecosystems. Agr. Syst. 1987, 24, 95–117. [Google Scholar] [CrossRef]
- Spencer, J.E.; Stewart, N.R. The nature of agricultural systems. Annals of the Association of American Geographers 1973, 63, 529–544. [Google Scholar] [CrossRef]
- NAS. Chemical Data for Soil Test. National Institute of Agricultural Sciences, Rural Development Administration 2023, www.data.go.kr/data/15073569/openapi.do (accessed on 01/30/2023).
- RDA. Precision soil maps. Rural Development Administration 2023. https://soil.rda.go.kr (accessed on 01/19/2023).
- NSDI. Degital Elevation Model (registered on: 08/11/2020). National Spatial Data Infrastructure 2020. https://data.nsdi.go.kr/dataset/20001 (accessed on.
- KMA. Climate Change Scenarios. 2023. https://www.climate.go.kr/home/CCS/contents_2021/35_download.php (accessed on 12-20-2023).
- NEO. Net Primary Productivity (1 year - TERRA/MODIS). NASA Earth Observations 2023. https://neo.gsfc.nasa.gov (accessed on 02/20/2023).
- Horn, B.K.P. Hill shading and the reflectance map. Proceedings of the IEEE 1981, 69, 14–47. [Google Scholar] [CrossRef]
- Quinn, P.F.; Beven, K.J.; Lamb, R. The in(a/tan/β) index: How to calculate it and how to use it within the topmodel framework. Hydrological Processes 1995, 9, 161–182. [Google Scholar] [CrossRef]
- EGIS. Land Cover Maps. Environmental Geographic Information Service 2023. https://egis.me.go.kr/intro/land.do (accessed on 01/13/2023).
- Statistics Korea. Arable land in Korea. Korean Statistical Information Service 2021. [Google Scholar]
- R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2023.
- Hastie, T.; Tibshirani, R.; Friedman, J. Unsupervised Learning. In The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Hastie, T., Tibshirani, R., Friedman, J., Eds.; Springer New York: New York, NY, 2009; pp. 485–585. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the KDD '16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016; pp. 785–794.
- Zhang, W.; Wu, C.; Zhong, H.; Li, Y.; Wang, L. Prediction of undrained shear strength using extreme gradient boosting and random forest based on Bayesian optimization. Geoscience Frontiers 2021, 12, 469–477. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017; pp. 4768–4777. [Google Scholar]
- Liu, J.; Li, C.; Ouyang, P.; Liu, J.; Wu, C. Interpreting the prediction results of the tree-based gradient boosting models for financial distress prediction with an explainable machine learning approach. Journal of Forecasting 2023, 42, 1112–1137. [Google Scholar] [CrossRef]
- Rodríguez-Pérez, R.; Bajorath, J. Interpretation of machine learning models using shapley values: application to compound potency and multi-target activity predictions. Journal of Computer-Aided Molecular Design 2020, 34, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. The Annals of Statistics 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Czerwinska, U. Interpretability of Machine Learning Models. In Applied Data Science in Tourism: Interdisciplinary Approaches, Methodologies, and Applications, Egger, R., Ed.; Springer International Publishing: Cham, 2022; pp. 275–303. [Google Scholar]
- Pittelkow, C.M.; Liang, X.Q.; Linquist, B.A.; van Groenigen, K.J.; Lee, J.; Lundy, M.E.; van Gestel, N.; Six, J.; Venterea, R.T.; van Kessel, C. Productivity limits and potentials of the principles of conservation agriculture. Nature 2015, 517, 365–U482. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.; Snapp, S. What is sustainable intensification? Views from experts. Land Use Policy 2015, 46, 1–10. [Google Scholar] [CrossRef]
- Adamtey, N.; Musyoka, M.W.; Zundel, C.; Cobo, J.G.; Karanja, E.; Fiaboe, K.K.M.; Muriuki, A.; Mucheru-Muna, M.; Vanlauwe, B.; Berset, E.; et al. Productivity, profitability and partial nutrient balance in maize-based conventional and organic farming systems in Kenya. Agriculture, Ecosystems & Environment 2016, 235, 61–79. [Google Scholar] [CrossRef]
- Petersen, E.H.; Hoyle, F.C. Estimating the economic value of soil organic carbon for grains cropping systems in Western Australia. Soil Research 2016, 54, 383–396. [Google Scholar] [CrossRef]
- Mikhailova, E.A.; Groshans, G.R.; Post, C.J.; Schlautman, M.A.; Post, G.C. Valuation of soil organic carbon stocks in the contiguous United States based on the avoided social cost of carbon emissions. Resources 2019, 8, 153. [Google Scholar] [CrossRef]
- Dube, B.; White, A.; Ricketts, T.; Darby, H. Valuation of soil health ecosystem services; The University of Vermont: 2022.
- Hacisalihoglu, S.; Toksoy, D.; Kalca, A. Economic valuation of soil erosion in a semi and area in Turkey. African Journal of Agricultural Research 2010, 5, 1–6. [Google Scholar] [CrossRef]
- Kane, D.A.; Bradford, M.A.; Fuller, E.; Oldfield, E.E.; Wood, S.A. Soil organic matter protects US maize yields and lowers crop insurance payouts under drought. Environmental Research Letters 2021, 16, 044018. [Google Scholar] [CrossRef]
- Sparling, G.P.; Wheeler, D.; Vesely, E.T.; Schipper, L.A. What is soil organic matter worth? Journal of Environmental Quality 2006, 35, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Henriksen, C.B.; Porter, J. Valuation of ecosystem services in organic cereal crop production systems with different management practices in relation to organic matter input. Ecosystem Services 2016, 22, 117–127. [Google Scholar] [CrossRef]
- Reyes, J.J.; Elias, E. Spatio-temporal variation of crop loss in the United States from 2001 to 2016. Environmental Research Letters 2019, 14, 074017. [Google Scholar] [CrossRef]
| Category | Variable (abbreviation) |
Unit | Resolution | Source |
|---|---|---|---|---|
| Soil | Organic matter (OM) | g kg-1 | Field | [31] |
| Available phosphate (AP) | mg kg-1 | Field | ||
| Available silicate (AS) | mg kg-1 | Field | ||
| Exchangeable magnesium (Mg) | cmol+ kg-1 | Field | ||
| Exchangeable potassium (K) | cmol+ kg-1 | Field | ||
| Exchangeable calcium (Ca) | cmol+ kg-1 | Field | ||
| pH (1:5 H2O) | Field | |||
| Electric conductivity (EC) | dS m-1 | Field | ||
| Soil map | Topsoil texture (TT) | class | 250 m | [32] |
| Drainage (DC) | class | 250 m | ||
| Soil order (OR) | group | 250 m | ||
| Soil structure (SS) | class | 250 m | ||
| Parent material (PM) | Type | 250 m | ||
| Erosion (EG) | grade | 250 m | ||
| Terrain | Elevation (DEM) | m | 90 m | [33] |
| Slope 1 | radians | 90 m | ||
| Aspect 1 | radians | 90 m | ||
| Flow direction (flowdir) 1 | m | 90 m | ||
| Roughness 1 | m | 90 m | ||
| Hill shade (hill) 2 | 90 m | |||
| Topographic position index (TPI) 1 | 90 m | |||
| Terrain ruggedness index (TRI) 1 | 90 m | |||
| Upslope contributing area (a) 1 | 90 m | |||
| Topographic wetness index (TWI) 1 | 90 m | |||
| Climate | Mean annual temperature (TA) | °C | 1 km | [34] |
| Maximum annual temperature (TAMAX) | °C | 1 km | ||
| Minimum annual temperature (TAMIN) | °C | 1 km | ||
| Mean annual precipitation (RN) | mm | 1 km | ||
| Solar irradiation (SI) | MJ m-2 | 1 km | ||
| Relative humidity (RHM) | % | 1 km | ||
| Wind speed (WS) | m s-1 | 1 km | ||
| Vegetation | Net primary productivity (NPP) | g C m-2 y-1 | 11 km | [35] |
| 1 Estimated based on DEM data. | ||||
| 2 Computed from slope and aspect values, assuming sun elevation and direction (azimuth) angles of 45° and 0°, respectively. | ||||
| Model parameter | Parameter grid | Decision Tree |
Random Forest |
Gradient Boosting |
|---|---|---|---|---|
| Maximum depth of a tree | [6,8,10,12] | 6 | 12 | 8 |
| Minimum samples per leaf | [8,12,18] | 12 | 8 | 18 |
| Minimum number of samples | [8,16,20] | 8 | 8 | 8 |
| Number of trees | [10,100] | - | 100 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





