Submitted:
26 January 2024
Posted:
29 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Foc isolates
2.2. Bioassay
2.3. Root bleaching and coloring
2.4. DNA extraction
2.5. PCR analysis
2.5. Data analysis
3. Results
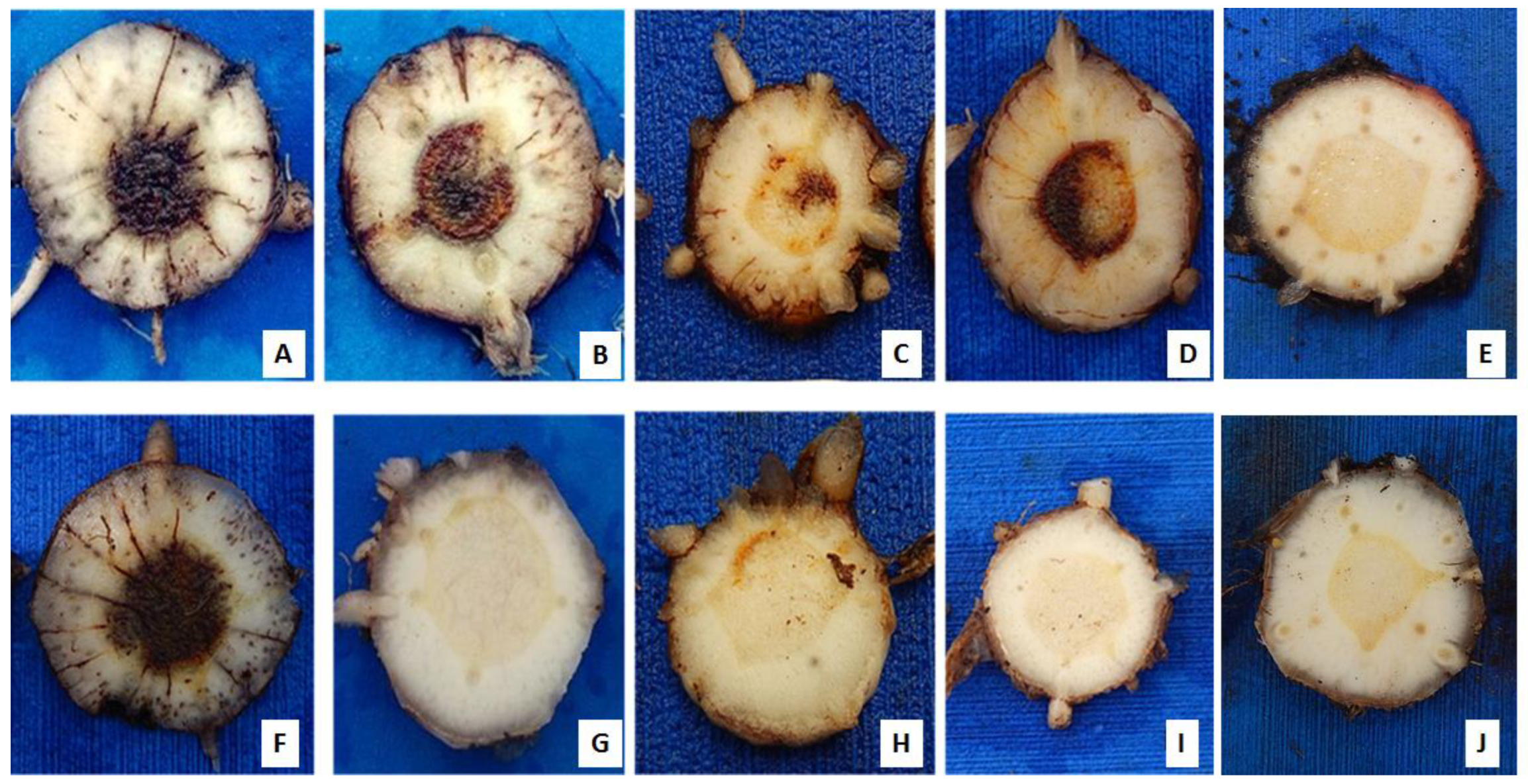
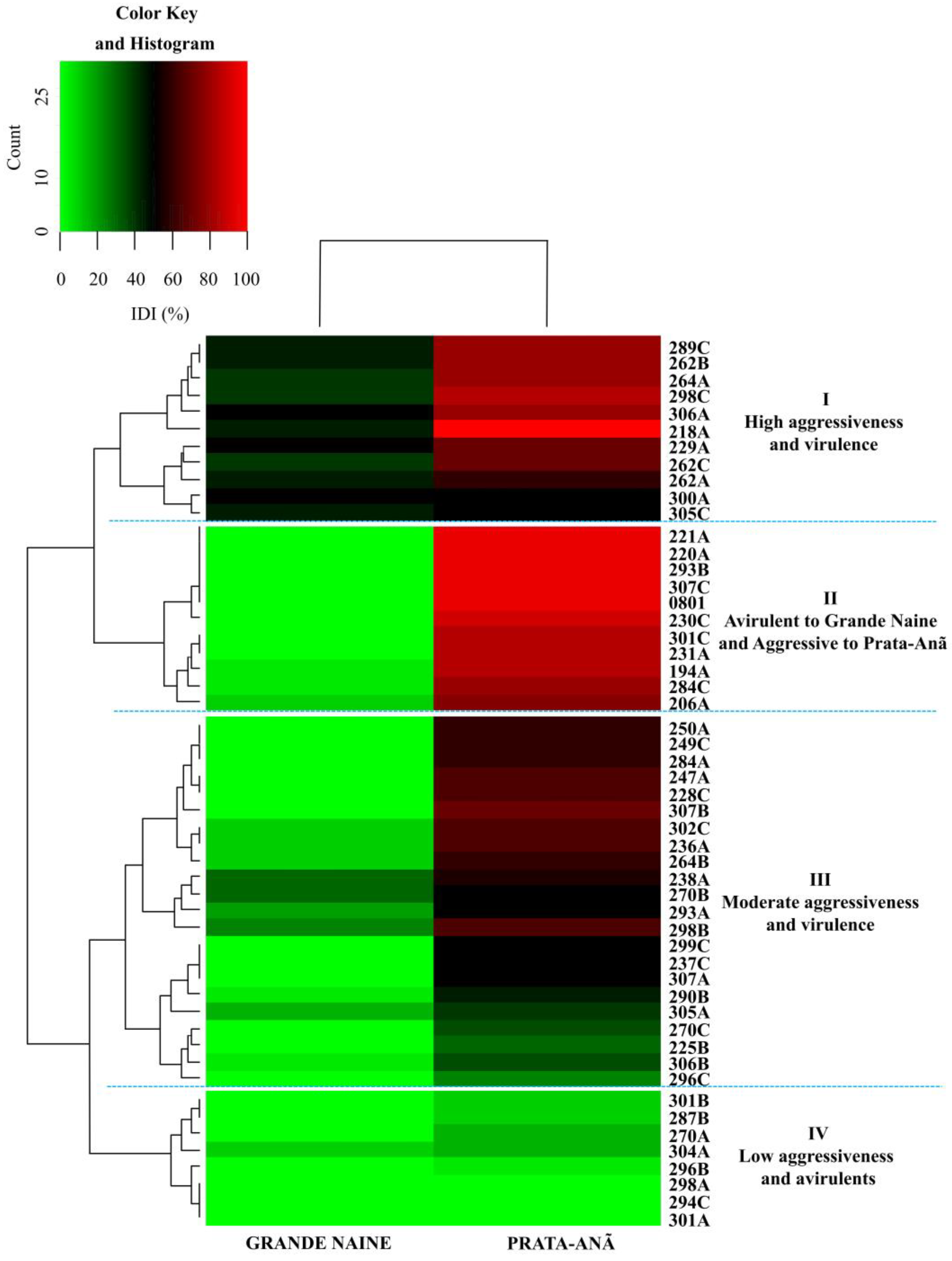
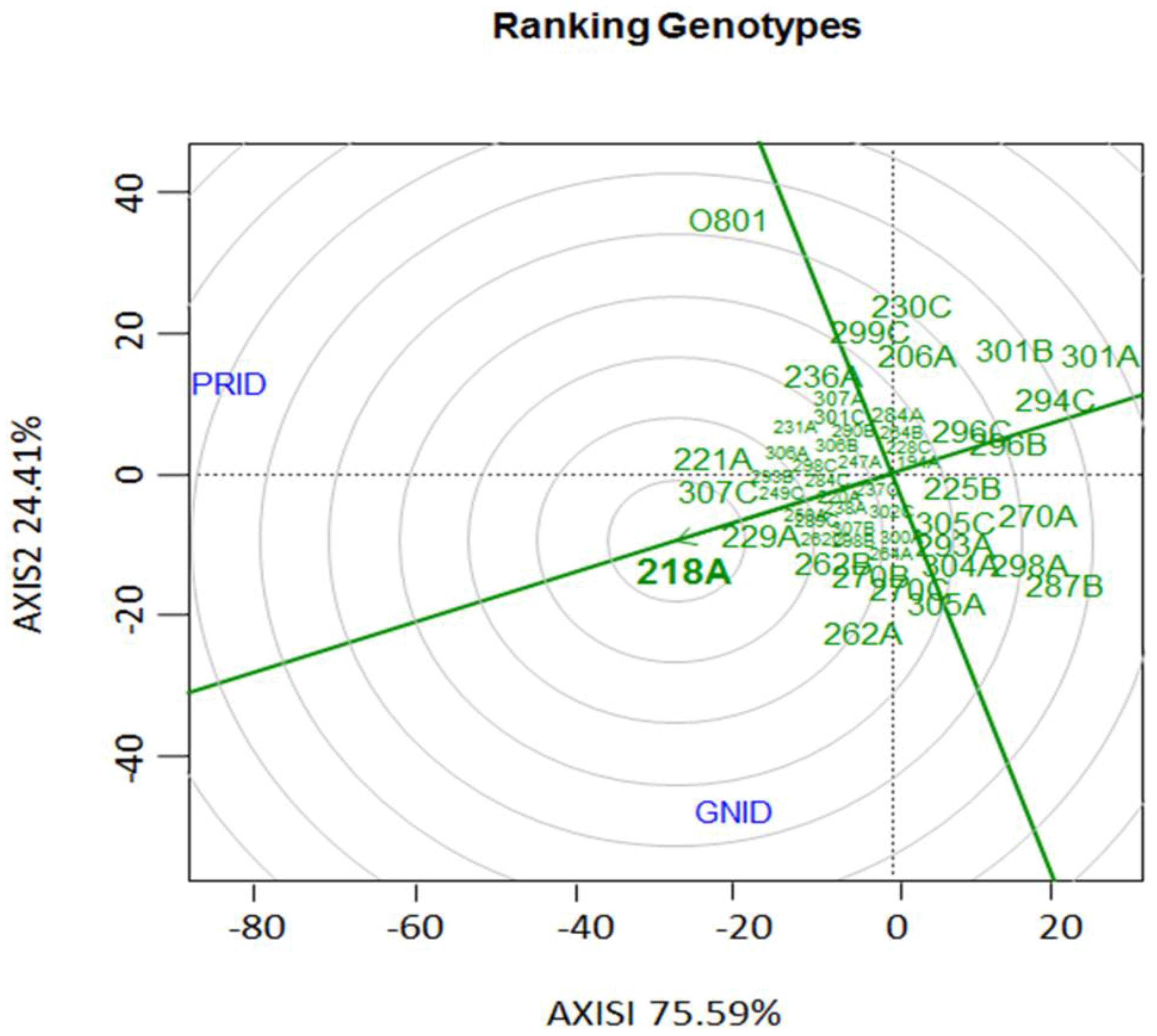
3.1. Evaluation of disease indexes
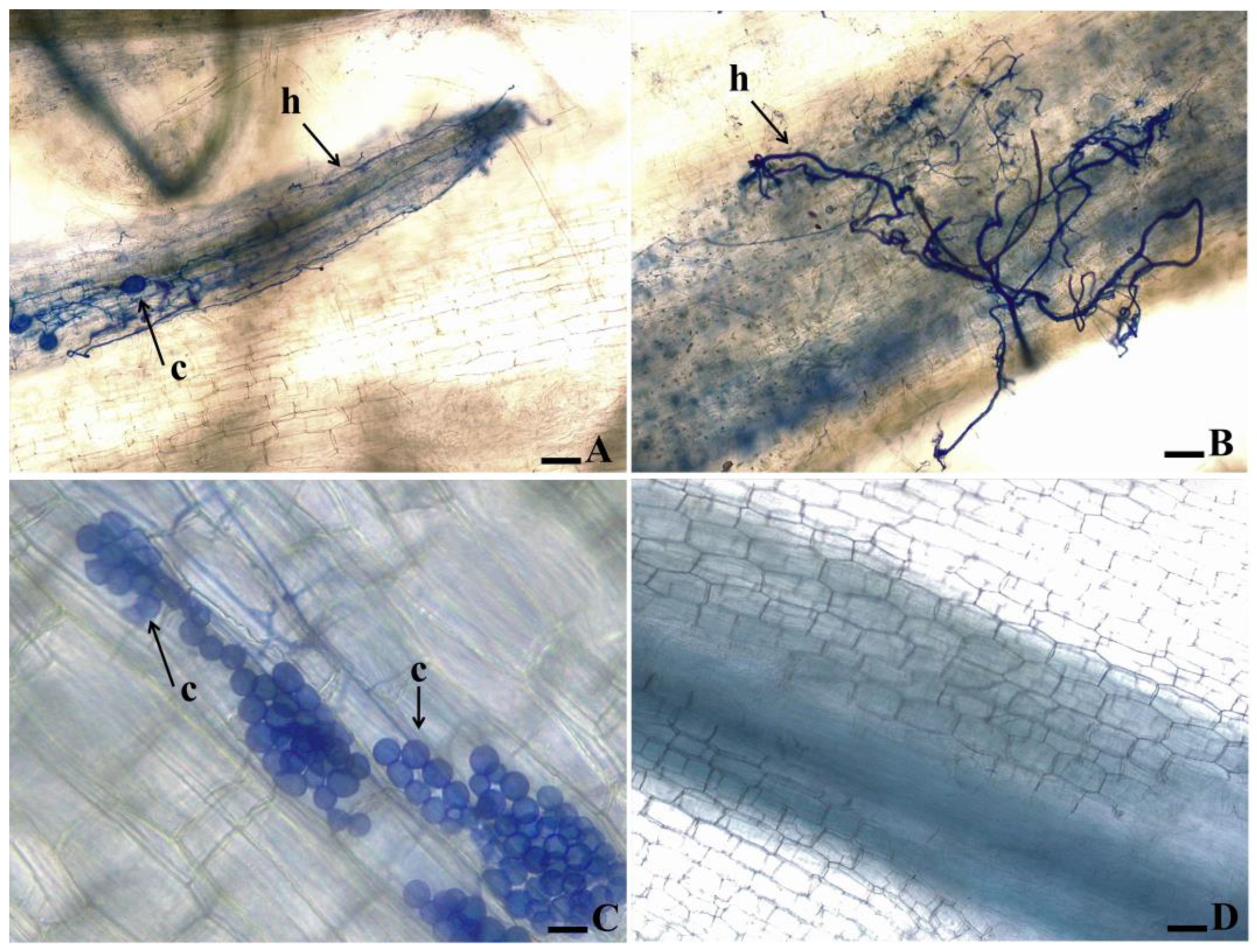
3.2. Root clarification and staining of fungal structures
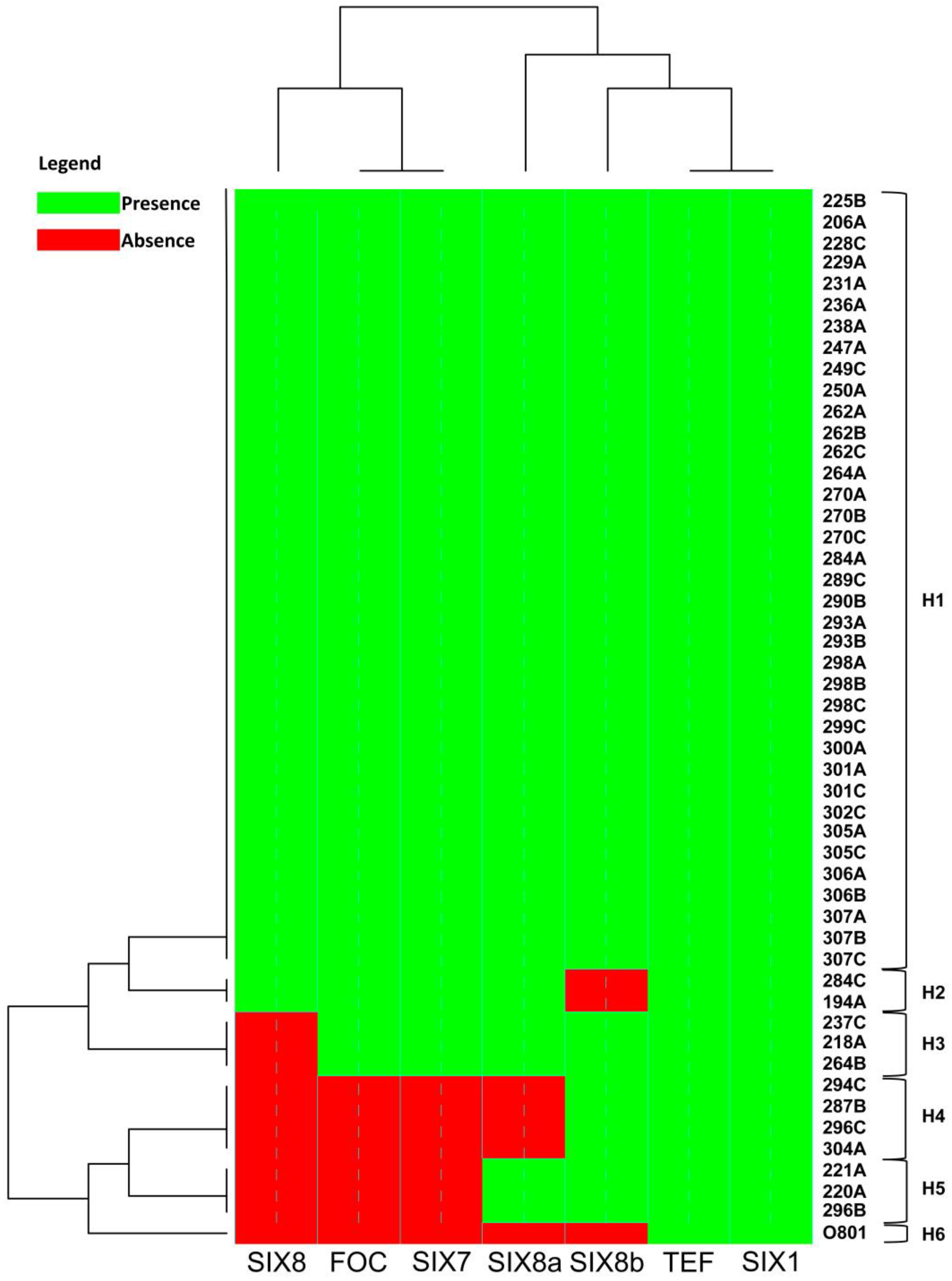
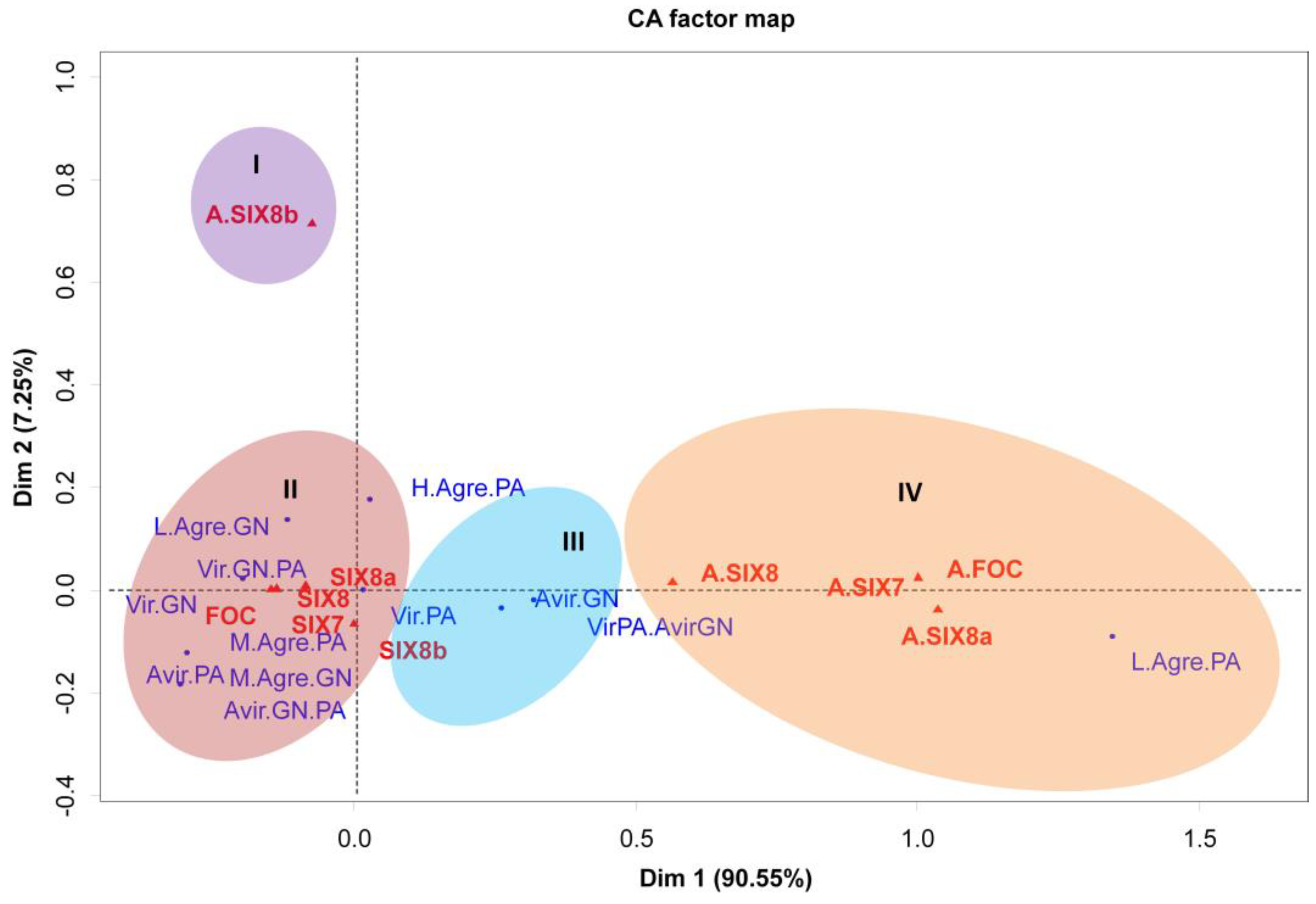
3.3. Presence of SIX genes by PCR analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. The global programme on banana fusarium wilt disease. Available online: https://www.fao.org/3/i7956e/i7956e.pdf. (accessed on 22 September 2022).
- Ploetz, R.C. Diseases and pests: A review of their importance and management. InfoMusa 2004, 13, 11–16. [Google Scholar]
- Ploetz, R.C. Fusarium Wilt of Banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef]
- Staver, C.; Pemsl, D.E.; Scheerer, L.; Perez Vicente, L.; Dita, M. Ex Ante Assessment of Returns on Research Investments to Address the Impact of Fusarium Wilt Tropical Race 4 on Global Banana Production. Front. Plant Sci. 2020, 11, 844. [Google Scholar] [CrossRef]
- Gonçalves, Z.S.; Da Invenção, D.R.S.; Ledo, C.A.D.S.; Ferreira, C.F.; Amorim, E.P. Research Article Agronomic Performance of Plantain Genotypes and Genetic Variability Using Ward-MLM Algorithm. Genet. Mol. Res. 2018, 17. [Google Scholar] [CrossRef]
- Waite, B.H.; Stover, R.H. Studies on Fusarium wilt of bananas: vi. Variability and the cultivar concept in Fusarium oxysporum f.sp. cubense. Can. J. Bot. 1960, 38, 985–994. [Google Scholar] [CrossRef]
- Pérez-Vicente, L.; Dita, M.A. Fusarium Wilt of Banana or Panama Disease by Fusarium oxysporum f. sp. cubense: A Review on History, Symptoms, Biology, Epidemiology and Management. In: Manual Prevention and Diagnostic of Fusarium Wilt (Panama Disease) of Banana Caused by Fusarium oxysporum f. sp. cubense Tropical Race 4 (TR4), Pérez-Vicente, L., Dita, M.A. Martínez-de la Parte., Eds.; FAO, Rome, 5-30.
- Dita, M.; Barquero, M.; Heck, D.; Mizubuti, E.S.G.; Staver, C.P. Fusarium Wilt of Banana: Current Knowledge on Epidemiology and Research Needs Toward Sustainable Disease Management. Front. Plant Sci. 2018, 9, 1468. [Google Scholar] [CrossRef] [PubMed]
- Ploetz, R.C. Fusarium Wilt of Banana Is Caused by Several Pathogens Referred to as Fusarium Oxysporum f. Sp. Cubense. Phytopathology® 2006, 96, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.; Fabregar, E.; Sinohin, V.G.; Herradura, L.; Fourie, G.; Viljoen, A. Confirmation of Tropical Race 4 of Fusarium oxysporum f. sp. cubense Infecting Cavendish Bananas in the Philippines. In Proceedings of the Centennial Meeting of the American Phytopathological Society, Minneapolis, MN, USA, 26–28 July 2008; Available online: https://cgspace.cgiar.org/handle/10568/674 (accessed on 2 August 2016).
- Buddenhagen, I. Understanding strain diversity in Fusarium oxysporum f. sp. cubense and history of introduction of “Tropical Race 4” to better manage banana production. Acta Hortic. 2009, No. 828, 193–204. [CrossRef]
- Ordoñez, N.; García-Bastidas, F.; Laghari, H.B.; Akkary, M.Y.; Harfouche, E.N.; Al Awar, B.N.; Kema, G.H.J. First Report of Fusarium Oxysporum f. Sp. Cubense Tropical Race 4 Causing Panama Disease in Cavendish Bananas in Pakistan and Lebanon. Plant Disease 2016, 100, 209. [Google Scholar] [CrossRef]
- Özarslandan, M.; Akgül, D.S. First Report of Fusarium Oxysporum f. Sp. Cubense Race 4 Causing Fusarium Wilt Disease of Banana in Turkey. Plant Disease 2020, 104, 974–974. [Google Scholar] [CrossRef]
- Thangavelu, R.; Mostert, D.; Gopi, M.; Devi, P.G.; Padmanaban, B.; Molina, A.B.; Viljoen, A. First Detection of Fusarium Oxysporum f. Sp. Cubense Tropical Race 4 (TR4) on Cavendish Banana in India. Eur. J. Plant Pathol. 2019, 154, 777–786. [Google Scholar] [CrossRef]
- Maryani, N.; Lombard, L.; Poerba, Y.S.; Subandiyah, S.; Crous, P.W.; Kema, G.H.J. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud. Myco. 2019, 92, 155–194. [Google Scholar] [CrossRef]
- O’Neill, W.T.; Henderson, J.; Pattemore, J.A.; O’Dwyer, C.; Perry, S.; Beasley, D.R.; Tan, Y.P.; Smyth, A.L.; Goosem, C.H.; Thomson, K.M.; Hobbs, R.L.; Grice, K.R.E.; Trevorrow, P.; Vawdrey, L.L.; Pathania, N.; Shivas, R.G. Detection of Fusarium Oxysporum f. Sp. Cubense Tropical Race 4 Strain in Northern Queensland. Australasian Plant Dis. Notes 2016, 11, 33. [Google Scholar] [CrossRef]
- García-Bastidas, F.A.; Quintero-Vargas, J.C.; Ayala-Vasquez, M.; Schermer, T.; Seidl, M.F.; Santos-Paiva, M.; Noguera, A.M.; Aguilera-Galvez, C.; Wittenberg, A.; Hofstede, R.; Sørensen, A.; Kema, G.H.J. First Report of Fusarium Wilt Tropical Race 4 in Cavendish Bananas Caused by Fusarium Odoratissimum in Colombia. Plant Disease 2020, 104, 994–994. [Google Scholar] [CrossRef]
- ICA. Instituto Colombiano Agropecuario. Available online: https://www.ica.gov.co/noticias/ica-amplia-y-refuerza-las-medidas-que-ya-venia-im. (accessed on 29 august 2019).
- Acuña, R.; Rouard, M.; Leiva, A.M.; Marques, C.; Olortegui, A.; Ureta, C.; Cabrera-Pintado, R.M.; Rojas, J.C.; López, D.; Cenci, A.; et al. First report of Fusarium oxysporum f. sp. cubense Tropical Race 4, causing Fusarium wilt in Cavendish bananas in Peru. Plant Dis. 2021, 106, 2268. [Google Scholar]
- Martínez, G.; Olivares, B.O.; Rey, J.C.; Rojas, J.; Cardenas, J.; Muentes, C.; Dawson, C. The Advance of Fusarium Wilt Tropical Race 4 in Musaceae of Latin America and the Caribbean: Current Situation. Pathogens 2023, 12, 277. [Google Scholar] [CrossRef]
- Takken, F.; Rep, M. The Arms Race between Tomato and Fusarium Oxysporum. Molecular Plant Pathology 2010, 11, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Meldrum, R.A.; Fraser-Smith, S.; Tran-Nguyen, L.T.T.; Daly, A.M.; Aitken, E.A.B. Presence of putative pathogenicity genes in isolates of Fusarium oxysporum f. sp. cubense from Australia. Australas. Plant Pathol. 2012, 41, 551–557. [Google Scholar] [CrossRef]
- Fraser-Smith, S.; Czislowski, E.; Meldrum, R.A.; Zander, M.; O’Neill, W.; Balali, G.R.; Aitken, E.A.B. Sequence Variation in the Putative Effector Gene SIX 8 Facilitates Molecular Differentiation of F Usarium Oxysporum f. Sp. Cubense. Plant Pathology 2014, 63, 1044–1052. [Google Scholar] [CrossRef]
- Czislowski, E.; Fraser-Smith, S.; Zander, M.; O’Neill, W.T.; Meldrum, R.A.; Tran-Nguyen, L.T.T.; Batley, J.; Aitken, E.A.B. Investigation of the Diversity of Effector Genes in the Banana Pathogen, Fusarium Oxysporum f. Sp. Cubense, Reveals Evidence of Horizontal Gene Transfer. Molecular Plant Pathology 2018, 19, 1155–1171. [Google Scholar] [CrossRef] [PubMed]
- Carvalhais, L.C.; Henderson, J.; Rincon-Florez, V.A.; O’Dwyer, C.; Czislowski, E.; Aitken, E.A.B.; Drenth, A. Molecular Diagnostics of Banana Fusarium Wilt Targeting Secreted-in-Xylem Genes. Front. Plant Sci. 2019, 10, 547. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.N.; Bragança, C.A.D.; Ribeiro, L.R.; Amorim, E.P.; Oliveira, S.A.S.; Dita, M.A.; Laranjeira, F.F.; Haddad, F. Genetic Structure of Fusarium Oxysporum f. Sp. Cubense in Different Regions from Brazil. Plant Pathology 2015, 64, 137–146. [Google Scholar] [CrossRef]
- Batista, I.C.; Heck, D.W.; Santos, A.; Alves, G.; Ferro, C.G.; Dita, M.; Mizubuti, E.S. The Brazilian population of Fusarium oxysporum f. sp. cubense is not structured by VCG or by geographic origin. bioRxiv 2022, 1–31. [Google Scholar]
- Ribeiro, L.R.; Oliveira, S.A.S.D.; Amorim, E.P.; Serejo, J.A.S.; Haddad, F. Sources of resistance to Fusarium oxysporum f. sp. cubense in banana germplasm. Rev. Bras. Frutic. 2018, 40, 1–8. [Google Scholar] [CrossRef]
- Rebouças, T.A.; Haddad, F.; Ferreira, C.F.; De Oliveira, S.A.S.; Da Silva Ledo, C.A.; Amorim, E.P. Identification of Banana Genotypes Resistant to Fusarium Wilt Race 1 under Field and Greenhouse Conditions. Scientia Horticulturae 2018, 239, 308–313. [Google Scholar] [CrossRef]
- Rocha, A.D.J.; Ferreira, M.D.S.; Rocha, L.D.S.; Oliveira, S.A.; Amorim, E.P.; Mizubuti, E.S.; Haddad, F. Interaction between Fusarium oxysporum f. sp. cubense and Radopholus similis can lead to changes in the resistance of banana cultivars to Fusarium wilt. Eur. J. Plant Pathol. 2020, 158, 403–417. [Google Scholar]
- Dita, M.A.; Garming, H.; Van den Bergh, I.; Staver, C.; Lescot, T. Banana in Latin America and the Caribbean: Current state, challenges and perspectives. Acta Hortic. 2011, 986, 365–380. [Google Scholar] [CrossRef]
- Dita, M.A.; Pérez Vicente, L.; Martínez, E. Inoculation of Fusarium oxysporum f. sp. cubense causal agent of fusarium wilt in banana. In Technical Manual: Prevention and Diagnostic of Fusarium Wilt of Banana Caused by Fusarium Oxysporum f. sp. cubense Tropical Race 4 (TR4); Pérez Vicente, L., Dita, M.A., de la Parte, E.M., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014; pp. 55–58. [Google Scholar]
- McKinney, H.H. Influence of Soil Temperature and Moisture on Infection of Wheat Seedlings by Helminthosporium sativum. J. Agric. Res. 1923, 26, 195. [Google Scholar]
- Phillips, J.M.; Hayman, D.S. Improved Procedures for Clearing Roots and Staining Parasitic and Vesicular-Arbuscular Mycorrhizal Fungi for Rapid Assessment of Infection. Transactions of the British Mycological Society 1970, 55, 158–IN18. [Google Scholar] [CrossRef]
- Brundrett, M.; Bougher, N.; Dell, B.; Grove, T.; Malajczuk, N. (Eds.) Working with Mycorrhizas in Forestry and Agriculture; ACIAR Monograph 32: Canberra, Australia, 2006; p. 374. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Dita, M.A.; Waalwijk, C.; Buddenhagen, I.W.; Souza, J.T.; Kema, G.H.J. A molecular diagnostic for tropical race 4 of the banana Fusarium wilt pathogen. Plant. Pathol. 2010, 59, 348–357. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Chang, J.-Y.; Liu, E.-T.; Chao, C.-P.; Huang, J.-W.; Chang, P.-F.L. Development of a Molecular Marker for Specific Detection of Fusarium Oxysporum f. Sp. Cubense Race 4. Eur. J. Plant Pathol. 2009, 123, 353–365. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple Evolutionary Origins of the Fungus Causing Panama Disease of Banana: Concordant Evidence from Nuclear and Mitochondrial Gene Genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; The R Foundation for Statistical Computing: Vienna, Austria, 2020; Volume 1. [Google Scholar]
- Vale, F.X.R.D.; Parlevliet, J.E.; Zambolim, L. Concepts in plant disease resistance. Fitopatol. Bras. 2001, 26, 577–589. [Google Scholar] [CrossRef]
- Ma, L.-J.; Van Der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Rocha, A.D.J.; Soares, J.M.D.S.; Nascimento, F.D.S.; Rocha, A.D.S.; Amorim, V.B.O.D.; Ramos, A.P.D.S.; Ferreira, C.F.; Haddad, F.; Amorim, E.P. Molecular, Histological and Histochemical Responses of Banana Cultivars Challenged with Fusarium Oxysporum f. Sp. Cubense with Different Levels of Virulence. Plants 2022, 11, 2339. [Google Scholar] [CrossRef]
- Fourie, G.; Steenkamp, E.T.; Ploetz, R.C.; Gordon, T.R.; Viljoen, A. Current Status of the Taxonomic Position of Fusarium Oxysporum Formae Specialis Cubense within the Fusarium Oxysporum Complex. Infection, Genetics and Evolution 2011, 11, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Czislowski, E.; Fraser-Smith, S.; Zander, M.; Aitken, E.A.B. Identifying Pathogenicity Genes in Fusarium Oxysporum f. Sp. Cubense. Acta Hortic. 2016, No. 1114, 101–106. [Google Scholar] [CrossRef]
- Maldonado, B.L.D.; Villarruel, O.J.L.; Calderón, O.M.A.; Sánchez, E.A.C. Secreted in Xylem (Six) genes in Fusarium oxysporum f. sp. cubense and their potential acquisition by horizontal transfer. Adv Biotech & Micro 2018, 10, 555779. [Google Scholar]
- Widinugraheni, S.; Niño-Sánchez, J.; Van Der Does, H.C.; Van Dam, P.; García-Bastidas, F.A.; Subandiyah, S.; Meijer, H.J.G.; Kistler, H.C.; Kema, G.H.J.; Rep, M. A SIX1 Homolog in Fusarium Oxysporum f.Sp. Cubense Tropical Race 4 Contributes to Virulence towards Cavendish Banana. PLoS ONE 2018, 13, e0205896. [Google Scholar] [CrossRef]
- An, B.; Hou, X.; Guo, Y.; Zhao, S.; Luo, H.; He, C.; Wang, Q. The effector SIX8 is required for virulence of Fusarium oxysporum f.sp. cubense tropical race 4 to Cavendish banana. Fungal. Biol. 2019, 123, 423–430. [Google Scholar] [PubMed]
- Stover, R.H. Fusarial Wilt (Panama Disease) of Bananas and Other Musa Species; Commonwealth Mycological Institute: Kew, UK, 1962; p. 117. [Google Scholar]
- Hall, C.; Heath, R.; Guest, D. The Infection Process of Fusarium Oxysporum f.Sp. Vasinfectum in Australian Cotton. Australasian Plant Pathol. 2013, 42, 1–8. [Google Scholar] [CrossRef]
- Li, C.; Chen, S.; Zuo, C.; Sun, Q.; Ye, Q.; Yi, G.; Huang, B. The Use of GFP-Transformed Isolates to Study Infection of Banana with Fusarium Oxysporum f. Sp. Cubense Race 4. Eur. J. Plant Pathol. 2011, 131, 327–340. [Google Scholar] [CrossRef]
- Li, C.; Yang, J.; Li, W.; Sun, J.; Peng, M. Direct Root Penetration and Rhizome Vascular Colonization by Fusarium Oxysporum f. Sp. Cubense Are the Key Steps in the Successful Infection of Brazil Cavendish. Plant Disease 2017, 101, 2073–2078. [Google Scholar] [CrossRef]
- Carlier, J.; Hayden, H.; Rivas, G.; Zapater, M. F. Genetic differentiation in Mycosphaerella leaf spot pathogens. In Mycosphaerella Leaf Spot Diseases of Bananas: Present Status and Outlook. Proceedings of the Workshop on Mycosphaerella Leaf Spot Diseases, San José, Costa Rica, 20-23 May 2002; Jacome L, Lepoivre P, Marin D, Ortiz R, et al., Eds. Montpellier: The International Network for the Improvement of Banana and Plantain 123-129.
- Groenewald, S.; Van Den Berg, N.; Marasas, W.F.O.; Viljoen, A. The Application of High-Throughput AFLP’s in Assessing Genetic Diversity in Fusarium Oxysporum f. Sp. Cubense. Mycological Research 2006, 110, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Truman, W.; Bennett, M.H.; Kubigsteltig, I.; Turnbull, C.; Grant, M. Arabidopsis Systemic Immunity Uses Conserved Defense Signaling Pathways and Is Mediated by Jasmonates. Proc. Natl. Acad. Sci. USA 2007, 104, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Karangwa, P.; Mostert, D.; Ndayihanzamaso, P.; Dubois, T.; Niere, B.; Zum Felde, A.; Schouten, A.; Blomme, G.; Beed, F.; Viljoen, A. Genetic Diversity of Fusarium Oxysporum f. Sp. Cubense in East and Central Africa. Plant Disease 2018, 102, 552–560. [Google Scholar] [CrossRef]
- Cunha, C.M.S.; Hinz, R.H.; Pereira, A.; Tcacenco, F.A.; Stadnik, M.J. Aggressiveness and genetic diversity of Fusarium oxysporum f. sp. cubense from Santa Catarina, southern Brazil. Trop. Plant Pathol. 2015, 40, 326–334. [Google Scholar]
| Primer | Amplicon length (bp) | Name | Sequence (5′ -3′) |
|---|---|---|---|
| SIX1* | 260 | SIX 1 – F | ATGGTACTCCTTGGCGCCCTC |
| SIX 1 – R | TGACAATGCGACCACGCCTCG | ||
| FOC** | 242 | FOC 1- F | CAGGGGATGTATGAGGAGGCT |
| FOC 2 – R | GTGACAGCGTCGTCTAGTTCC | ||
| SIX2* | 660 | SIX 2 – F | ACGACCTGGGCCATCTCGGT |
| SIX 2 – R | ACACCTTGACTGCGACGCAACG | ||
| SIX3* | 555 | SIX 3 – F | ACCGACCATCTTGCCTAAACATTTACC |
| SIX 3 – R | TTAACCACTCTGCCAAGGGGAACT | ||
| SIX4* | 808 | SIX 4 – F | TGCTTCGGTGGCTGTTACATCTGC |
| SIX 4 – R | CCTAACCTAAGCTCACCCTCAGGAA | ||
| SIX5* | 326 | SIX 5 – F | TGCGCTTCGAGTACATCTCTGTTC |
| SIX 5 – R | CTGGTGAGATTTAGAGCAGTCAAAGCA | ||
| SIX6* | 611 | SIX 6 – F | GGCTGCGTAGCTGGTCCCCT |
| SIX 6 – R | CATGTCATGAATGTACGCATGTCCCT | ||
| SIX7* | 610 | SIX 7 – F | ACCTTTACCTCCTTTTCCATTTCGCCC |
| SIX 7 – R | CGAAAGTCAGCAAGGCCCCTGG | ||
| SIX8* | 250 | SIX 8 – F | TCGCCTGCATAACAGGTGCCG |
| SIX 8 – R | TTGTGTAGAAACTGGACAGTCGATGC | ||
| SIX8a*** | 770 | Foc-SIX8-F | CGAAGTGCGCCATATAAGACT |
| Foc-SIX8-R | CACCTGCTTGCTCCTTATCC | ||
| SIX8b*** | 595 | Foc-SIX8b-F | CGTCCTTACTTATATACCCTCTCAA |
| Foc-SIX8b-R | GGCCTAATCCACACAACA | ||
| FocTR4**** | 463 | FocTR4 – F | CACGTTTAAGGTGCCATGAGAG |
| FocTR4 – R | GCCAGGACTGCCTCGTGA | ||
| TEF-1α | 650 | EF1 | ATGGGTAAGGARGACAAGAC |
| EF2 | GGARGTACCAGTSATCATGTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





