Submitted:
26 January 2024
Posted:
26 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Patients and Tumors
2.2. Antibodies and Reagents
2.3. RNA Extraction and Reverse Transcriptase Polymerase Chain Reaction
2.4. Tumor DNA purification
2.5. DNA Sequencing
2.6. Indirect IF
2.7. Statistical Analysis
3. Results
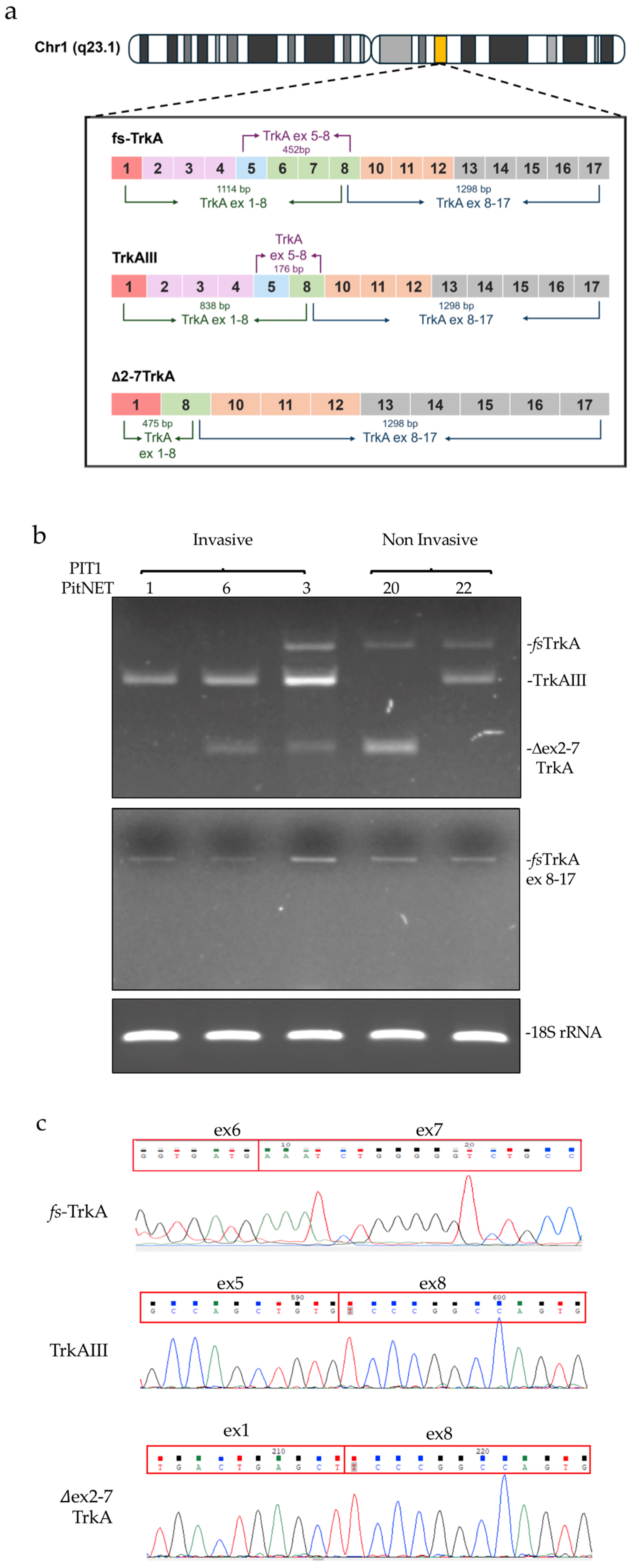
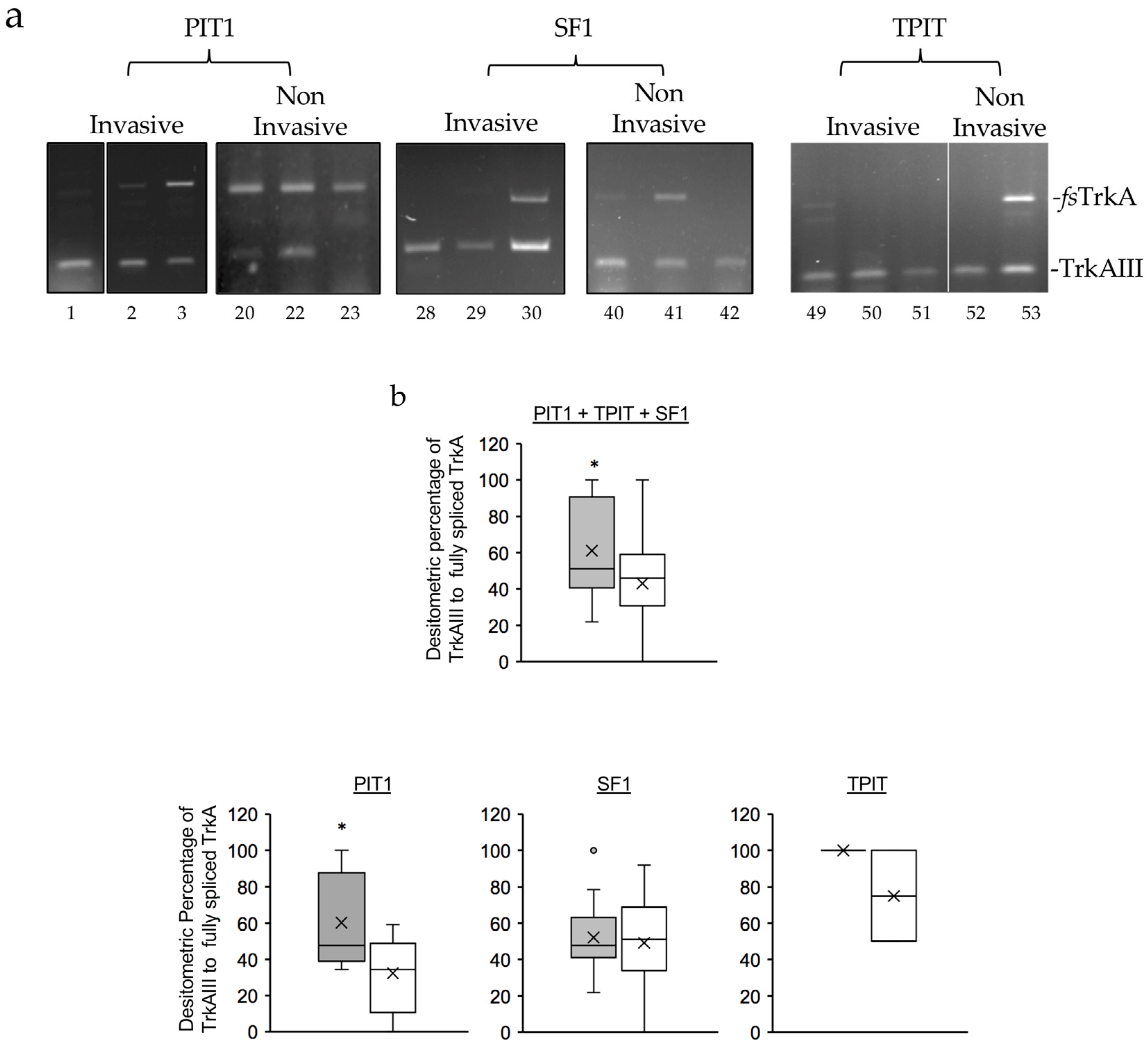
3.1. TrkAIII was the only in-frame alternative TrkA splice variant expressed in PitNETs
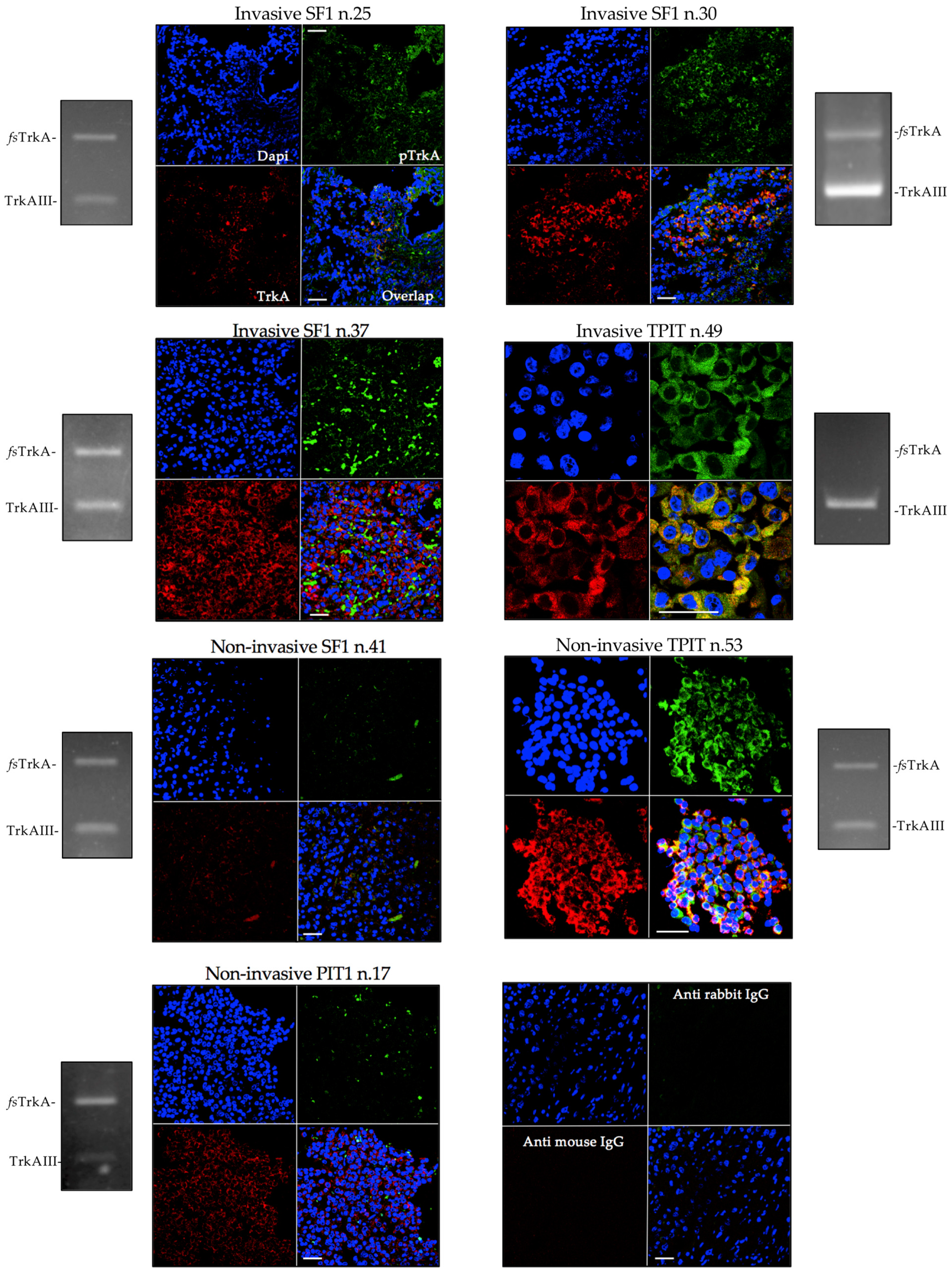
3.2. TrkAIII mRNA expression associates with IF immunoreactivity consistent with intracellular TrkAIII activation.
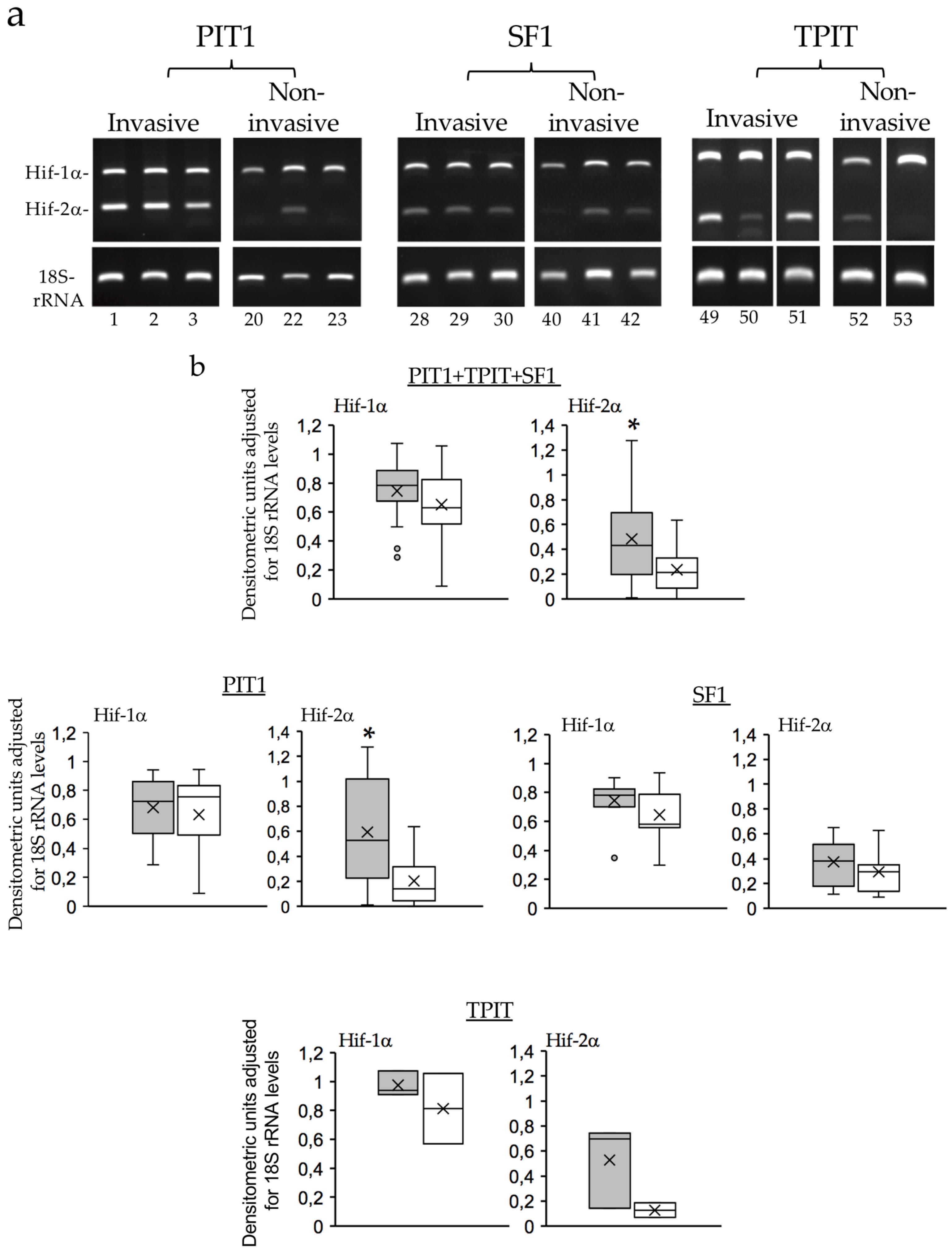
3.3. Enhanced alternative TrkAIII splicing in invasive PIT1 PitNETs associates with increased HIF2a mRNA expression
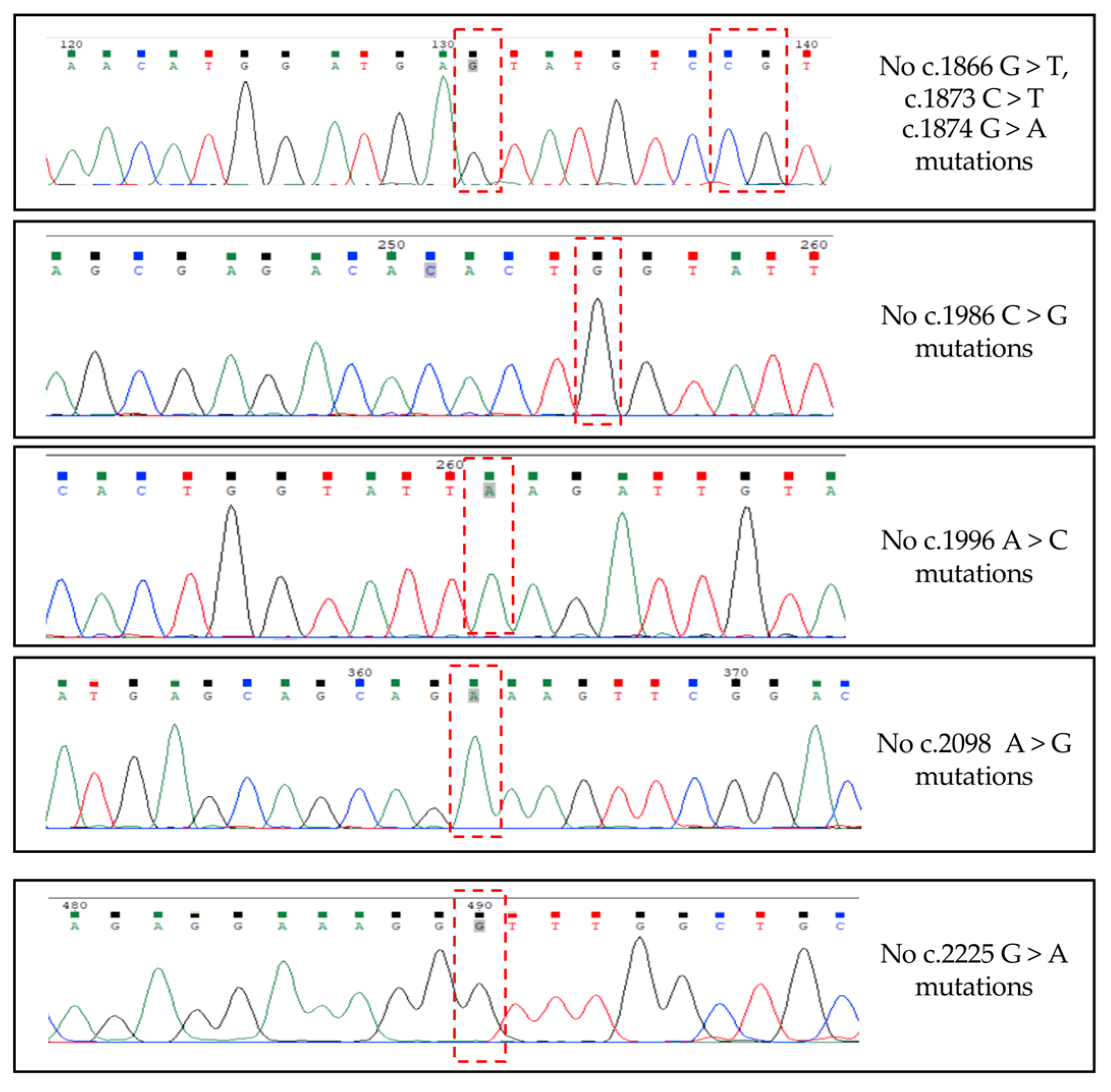
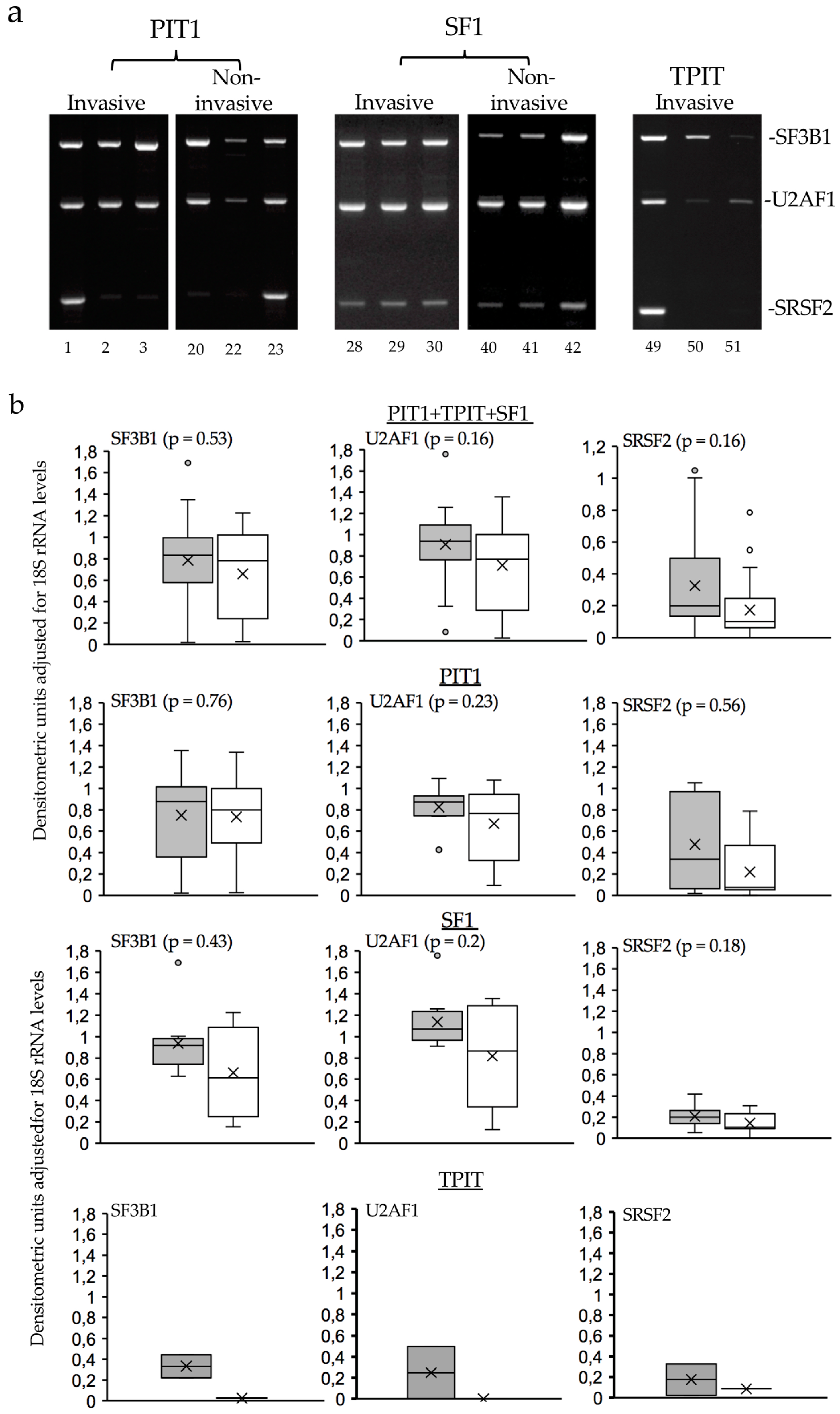
3.3. Alternative TrkAIII splicing in PitNETs does not associate with hotspot SF3B1 mutations or de-regulated SF3B1, SRSF2, U2AF1 expression.
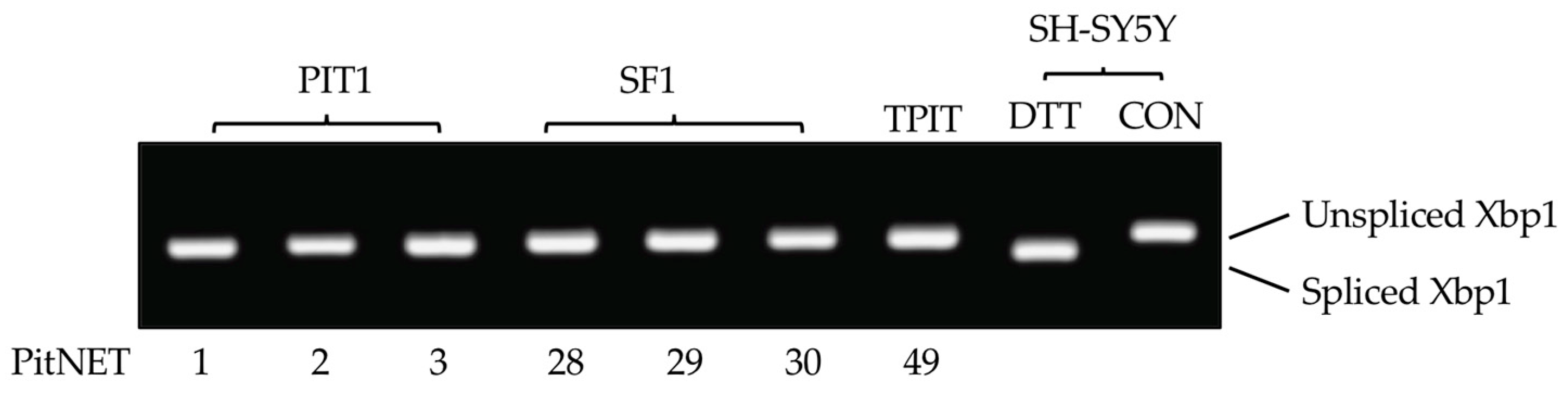
3.4. PitNET alternative TrkAIII splicing does not associate with unconventional Xbp1 splicing or JCPyV large T antigen mRNA expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Osamura, R.Y.; Grossman, A.; Korbonits, M.; Kovacs, K.; Lopes, M.B.S.; Matsuno, A.; Trouillas, J. Pituitary adenoma. In: WHO Classification of Tumours of Endocrine Organs. Editors. Lloyd, R.V.; Osamura, R.Y.; Koppel, G.R. WHO, OMS, International Agency for Research on Cancer. Lyon France, 2017 pp. 14–18.
- Trouillas, J.; Jaffrain-Rea, M.L.; Vasiljevic, A.; Raverot, G.; Rioncaroli, F.; Villa, C. How to classify pituitary neuroendocrine tumors (PitNET)s in 2020. Cancers 2020, 12, 514. [CrossRef]
- Asa, S.L.; Mete, O.; Perry, A.; Osamura, R.Y. Overview of the 2022 WHO Classification of Pituitary Tumors. Endocr Pathol 2022, 33, 6-26. [CrossRef]
- Tsukamoto,T.; Miki,Y. Imaging of pituitary tumors: an update with the 5th WHO classifications: part 1. Pituitary neuroendocrine tumor (PitNET) pituitary adenoma. Jap J Radiol 2023, 41, 789-806. [CrossRef]
- Neou, M., Villa, C., Armignacco, R., Jouinot, A., Raffin-Sanson, M.L., Septier, A., Letoumeur, F., Diry, S., Diedisheim, M., Izac, B., Gaspar, C., Perlemione, K., Verjus,, V., Bernier M., Boulin, A., Emile, J.F., Bertagna, X., Jaffrezic, F., Laloe, D., Baussart, B., Bertherat, J., Gaillard, S., Assie, G. Pangenomic classification of pituitary neuroendocrine tumors. Cancer Cell 2020, 37, 123-134. [CrossRef]
- Raverot, G.; Ilie, M.D.; Lasolle, H.; Amodru, V.; Trouillas, J.; Castinetti, F.; Brue, T. Aggressive pituitary tumours and pituitary carcinomas. Nat Rev Endocrinol 2021, 17, 671-684. [CrossRef]
- Burman P, Trouillas J, Losa M, McCormack A, Petersenn S, Popovic V, Theodoropoulou M, Raverot G, Dekkers OM; ESE survey collaborators. Aggressive pituitary tumours and carcinomas, characteristics and management of 171 patients. Eur J Endocrinol 2022, 187, 593-605. [CrossRef]
- Trouillas, J.; Jaffrain-Rea, M.L.; Vasiljevic, A.; Dekkers, O.; Popovic, V.; Wierinckx, A.; McCormack, A.; Petersenn, S.; Burman, P.; Raverot, G., Villa, C. Are aggressive pituitary tumors and carcinomas two sides of the same coin? Pathologists reply to clinician's questions. Rev Endocr Metab Disord 2020, 21, 243-251. [CrossRef]
- Guaraldi, F.; Morandi, L.; Zoli, M.; Mazzatenta, D.; Righi, A.; Evangelisti, S.; Ambrosi, F.; Tonon, C.; Giannini, C.; Lloyd, R.V.; Asioli, S. Epigenomic and somatic mutations in pituitary tumors with clinical and pathological correlations in 111 patients. Clin Endocrinol (Oxf) 2022, 97, 763-772. [CrossRef]
- Srirangam Nadhamuni, V.; Korbonits, M. Novel Insights into Pituitary Tumorigenesis: Genetic and Epigenetic Mechanisms. Endocr Rev 2020, 41, 821-846. [CrossRef]
- Melmed, S.; Kaiser, U.B.; Lopes, M.B.; Bertherat, J.; Syro, L.V.; Raverot, G.; Reincke, M.; Johannsson, G.; Beckers, A.; Fleseriu, M.; Giustina, A.; Wass, J.A.H.; Ho, K.K.Y. Clinical Biology of the Pituitary Adenoma. Endocr Rev 2022, 43, 1003-1037. [CrossRef]
- Bao, Y.; Yoshida, D.; Morimoto, M.D.; Teramoto, A. Expression of laminin B2: a novel marker of hypoxia in pituitary adenomas. Endocr Pathol 2006, 17, 251-261. [CrossRef]
- Zhou, Y.; Zhang, A.; Fang, C.; Yuan, L.; Shao, A.; Xu, Y.; Zho,u D. Oxidative stress in pituitary neuroendocrine tumors: affecting the tumor microenvironment and becoming a new target for pituitary neuroendocrine tumor therapy. CNS Neurosci Ther 2023, 29, 2744-2759. [CrossRef]
- Li, C.; Xie, W.; Rosenblum, J.S.; Zhou, J.; Guo, J.; Miao, Y.; Shen,Y.; Wang, H.; Gong, L.; Li, M.; Zhao, S.; Cheng, S.; Zhu, H.; Jiang, T.; Ling, S.; Wang, F.; Zhang, H.; Zhang, M.; Qu, Y.; Zhang, Q.; Li, G.; Wang, J.; Ma, J.; Zhuang, Z.; Zhang, Y. Somatic SF3B1 hotspot mutation in prolactinomas. Nature Commun 2020, 11, 2056. [CrossRef]
- Torres-Moran, M.; Franco-Alvarez, A.; Rebollar-Vega, R.G.; Hernandez-Ramirez, L.C. Hotspots of somatic genetic variation in pituitary neuroendocrine tumors. Cancers (Basel) 2023, 15, 5685. [CrossRef]
- Simon, J.; Perez-Rivas, L.G.; Zhao, Y.; Chasseloup, F.; Lasolle, H., Cortet, C.; Descotes, F.; Villa, C., Baussart, B.; Burman, P., Maiter, D.; von Selzam, V.; Rotermund, R.; Flitsch J.; Thorsteinsdottir, J.; Jouanneau, E.; Buchfelder, M.; Chanson, P.; Raverot, G.; Theodoropoulou, M. Prevalence and clinical correlations of SF3B1 variants in lactotroph tumours. Eur J Endocrinol 2023, 189, 372-378. [CrossRef]
- Vazquez-Borrego, M.C.; Fuentes-Fayos, A.C.; Venegas-Moreno, E.; Rivero-Cortes, E.; Dios, E.; Moreno-Moreno, P.; Madrazo-Atutxa, A.; Remon, P.; Solivera, J.; Wildemberg, L.E.; Kasuli, L.; Lopez-Fernandez, J.M.; Gadelha, M.R.; Galvez-Moreno, M.A.; Soto-Moreno, A.; Gahete, M.D.; Castano, J.P.; Luque, R.M. Splicing machinery is dysregulated in pituitary neuroendocrine tumors and is associated with aggressive features. Cancers (Basel) 2019, 11, 1439. [CrossRef]
- Gordon, J.; Del Valle, L.; Otte, J.; Khalili, K. Pituitary neoplasia induced by expression of human neurotropic polyomavirus, JCV, early genome in transgenic mice. Oncogene 2000, 19, 4840-4846. [CrossRef]
- Del Valle, L.; Khalili, K. Induction of brain tumors by the archetypal strain of human neurotropic JCPyV in a transgenic mouse model. Viruses 2021, 13, 162. [CrossRef]
- Farina, A.R., Cappabianca, L., Sebastiano, M., Zelli, V., Guadagn,i S., Mackay, A.R. Hypoxia-induced alternative splicing: The 11th hallmark of cancer. J Clin Exp Cancer Res 2020, 39:110. [CrossRef]
- Siddaway R, Milos S, Kumaran Anguraj Vadivel A, Dobson THW, Swaminathan J, Ryall S, Pajovic S, Patel PG, Nazarian J, Becher O, Brudno M, Ramani A, Gopalahrishnan V, Hawkins C. Splicing is an alternative oncogenic pathway activation mechanism in glioma. Nature Commun 2022, 13, 588. [CrossRef]
- Bonomi, S.; Gallo, S.; Catillo, M.; Pignataro, D.; Biamonti, G.; Ghigna, C. Oncogenic alternative splicing switches: role in cancer progression and prospects for therapy. Int J Cell Biol 2013, 2013, 962038. [CrossRef]
- Patterson, J.C.; Childs, G.V. Nerve growth factor and its receptor in the anterior pituitary. Endocrinology 1994, 135, 1689-9166. [CrossRef]
- Assimakopoulou, M.; Zolota, V.; Chondrogianni, C.; Gatzounis, G.; Varakis, J. p75 and TrkC neurotrophin receptors demonstrate a different immunoreactivity profile in comparison to TrkA and TrkB receptors in human normal pituitary gland and adenomas. Neuroendocrinology 2008, 88, 127-134. [CrossRef]
- Tacconelli, A., Farina, A.R., Cappabianca, L., DeSantis, G., Tessitore, A., Vetuschi, A., Sferra, R., Rucci, N., Argenti, B., Screpanti, I., Gulino, A, Mackay, A.R. TrkA alternative splicing: A regulated tumor-promoting switch in human neuroblastoma. Cancer Cell 2004, 6, 347–360. [CrossRef]
- Cappabianca, L., Guadagni, S., Maccarone, R., Sebastiano, M., Chiominto, A., Farina, A.R., Mackay, A.R. A pilot study of alternative TrkAIII splicing in Merkel cell carcinoma: A potential oncogenic mechanism and novel therapeutic target. J Exp Clin Cancer Res 2019, 38, 424. [CrossRef]
- Cappabianca, L., Zelli, V., Pellegrini, C., Sebastiano, M., Maccarone, R., Clementi, M., Chiominto, A., Ruggeri, P., Cardelli, L., Ruggieri, M., Sbaffone, M., Fargnoli, M.C., Guadagni, S., Farina, A.R., Mackay, A.R. The alternative TrkAIII splice variant, a targetable oncogenic participant in human cutaneous malignant melanoma. Cells 2023, 12, 237. [CrossRef]
- Schramm, A., Schowe, B., Fielitz, K., Hweilman, M., Martin, M., Marschall, T., Koster, J., Vandenstompele, J., Vermeulen, J., de Pterter, K., Koster, J., Versteeg, R., Noguera, R., Speleman, F., Rahmann, S., Eggert, A., Morik, K., Schulte, J.H. Exon-level expression analyses identify MYCN and NTRK1 as major determinants of alternative exon usage and robustly predict primary neuroblastoma outcome. Br J Cancer 2012, 107, 1409-1417. [CrossRef]
- Lebedev, T.D., Vagapova, E.R., Popenko, V.I., Leonova, O.G., Spirin, P.V., Prassolov, V.S. Two receptors, two isoforms, two cancers: comprehensive analysis of KIT and TrkA expression in neuroblastoma and acute myeloid leukemia. Front Oncol 2019, 9, 1046. [CrossRef]
- Farina, A.R., Di Ianni, N., Cappabianca, L., Ruggeri, P., Gneo, L., Pellegrini, C., Fargnoli, M.C., Mackay, A.R. The oncogenic neurotrophin receptor tropomyosin-related kinase variant, TrkAIII. J Clin Exp Cancer Res 2018, 37, 119. [CrossRef]
- Arevalo, J.C., Conde, B., Hempstead, B.L., Chao, M.V., Martin-Zanca, D., Perez, P. TrkA immunoglobulin-like ligand binding domains inhibit spontaneous activation of the receptor. Mol Cell Biol 2000, 20, 5908–5916. [CrossRef]
- Watson, F.L., Porcionatto, M.A., Battacharyya, A., Stiles, C.D., Segal, R.A. TrkA glycosylation regulates receptor localization and activity. J Neurobiol 1999, 39, 323-336. [CrossRef]
- Farina, A.R.; Cappabianca, L.; Ruggeri, P.; Gneo, L.; Maccarone, R.; Mackay, A.R. Retrograde TrkAIII transport from ERGIC to ER: A re-localization mechanism for oncogenic activity. Oncotarget 2015, 34, 35636-35651. [CrossRef]
- Farina, A.R.; Tacconelli, A.; Cappabianca L.; Cea, G.; Panella, S.; Chioda, A.; Romanelli, A.; Pedone, C.; Gulino, A.; Mackay, A. R. The alternative TrkA splice variant targets the centrosome and promotes genetic instability. Mol Cell Biol 2009, 17, 4182-4130. [CrossRef]
- Farina, A.R.; Cappabianca, L.; Gneo, L.; Ruggeri, P.; Mackay, A.R. TrkAIII signals endoplasmic reticulum stress to the mitochondria in neuroblastoma cells, resulting in glycolytic metabolic adaptation. Oncotarget 2017, 9, 8368-8390. [CrossRef]
- Cappabianca, L.; Sebastiano, M.; Ruggieri, M.; Sbaffone, M.; Zelli, V.; Farina, A.R.; Mackay, A.R. Doxorubicin-induced TrkAIII activation: A selection mechanism for resistant dormant neuroblastoma cells. Int J Mol Sci 2022, 18, 10895. [CrossRef]
- Shulman, D.S., DuBois, S.G. The evolving diagnostic and treatment landscape of NTRK-fusion driven pediatric cancers. Pediatric Drugs 2020, 22, 189-197. [CrossRef]
- Rohrberg, K.S., Lassen, U. Detecting and targeting NTRK fusions in cancer in the era of tumor agnostic oncology. Drugs 2021, 81, 445-452. [CrossRef]
- Villa C, Vasiljevic A, Jaffrain-Rea ML, Ansorge O, Asioli S, Barresi V, Chinezu L, Gardiman MP, Lania A, Lapshina AM, Poliani L, Reiniger L, Righi A, Saeger W, Soukup J, Theodoropoulou M, Uccella S, Trouillas J, Roncaroli F. A standardised diagnostic approach to pituitary neuroendocrine tumours (PitNETs): a European Pituitary Pathology Group (EPPG) proposal. Virchows Arch 2019, 475, 687-692. [CrossRef]
- Singh R, Green MR. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 1993, 259, 365-368. [CrossRef]
- Feola, T.; Carbonara, F.; Verrico, M.; Di Crescenzo, R.M.; Gianno, F.; Colonnese, C.; Arcella, A.; de Alcubierre, D.; Tomao, S.; Esposito, V.; Giangaspero, F.; Minniti, G.; Jaffrain-Rea, M.L. Immunotherapy for Aggressive and Metastatic Pituitary Neuroendocrine Tumors (PitNETs): State-of-the Art. Cancers (Basel) 2022, 14, 4093. [CrossRef]
- Yoshida, H. Unconventional splicing of XBP-1 mRNA in the unfolded protein response. Antioxid Redoc Signal 2007, 9, 2323-2333. [CrossRef]
- Yang, Q.; Li X. Molecular Network Basis of Invasive Pituitary Adenoma: A Review. Front Endocrinol (Lausanne) 2019, 10, 7. [CrossRef]
- Artico, M.; Bianchi, E.; Magliulo, G.; De Vincentiis, M.; De Santis, E.; Orlandi, A.; Santoro, A.; Pastore, F.S.; Giangaspero, F.; Caruso, R.; Re, M.; Fumagalli, L. Neurotrophins, their receptors and KI-67 in human GH-secreting pituitary adenomas: an immunohistochemical analysis. Int J Immunopathol Pharmacol 2012, 25, 117-125. [CrossRef]
- Tebani, A., Jotanovic, J., Hekmati, N., Sivertsson, A., Gudjonsson, O., Engstrom, B.E., Wikstrom, J., Uhlen, M., Casar-Borota, O., Ponten, F. Annotation of pituitary neuroendocrine tumor with genome-wide expression analysis. Acta Neuropathol Commun 2021, 9, 181. [CrossRef]
- Lucia, K.; Wu, Y.; Garcia, J.M.; Barlier, A.; Buchfelder, M.; Saeger, W.; Renner, U.; Stalla, G.K.; Theodoropoulou, M. Hypoxia and the hypoxia inducible factor 1α activate protein kinase A by repressing RII beta subunit transcription. Oncogene 2020, 39, 3367-3380. [CrossRef]
- Hamidian, A., von Stedingk, K., Thoren, M.M., Mohlin, S., Pahlman, S. Differential regulation of HIF-1a and HIF-2a in neuroblastoma, estrogen related receptor alpha (ERRa) regulates HIF2A transcription and correlates to poor outcome. BBRC 2015, 461, 560-567. [CrossRef]
- Johnsen, J.I., Dyberg, C., Wickstrom, M. Neuroblastoma-a neural crest derived embryonal malignancy. Front Mol Neurosci 2019, 12, 9. [CrossRef]
- Ferrand, R., Pearse, A.G.E., Polak, J.M., Le Douarin, N.M. Immunohistochemical studies on the development of avian embryo pituitary corticotrophs under normal and experimental conditions. Histochemistry 1974, 38, 133-141. [CrossRef]
- Eagleson, G.W., Jenks, B.G., Van Overbeeke, A.P. The pituitary adrenocorticotropes originate from neural ridge tissue in xenopus laevis. J Embryol Exp Morph 1986, 95, 1-14. [CrossRef]
- Ueharu, H., Yoshida, S., Kikkawa, T., Kanno, N., Higuchi, M., Kato, T., Osumi, N., Kato, Y. Gene tracing analysis reveals the contribution of neural crest-derived cells in pituitary development. J Anat 2017, 230, 373-380. [CrossRef]
- Ueharu, H., Yoshida, S., Kanno, N., Horiguchi, K., Nishimura, N., Kato, T., Kato, Y. SOX10-positive cells emerge in the rat pituitary gland during late embryogenesis and start to express S100β. Cell Tissue Res 2018, 372, 77-90. [CrossRef]
- Duan, S., Sawyer, T.W., Sontz, R.A., Wieland, B.A., Diaz, A.F., Merchant, J.L. GFAP-directed inactivation of men1 exploits glail cell plasticity in favor of neuroendocrine reprogramming. Cell Mol Gastroent Hepatol 2022, 14, 1025-1051. [CrossRef]
- Albanese, A.; Daly, L.A.; Mennerich, D.; Kietzmann, T.; Sée, V. The Role of Hypoxia-Inducible Factor Post-Translational Modifications in Regulating Its Localisation, Stability, and Activity. Int J Mol Sci 2020, 22, 268. [CrossRef]
- Gupta, P.; Dutta, P. Landscape of Molecular Events in Pituitary Apoplexy. Front Endocrinol (Lausanne) 2018, 9, 107. [CrossRef]
- Chipurupalli, S., Kannan, E., Tergaonkar, V., D’Andrea, R., Robinson, N. Hypoxia induced ER stress response as an adaptive mechanism in cancer. Int J Mol Sci 2019, 20, 749. [CrossRef]
- Cocco, E., Scaltrtiti, M., Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018, 15, 731-747. [CrossRef]
- Treis, D., Umapoathy, G., Fransson, S., Guan, J., Mendoza-Garcia, P., Siaw, J.T., Wessman, S., Gordon Murkes, L., Stenman, J.J.E., Djos, A., Elfman, L.H.M., Inge Johnsen, J., Hallberg, B., Palmer, R.H., Martinsson, T., Kogner, P. Sustained response to entrectinib in an infant with a germline ALKAL2 variant and refractory metastatic neuroblastoma with chromosomal 2p gain and anaplastic lymphoma kinase and tropomyosin receptor kinase activation. JCO Precision Oncol 2022, 6. [CrossRef]
| PIT1 PitNETs | |||||||||||||||||||||||||||||||
| INVASIVE (n = 11) | NON-INVASIVE (n= 13) | ||||||||||||||||||||||||||||||
| Pt | Age | Sex | IHC |
Clinical Status |
Rec |
Ki67 (%) |
Pt | Age | Sex | IHC |
Clinical Status |
Rec |
Ki67 (%) |
||||||||||||||||||
| 1 | 53 | F | PRL | F | Y(A) | ≥3 | 12* | 37 | F | GH | F | N | ≥3 | ||||||||||||||||||
| 2 | 19 | M | GH | F | N | <3 | 13 | 52 | F | GH | F | N | <3 | ||||||||||||||||||
| 3 | 16 | M | PRL | NF | N | ≥3 | 14 | 52 | M | GH | F | N | <3 | ||||||||||||||||||
| 4 | 18 | F | GH | NF | N | ≥3 | 15 | 34 | F | PRL | F | N | n/a | ||||||||||||||||||
| 5 | 74 | M | TSH | F | N | ≥3 | 16* | 49 | M | GH/PRL | F | N | <3 | ||||||||||||||||||
| 6 | 37 | M | TSH | F | N | ≥3 | 17 | 40 | F | GH/PRL | F | N | ≥3 | ||||||||||||||||||
| 7 | 25 | F | GH | F | N | ≥3 | 18 | 55 | M | GH/PRL | F | N | ≥3 | ||||||||||||||||||
| 8 | 21 | M | PRL | F | N | n/a | 19 | 36 | F | PRL | F | N | n/a | ||||||||||||||||||
| 9 | 76 | F | GH | F | N | <3 | 20 | 26 | M | PRL | F | N | n/a | ||||||||||||||||||
| 10 | 14 | M | GH/PRL | F | Y | ≥3 | 21* | 50 | M | GH | F | N | ≥3 | ||||||||||||||||||
| 11 | 62 | M | Pit1 only | NF | Y(M) | ≥3 | 22 | 43 | F | GH/PRL | NF | N | <3 | ||||||||||||||||||
| 23 | 49 | M | PRL | F | N | n/a | |||||||||||||||||||||||||
| 24 | 32 | F | GH | F | N | <3 | |||||||||||||||||||||||||
| SF1 PitNETs | |||||||||||||||||||||||||||||||
| INVASIVE (n = 12) | NON-INVASIVE (n = 12) | ||||||||||||||||||||||||||||||
| Pt | Age | Sex | IHC | Clinical status | Rec |
Ki67 (%) |
Pt | Age | Sex | IHC | Clinical status | Rec |
Ki67 (%) |
||||||||||||||||||
| 25 | 45 | M | FSH/LH | NF | N | ≥3 | 37 | 68 | M | FSH/LH | NF | N | ≥3 | ||||||||||||||||||
| 26 | 56 | M | FSH/LH | NF | N | ≥3 | 38 | 71 | F | SF1 only | NF | Y | <3 | ||||||||||||||||||
| 27 | 73 | F | FSH/LH | NF | N | ≥3 | 39* | 71 | M | FSH/LH | NF | N | <3 | ||||||||||||||||||
| 28 | 49 | F | SF1 only | NF | N | ≥3 | 40 | 67 | M | SF1 only | NF | Y | ≥3 | ||||||||||||||||||
| 29 | 55 | F | SF1 only | NF | N | ≥3 | 41 | 61 | M | FSH/LH | NF | N | <3 | ||||||||||||||||||
| 30 | 48 | M | FSH/LH | NF | N | ≥3 | 42 | 46 | M | FSH/LH | NF | N | ≥3 | ||||||||||||||||||
| 31 | 53 | M | FSH/LH | NF | N | ≥3 | 43 | 75 | M | FSH/LH | NF | N | <3 | ||||||||||||||||||
| 32 | 47 | F | FSH/LH | NF | N | <3 | 44 | 74 | M | SF1 only | NF | N | <3 | ||||||||||||||||||
| 33 | 69 | M | FSH/LH | NF | Y | <3 | 45 | 66 | M | FSH/LH | NF | N | <3 | ||||||||||||||||||
| 34 | 70 | F | FSH/LH | NF | N | <3 | 46 | 39 | M | FSH/LH | NF | N | <3 | ||||||||||||||||||
| 35 | 55 | M | FSH/LH | NF | N | ≥3 | 47 | 46 | M | FSH/LH | NF | N | ≥3 | ||||||||||||||||||
| 36 | 73 | M | SF1 only | NF | N | ≥3 | 48 | 69 | F | FSH/LH | NF | N | <3 | ||||||||||||||||||
| TPIT PitNETs | |||||||||||||||||||||||||||||||
| INVASIVE (n = 3) | NON-INVASIVE (n = 2) | ||||||||||||||||||||||||||||||
| Pt | Age | Sex | IHC | Clinical status | Rec |
Ki67 (%) |
Pt | Age | Sex | IHC | Clinical status | Rec |
Ki67 (%) |
||||||||||||||||||
| 49 | 57 | M | ACTH | F | Y(A) | ≥3 | 52 | 78 | F | ACTH | F | N | <3 | ||||||||||||||||||
| 50 | 52 | F | ACTH | NF | N | ≥3 | 53 | 36 | F | ACTH | F | N | ≥3 | ||||||||||||||||||
| 51 | 26 | F | ACTH | F | N | ≥3 | |||||||||||||||||||||||||
| Target | Sequence | Denat | Ann | Ext | Amplicon |
|---|---|---|---|---|---|
| 18S rRNA**** |
F: 5’-AAACGGCTACCACATCCAAG-3’ R: 5’-CCTCGAAAGAGTCCTGTATTG-3’ |
30s - 94°C | 30s - 58°C | 30s-72°C | 100bp |
| TrkA ex 8-17* |
F: 5’-AACCCCTTCGGCCAGGCCTCC-3’ R: 5’-CTAGCCCAGGACATCCAGGTA-3’ |
1 min - 94°C | 30s - 65°C | 1 min -72°C | 1298bp TrkA |
| TrkA ex 1-8* |
F: 5’-ATGCTGCGAGGCGGACGGCGC-3’ R: 5’-GGAGGCCTGGCCGAAGGGGTT-3’ |
1 min -94°C | 30s - 68°C | 1 m- 72°C | 1114bp TrkA, 838bp TrkAIII, 475bp Dex2-7 TrkA |
| TrkA ex 5-8* |
F: 5’-AGAAGCTGCAGTGTCATGGG-3’ R: 5’-ATTGAGCACGGAGCCATTGA-3’ |
40s - 94°C | 30s - 58°C | 40s -72°C | 452bp TrkA 176bp TrkAIII |
| SRSF2*** |
F: 5’-CTCCCGATGTGGAGGGTATG-3’ R: 5’-GAGATCGGCTGCGAGACC-3’ |
40s - 94°C | 30s - 58°C | 40 s - 72°C | 408 bp |
| SF3B1** |
F: 5’-TGTGCATAAGATCCTCGTGGT-3’ R: 5’-ACACCATCTGTCCCACAACA-3’ |
40s - 94°C | 30s - 58°C | 4s - 72°C | 693 bp |
| SF3B1 ( tDNA) |
F: 5’-TAGGCTGCTGGTCTGGCTAC-3’ R: 5’-ATGGCACAGCCCATAAGAATAG-3’ |
30s - 95°C | 30s - 60°C | 1m -72°C | 233 bp |
| U2AF1** |
F: 5’-CGGAGTATCTGGCCTCCATC-3’ R: 5’-GCAGCTCTCTGGAAATGGGCT-3’ |
40s - 94°C | 30s - 60°C | 40s -72°C | 606 bp |
| HIF-1a** |
F: 5’-TTCACCTGAGCCTAATAGTCC-3’ R: 5’-AAGTCTAAATCTGTGTCCTG-3’ |
30s - 94°C | 30s - 50°C | 30s - 72°C | 150 bp |
| HIF-2a*** |
F: 5’-AGCCTCCATCTGCCATCAGTC-3’ R: 5’-CTTGCCATGCCTGACACCTTG-3’ |
30s - 94°C | 30s - 55°C | 30s - 72°C | 121 bp |
| JCPyV T-Ag* |
F: 5’-ATATTATGACCCCCAAAACCATG-3’ R: 5’-GGTAGAAGACCCTAAGGACTTTCC-3’ |
40s -94°C | 30s - 58°C | 40s - 68°C | 189 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





