Submitted:
25 January 2024
Posted:
26 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. State of the Art
2.1. Mathematical/Statistical Tools
2.1.1. Root Mean Squared Error (RMSE)
2.1.2. Correlation Coefficient
2.1.3. Mean Absolute Error
2.2. Clinical Tools
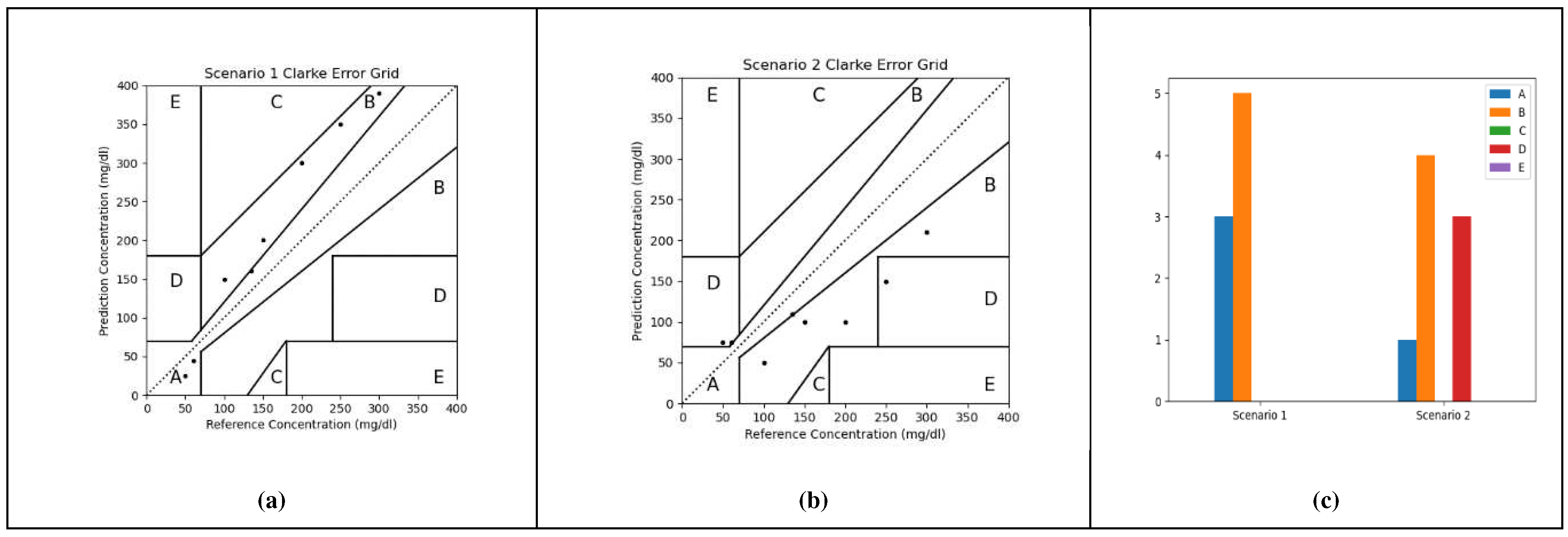
2.2.1. Clarke’s Error Grid Analysis (C-EGA)
- Region A: points within 20% deviation from of the reference value.
- Region B: points outside 20% deviation but would not lead to inappropriate treatment.
- Region C: points leading to unnecessary treatment.
- Region D: points indicating a potentially dangerous failure to detect hypoglycemia or hyperglycemia.
- Region E: points that would confuse the treatment of hypoglycemia for hyperglycemia and vice versa.
2.2.2. Parkes’ Error Grid Analysis
3. Methods
3.1. Derivation of the Metric Equation
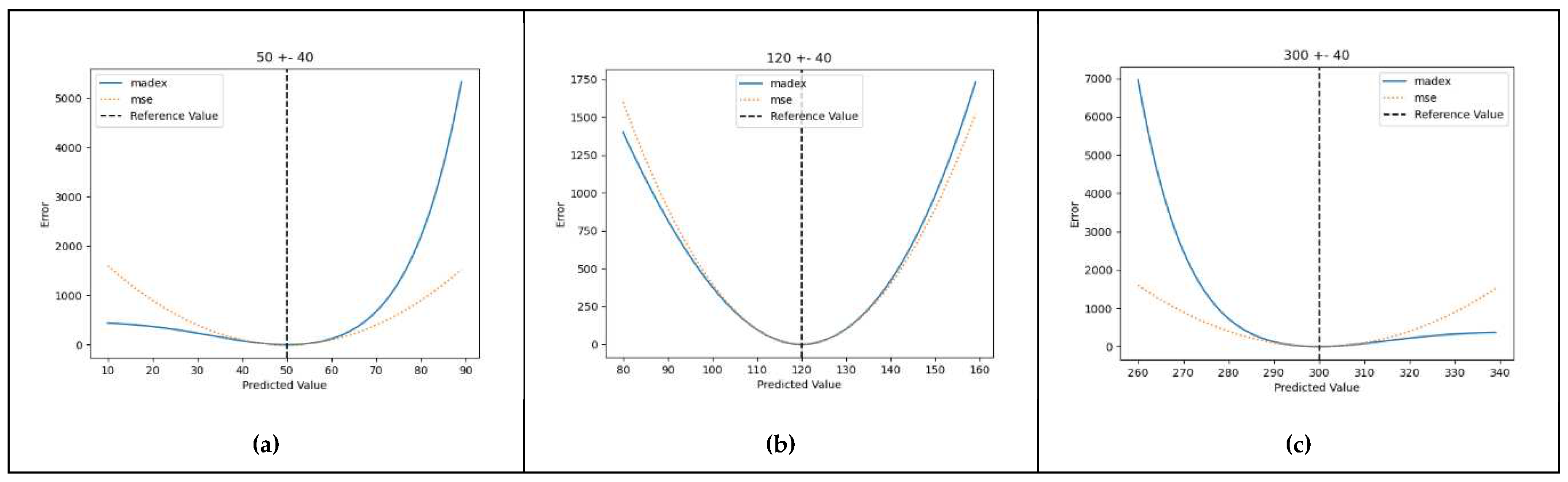
- The penalizing power applied to the residuals should vary, getting bigger in proportion to the distance of the reference value from a certain point on the reference axis (the arithmetical center of the normal range). In this manner, the calculated error gets more prominent as the reference value moves away from the normal levels (euglycaemia).
- The penalizing factor should be specific and behave differently based on the direction of the error. More specifically it should penalize more heavily values that provide a distorted evaluation of the reference glycaemic status, that is high predicted values when the true value is in the hypoglycaemic range, as well as low values when the true value is in the hyperglycaemic range.
3.2. Proof-Of-Concept Marginal Test
3.3. Preliminary Application on Real-World Data
4. Results
5. Discussion
- It is a continuous function assuring smooth value progression and avoiding stepping phenomena
- It is not dependent on clinical interpretations (a center of euglycemia is easier to agree upon)
- It is computationally more efficient compared to algorithms with conditional (if_then_else, switch case) arguments
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- D. J. Magliano, E. J. Boyko, and I. D. A. 10th edition scientific committee, “IDF DIABETES ATLAS,” pp. 54–55, 2021.
- P. Saeedi et al., “Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition,” Diabetes Res. Clin. Pract., vol. 157, p. 107843, Nov. 2019, doi: 10.1016/J.DIABRES.2019.107843.
- X. Lin et al., “Global, regional, and national burden and trend of diabetes in 195 countries and territories: an analysis from 1990 to 2025,” Sci. Rep. 2020 101, vol. 10, no. 1, pp. 1–11, Sep. 2020. [CrossRef]
- M. Greenwood and F. Wood, “The Relation between the Cancer and Diabetes Death-rates,” J. Hyg. (Lond.), vol. 14, no. 1, pp. 83–118, Apr. 1914. [CrossRef]
- Cignarelli et al., “Diabetes and cancer: Pathophysiological fundamentals of a ‘dangerous affair,’” Diabetes Res. Clin. Pract., vol. 143, pp. 378–388, Sep. 2018. [CrossRef]
- Zhu and S. Qu, “The Relationship Between Diabetes Mellitus and Cancers and Its Underlying Mechanisms,” Front. Endocrinol., vol. 13, 2022, Accessed: Dec. 04, 2023. [Online]. Available: https://www.frontiersin.org/articles/10.3389/fendo.2022.800995.
- S. J. Livingstone et al., “Estimated life expectancy in a Scottish cohort with type 1 diabetes, 2008-2010,” JAMA, vol. 313, no. 1, pp. 37–44, Jan. 2015. [CrossRef]
- K. K. Tsilidis, J. C. Kasimis, D. S. Lopez, E. E. Ntzani, and J. P. A. Ioannidis, “Type 2 diabetes and cancer: umbrella review of meta-analyses of observational studies,” BMJ, vol. 350, p. g7607, Jan. 2015. [CrossRef]
- Y. Chen et al., “Association between type 2 diabetes and risk of cancer mortality: a pooled analysis of over 771,000 individuals in the Asia Cohort Consortium,” Diabetologia, vol. 60, no. 6, pp. 1022–1032, Jun. 2017. [CrossRef]
- S. Ellahham, “Artificial Intelligence: The Future for Diabetes Care,” Am. J. Med., vol. 133, no. 8, pp. 895–900, Aug. 2020. [CrossRef]
- Dankwa-Mullan, M. Rivo, M. Sepulveda, Y. Park, J. Snowdon, and K. Rhee, “Transforming Diabetes Care Through Artificial Intelligence: The Future Is Here,” Popul. Health Manag., vol. 22, no. 3, pp. 229–242, Jun. 2019. [CrossRef]
- Nomura, M. Noguchi, M. Kometani, K. Furukawa, and T. Yoneda, “Artificial Intelligence in Current Diabetes Management and Prediction,” Curr. Diab. Rep., vol. 21, no. 12, Dec. 2021. [CrossRef]
- Z. Woldaregay et al., “Data-driven modeling and prediction of blood glucose dynamics: Machine learning applications in type 1 diabetes,” Artif. Intell. Med., vol. 98, pp. 109–134, Jul. 2019. [CrossRef]
- W. L. Clarke, D. Cox, L. A. Gonder-Frederick, W. Carter, and S. L. Pohl, “Evaluating clinical accuracy of systems for self-monitoring of blood glucose,” Diabetes Care, vol. 10, no. 5, pp. 622–628, 1987. [CrossRef]
- L. Parkes, S. L. Slatin, S. Pardo, and B. H. Ginsberg, “A new consensus error grid to evaluate the clinical significance of inaccuracies in the measurement of blood glucose,” Diabetes Care, vol. 23, no. 8, pp. 1143–1148, 2000. [CrossRef]
- “Sigmoid Function Definition | DeepAI.” Accessed: Apr. 18, 2022. [Online]. Available: https://deepai.org/machine-learning-glossary-and-terms/sigmoid-function.
- S. Pitoglou, PhD thesis 2023. Accessed: Dec. 23, 2023. [Online]. Available: http://artemis.cslab.ece.ntua.gr:8080/jspui/handle/123456789/18960.
- Marling and R. Bunescu, “The OhioT1DM dataset for blood glucose level prediction,” in CEUR Workshop Proceedings, 2018.
- “PyCaret 3.0.” Accessed: Dec. 01, 2023. [Online]. Available: https://pycaret.gitbook.io/docs/.
- “Linear regression,” Wikipedia. Oct. 23, 2023. Accessed: Oct. 24, 2023. [Online]. Available: https://en.wikipedia.org/w/index.php?title=Linear_regression&oldid=1181530004.
- “Linear regression | Definition, Formula, & Facts | Britannica.” Accessed: Oct. 24, 2023. [Online]. Available: https://www.britannica.com/topic/linear-regression.
- Zach, “Introduction to Lasso Regression,” Statology. Accessed: Oct. 24, 2023. [Online]. Available: https://www.statology.org/lasso-regression/.
- G. L. Team, “A Complete understanding of LASSO Regression,” Great Learning Blog: Free Resources what Matters to shape your Career! Accessed: Oct. 24, 2023. [Online]. Available: https://www.mygreatlearning.com/blog/understanding-of-lasso-regression/.
- “Ridge regression,” Wikipedia. Aug. 29, 2023. Accessed: Oct. 24, 2023. [Online]. Available: https://en.wikipedia.org/w/index.php?title=Ridge_regression&oldid=1172832572.
- “Tikhonov Regularization - an overview | ScienceDirect Topics.” Accessed: Oct. 24, 2023. [Online]. Available: https://www.sciencedirect.com/topics/computer-science/tikhonov-regularization.
- Friedman, T. Hastie, and R. Tibshirani, “Regularization Paths for Generalized Linear Models via Coordinate Descent,” J. Stat. Softw., vol. 33, no. 1, pp. 1–22, 2010.
- S.-J. Kim, K. Koh, M. Lustig, S. Boyd, and D. Gorinevsky, “An Interior-Point Method for Large-Scale \ell_1-Regularized Least Squares,” IEEE J. Sel. Top. Signal Process., vol. 1, no. 4, pp. 606–617, Dec. 2007. [CrossRef]
- Efron, T. Hastie, I. Johnstone, and R. Tibshirani, “Least angle regression,” Ann. Stat., vol. 32, no. 2, Apr. 2004. [CrossRef]
- S. G. Mallat and Z. Zhang, “Matching pursuits with time-frequency dictionaries,” IEEE Trans. Signal Process., vol. 41, no. 12, pp. 3397–3415, Dec. 1993. [CrossRef]
- R. Rubinstein, M. Zibulevsky, and M. Elad, “Efficient Implementation of the K-SVD Algorithm using Batch Orthogonal Matching Pursuit,” 2008. Accessed: Oct. 24, 2023. [Online]. Available: https://www.semanticscholar.org/paper/Efficient-Implementation-of-the-K-SVD-Algorithm-Rubinstein-Zibulevsky/7f98bd398a6b422e140b9cd83f0d64444f8dbca5.
- H. Zhu, W. Chen, and Y. Wu, “Efficient Implementations for Orthogonal Matching Pursuit,” Electronics, vol. 9, no. 9, Art. no. 9, Sep. 2020. [CrossRef]
- Pattern Recognition and Machine Learning. Accessed: Oct. 24, 2023. [Online]. Available: https://link.springer.com/book/9780387310732.
- J. C. MacKay, “Bayesian Interpolation,” Neural Comput., vol. 4, no. 3, pp. 415–447, 1992. [CrossRef]
- E. Tipping, “Sparse bayesian learning and the relevance vector machine,” J. Mach. Learn. Res., vol. 1, pp. 211–244, Sep. 2001. [CrossRef]
- Wipf and S. Nagarajan, “A new view of automatic relevance determination,” in Proceedings of the 20th International Conference on Neural Information Processing Systems, in NIPS’07. Red Hook, NY, USA: Curran Associates Inc., Dec. 2007, pp. 1625–1632.
- K. Crammer, O. Dekel, J. Keshet, S. Shalev-Shwartz, and Y. Singer, “Online Passive-Aggressive Algorithms,” J. Mach. Learn. Res., vol. 7, pp. 551–585, Dec. 2006.
- S. Choi, T. Kim, and W. Yu, “Performance Evaluation of RANSAC Family,” in Procedings of the British Machine Vision Conference 2009, London: British Machine Vision Association, 2009, p. 81.1-81.12. doi: 10.5244/C.23.81.
- A. Fischler and R. C. Bolles, “Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography,” Commun. ACM, vol. 24, no. 6, pp. 381–395, Jun. 1981. [CrossRef]
- “Random sample consensus,” Wikipedia. Oct. 11, 2023. Accessed: Oct. 24, 2023. [Online]. Available: https://en.wikipedia.org/w/index.php?title=Random_sample_consensus&oldid=1179652738.
- “Theil–Sen estimator,” Wikipedia. Sep. 20, 2023. Accessed: Oct. 24, 2023. [Online]. Available: https://en.wikipedia.org/w/index.php?title=Theil%E2%80%93Sen_estimator&oldid=1176301934.
- X. Wang, X. Dang, H. Peng, and H. Zhang, “THE THEIL-SEN ESTIMATORS IN A MULTIPLE LINEAR REGRESSION MODEL,” 2009. Accessed: Oct. 24, 2023. [Online]. Available: https://www.semanticscholar.org/paper/THE-THEIL-SEN-ESTIMATORS-IN-A-MULTIPLE-LINEAR-MODEL-Wang-Dang/63167c5dbb9bae6f0a269237a9b6a28fa7e1ac20.
- T. Kärkkäinen and S. Äyrämö, “ON COMPUTATION OF SPATIAL MEDIAN FOR ROBUST DATA MINING,” Accessed: Oct. 24, 2023. [Online]. Available: https://www.semanticscholar.org/paper/ON-COMPUTATION-OF-SPATIAL-MEDIAN-FOR-ROBUST-DATA-K%C3%A4rkk%C3%A4inen-%C3%84yr%C3%A4m%C3%B6/4db73e6aac3c7b7390a912ec0a6bbd9344622586.
- P. J. Huber and E. M. Ronchetti, Robust Statistics. John Wiley & Sons, 2011.
- K. P. Murphy, Machine Learning: A Probabilistic Perspective, Illustrated edition. Cambridge, MA: The MIT Press, 2012.
- J. Platt and N. Karampatziakis, “Probabilistic Outputs for SVMs and Comparisons to Regularized Likelihood Methods,” 2007. Accessed: Oct. 24, 2023. [Online]. Available: https://www.semanticscholar.org/paper/Probabilistic-Outputs-for-SVMs-and-Comparisons-to-Platt-Karampatziakis/18a72c64859a700c16685386514c30d70765a63e.
- “A tutorial on support vector regression | Statistics and Computing.” Accessed: Oct. 24, 2023. [Online]. Available: https://dl.acm.org/doi/10.1023/B%3ASTCO.0000035301.49549.88.
- “k-nearest neighbors algorithm,” Wikipedia. Sep. 27, 2023. Accessed: Oct. 24, 2023. [Online]. Available: https://en.wikipedia.org/w/index.php?title=K-nearest_neighbors_algorithm&oldid=1177452500.
- L. Breiman, J. H. Friedman, R. A. Olshen, and C. J. Stone, Classification and Regression Trees, vol. 19. 1984. doi: 10.1371/journal.pone.0015807.
- L. Breiman, “Random forests,” Mach. Learn., vol. 45, no. 1, pp. 5–32, 2001. [CrossRef]
- P. Geurts, D. Ernst, and L. Wehenkel, “Extremely randomized trees,” Mach. Learn., vol. 63, no. 1, pp. 3–42, Apr. 2006. [CrossRef]
- Y. Freund and R. E. Schapire, “A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting,” J. Comput. Syst. Sci., vol. 55, no. 1, pp. 119–139, Aug. 1997. [CrossRef]
- H. Drucker, “Improving Regressors using Boosting Techniques,” presented at the International Conference on Machine Learning, Jul. 1997. Accessed: Oct. 24, 2023. [Online]. Available: https://www.semanticscholar.org/paper/Improving-Regressors-using-Boosting-Techniques-Drucker/6d8226a52ebc70c8d97ccae10a74e1b0a3908ec1.
- J. H. Friedman, “Greedy function approximation: A gradient boosting machine.,” Ann. Stat., vol. 29, no. 5, pp. 1189–1232, Oct. 2001. [CrossRef]
- J. H. Friedman, “Stochastic gradient boosting,” Comput. Stat. Data Anal., vol. 38, no. 4, pp. 367–378, Feb. 2002. [CrossRef]
- T. Hastie, R. Tibshirani, and J. Friedman, The Elements of Statistical Learning. in Springer Series in Statistics. New York, NY: Springer, 2009. doi: 10.1007/978-0-387-84858-7.
- E. Hinton, “Connectionist learning procedures,” Artif. Intell., vol. 40, no. 1–3, pp. 185–234, Sep. 1989. [CrossRef]
- “Extreme Gradient Boosting - an overview | ScienceDirect Topics.” Accessed: Oct. 24, 2023. [Online]. Available: https://www.sciencedirect.com/topics/computer-science/extreme-gradient-boosting.
- Ke et al., “LightGBM: A Highly Efficient Gradient Boosting Decision Tree,” in Advances in Neural Information Processing Systems, Curran Associates, Inc., 2017. Accessed: Aug. 30, 2023. [Online]. Available: https://proceedings.neurips.cc/paper_files/paper/2017/hash/6449f44a102fde848669bdd9eb6b76fa-Abstract.html.
- L. Ostroumova, G. Gusev, A. Vorobev, A. V. Dorogush, and A. Gulin, “CatBoost: unbiased boosting with categorical features,” presented at the Neural Information Processing Systems, Jun. 2017. Accessed: Oct. 26, 2023. [Online]. Available: https://www.semanticscholar.org/paper/CatBoost%3A-unbiased-boosting-with-categorical-Ostroumova-Gusev/ee0a0f04d45f86bf50b24d7258e884725fcaa621.
- “CatBoost.” Accessed: Oct. 26, 2023. [Online]. Available: https://catboost.ai/en/docs/.
| Symbol | Explanation |
| Reference("real"/expected) value | |
| Mean value of y | |
| Predicted value of y | |
| Number of observations/measurements |
| Reference Values | Scenario 1 predictions | Scenario 2 predictions |
| 50 | 25 | 75 |
| 60 | 45 | 75 |
| 100 | 150 | 50 |
| 135 | 160 | 110 |
| 150 | 200 | 100 |
| 200 | 300 | 100 |
| 250 | 350 | 150 |
| 300 | 390 | 210 |
| Algorithm | Reference |
| Linear Regression | [20,21] |
| Lasso Regression | [22,23] |
| Ridge Regression | [24,25] |
| Elastic Net | [26,27] |
| Least Angle Regression | [28] |
| Lasso Least Angle Regression | [28] |
| Orthogonal Matching Pursuit | [29,30,31] |
| Bayesian Ridge | [32,33,34] |
| Automatic Relevance Determination | [32,34,35] |
| Passive Aggressive Regressor | [36] |
| Random Sample Consensus | [37,38,39] |
| TheilSen Regressor | [40,41,42] |
| Huber Regressor | [43] |
| Kernel Ridge | [44] |
| Support Vector Regression | [32,45,46] |
| K Neighbors Regressor | [47] |
| Decision Tree Regressor | [48] |
| Random Forest Regressor | [49] |
| Extra Trees Regressor | [50] |
| AdaBoost Regressor | [51,52] |
| Gradient Boosting Regressor | [53,54,55] |
| MLP Regressor | [56] |
| Extreme Gradient Boosting | [57] |
| Light Gradient Boosting Machine | [58] |
| CatBoost Regressor | [59,60] |
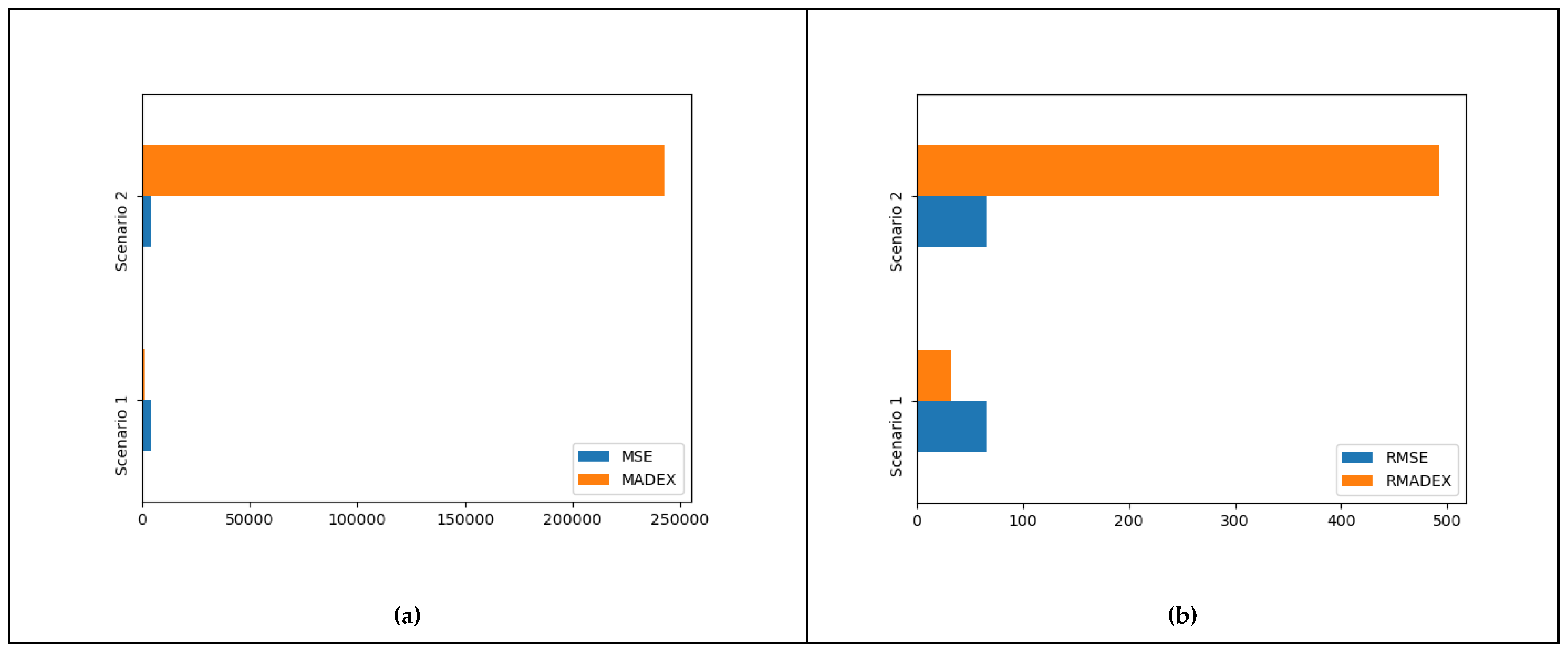
| A | B | C | D | E | MSE | RMSE | MADEX | RMADEX | |
| Scenario 1 | 3 | 5 | 0 | 0 | 0 | 4321.88 | 65.74 | 1034.32 | 32.16 |
| Scenario 2 | 1 | 4 | 0 | 3 | 0 | 4321.88 | 65.74 | 242936.4 | 492.89 |
| Model Name | MAE | MSE | RMSE | MAPE | MADEX | RMADEX |
| Light Gradient Boosting Machine | 1.70E+01 | 5.72E+02 | 2.39E+01 | 1.14E-01 | 6.39E+04 | 1.60E+02 |
| Decision Tree Regressor | 2.35E+01 | 1.15E+03 | 3.39E+01 | 1.57E-01 | 7.97E+05 | 7.37E+02 |
| Lasso Least Angle Regression | 1.79E+01 | 6.58E+02 | 2.56E+01 | 1.21E-01 | 1.41E+06 | 8.48E+02 |
| Elastic Net | 1.79E+01 | 6.59E+02 | 2.57E+01 | 1.21E-01 | 1.51E+06 | 8.74E+02 |
| Lasso Regression | 1.80E+01 | 6.59E+02 | 2.56E+01 | 1.21E-01 | 1.66E+06 | 9.08E+02 |
| Bayesian Ridge | 1.79E+01 | 6.64E+02 | 2.57E+01 | 1.20E-01 | 2.01E+06 | 9.59E+02 |
| Orthogonal Matching Pursuit | 1.92E+01 | 7.19E+02 | 2.68E+01 | 1.29E-01 | 2.38E+06 | 9.88E+02 |
| K Neighbors Regressor | 2.36E+01 | 1.16E+03 | 3.24E+01 | 1.58E-01 | 4.00E+06 | 1.07E+03 |
| Linear Regression | 1.77E+01 | 6.59E+02 | 2.56E+01 | 1.19E-01 | 1.92E+07 | 1.86E+03 |
| Ridge Regression | 1.78E+01 | 6.60E+02 | 2.57E+01 | 1.19E-01 | 1.70E+07 | 1.87E+03 |
| Dummy Regressor | 5.76E+01 | 4.92E+03 | 7.02E+01 | 4.32E-01 | 7.41E+07 | 8.33E+03 |
| Huber Regressor | 1.82E+01 | 7.59E+02 | 2.74E+01 | 1.17E-01 | 8.80E+08 | 1.04E+04 |
| Passive Aggressive Regressor | 3.94E+01 | 5.88E+03 | 6.44E+01 | 2.40E-01 | 3.10E+57 | 1.76E+28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





