Submitted:
09 February 2024
Posted:
14 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Functions of Cholesterol in the Retina
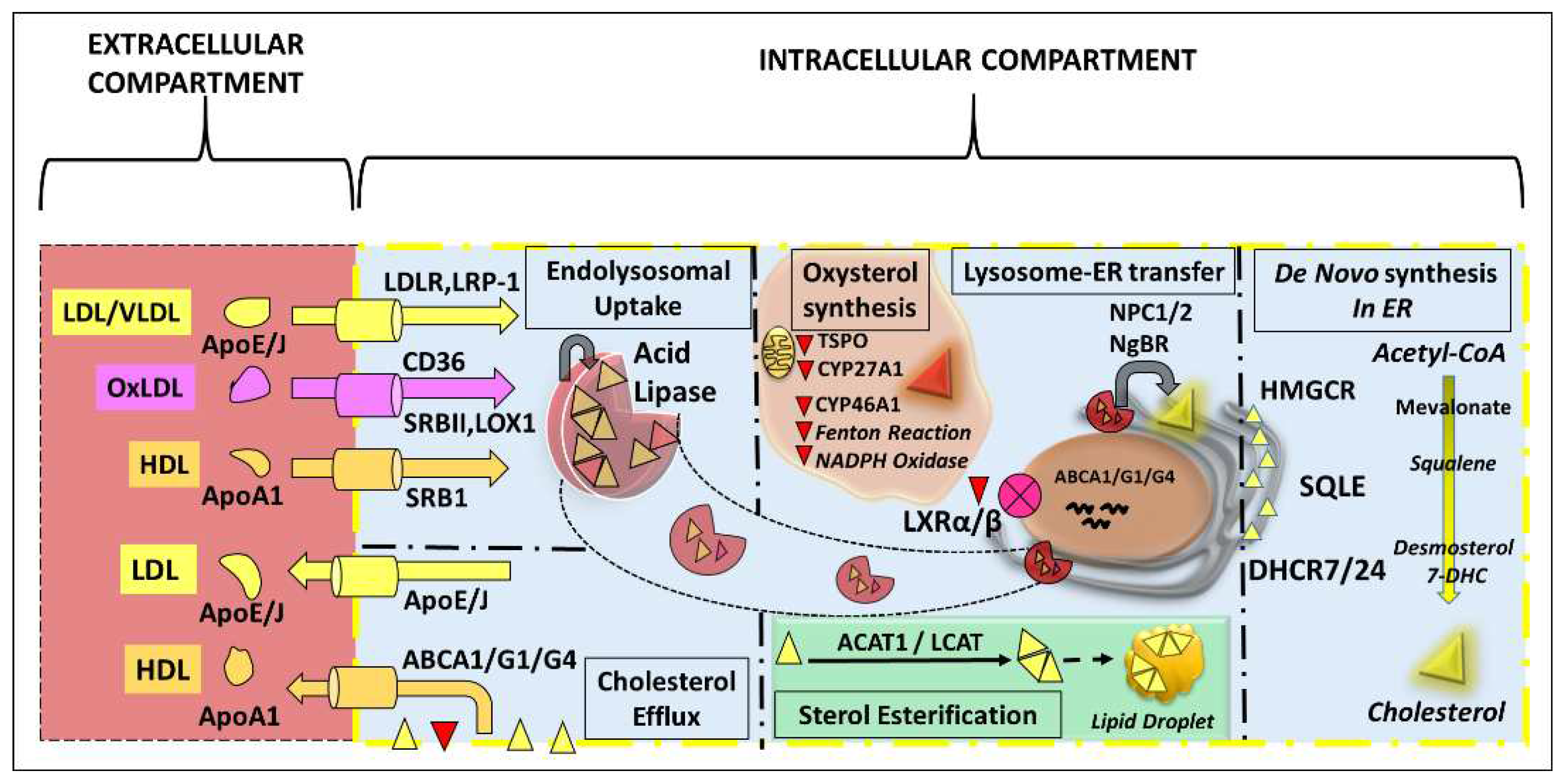
3. De Novo Sterol Synthesis in the Retina
4. Biological Considerations in Measuring Tissue Sterol Synthetic Rates Using Radioisotope Approach
5. Technical and Biological Challenges in Achieving Tissue/Cell Type-Targeted Inhibition of Cholesterol Synthesis
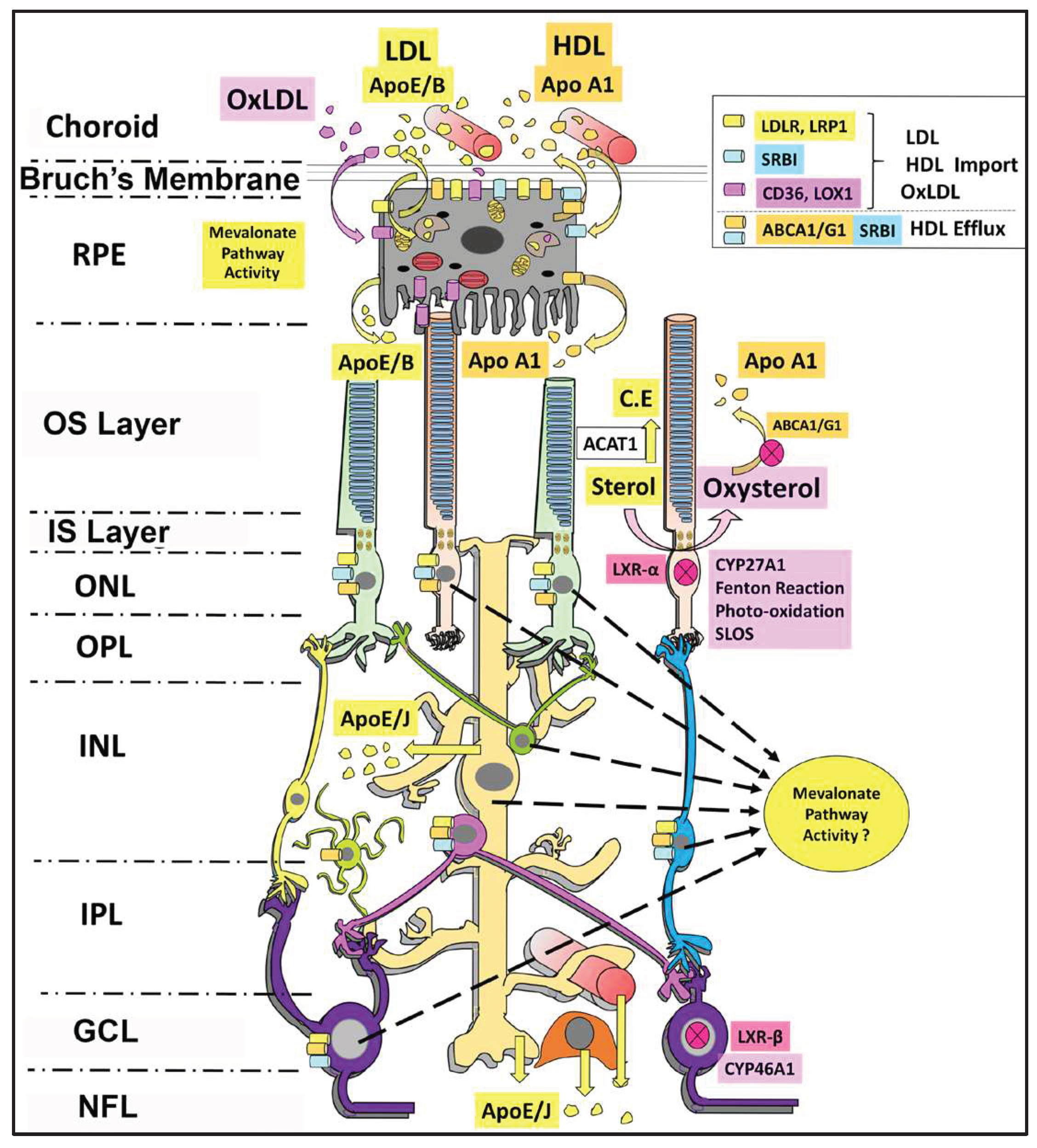
6. Sterol Uptake by the Retina
7. Sterol Efflux from the Retina
8. Strategies to Measure Sterol Uptake and Efflux Rates in the Neuronal Retina
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fliesler, S.J.; Anderson, R.E. Chemistry and metabolism of lipids in the vertebrate retina. Prog Lipid Res 1983, 22, 79–131. [Google Scholar] [CrossRef] [PubMed]
- Porter, F.D. Human malformation syndromes due to inborn errors of cholesterol synthesis. Curr Opin Pediatr. 2003, 15, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Haas, D.; Kelley, R.I.; Hoffmann, G.F. Inherited disorders of cholesterol biosynthesis. Neuropediatrics 2001, 32, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Skeyni, A.; Pradignac, A.; Matz, R.L.; Terrand, J.; Boucher, P. Cholesterol Trafficking, Lysosomal Function and Atherosclerosis. Am J Physiol Cell Physiol 2023. [Google Scholar] [CrossRef]
- Li, B.; Goss, D.; Miller, J.W.; Lin, J.B.; Vavvas, D.G. Systemic Dyslipidemia in Age-related Macular Degeneration: An Updated Systematic Review and Meta-analysis. Ophthalmol Sci 2024, 4, 100341. [Google Scholar] [CrossRef]
- Busik, J.V. Lipid metabolism dysregulation in diabetic retinopathy. J Lipid Res 2021, 62, 100017. [Google Scholar] [CrossRef]
- Fliesler, S.J.; Bretillon, L. The ins and outs of cholesterol in the vertebrate retina. J Lipid Res 2010, 51, 3399–3413. [Google Scholar] [CrossRef]
- Ramachandra Rao, S.; Fliesler, S.J. Cholesterol homeostasis in the vertebrate retina: biology and pathobiology. J Lipid Res 2021, 62, 100057. [Google Scholar] [CrossRef]
- Pikuleva, I.A.; Curcio, C.A. Cholesterol in the retina: the best is yet to come. Prog Retin Eye Res 2014, 41, 64–89. [Google Scholar] [CrossRef]
- Cavodeassi, F.; Creuzet, S.; Etchevers, H.C. The hedgehog pathway and ocular developmental anomalies. Hum Genet 2019, 138, 917–936. [Google Scholar] [CrossRef] [PubMed]
- Elliott, M.H.; Nash, Z.A.; Takemori, N.; Fliesler, S.J.; McClellan, M.E.; Naash, M.I. Differential distribution of proteins and lipids in detergent-resistant and detergent-soluble domains in rod outer segment plasma membranes and disks. J Neurochem 2008, 104, 336–352. [Google Scholar] [CrossRef] [PubMed]
- Elliott, M.H.; Fliesler, S.J.; Ghalayini, A.J. Cholesterol-dependent association of caveolin-1 with the transducin alpha subunit in bovine photoreceptor rod outer segments: disruption by cyclodextrin and guanosine 5'-O-(3-thiotriphosphate). Biochemistry 2003, 42, 7892–7903. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, X.; Zhang, L.; He, F.; Zhang, G.; Jamrich, M.; Wensel, T.G. Activation-dependent hindrance of photoreceptor G protein diffusion by lipid microdomains. J Biol Chem 2008, 283, 30015–30024. [Google Scholar] [CrossRef]
- Zheng, W.; Reem, R.E.; Omarova, S.; Huang, S.; DiPatre, P.L.; Charvet, C.D.; Curcio, C.A.; Pikuleva, I.A. Spatial distribution of the pathways of cholesterol homeostasis in human retina. PLoS One 2012, 7, e37926. [Google Scholar] [CrossRef]
- Boesze-Battaglia, K.; Hennessey, T.; Albert, A.D. Cholesterol heterogeneity in bovine rod outer segment disk membranes. J Biol Chem 1989, 264, 8151–8155. [Google Scholar] [CrossRef]
- Lakkaraju, A.; Umapathy, A.; Tan, L.X.; Daniele, L.; Philp, N.J.; Boesze-Battaglia, K.; Williams, D.S. The cell biology of the retinal pigment epithelium. Prog Retin Eye Res 2020, 100846. [Google Scholar] [CrossRef]
- Toops, K.A.; Tan, L.X.; Jiang, Z.; Radu, R.A.; Lakkaraju, A. Cholesterol-mediated activation of acid sphingomyelinase disrupts autophagy in the retinal pigment epithelium. Mol Biol Cell 2015, 26, 1–14. [Google Scholar] [CrossRef]
- Caldwell, R.B.; McLaughlin, B.J. Freeze-fracture study of filipin binding in photoreceptor outer segments and pigment epithelium of dystrophic and normal retinas. J Comp Neurol 1985, 236, 523–537. [Google Scholar] [CrossRef]
- Rai, A.; Pathak, D.; Thakur, S.; Singh, S.; Dubey, A.K.; Mallik, R. Dynein Clusters into Lipid Microdomains on Phagosomes to Drive Rapid Transport toward Lysosomes. Cell 2016, 164, 722–734. [Google Scholar] [CrossRef]
- Ramachandra Rao, S.; Pfeffer, B.A.; Mas Gomez, N.; Skelton, L.A.; Keiko, U.; Sparrow, J.R.; Rowsam, A.M.; Mitchell, C.H.; Fliesler, S.J. Compromised phagosome maturation underlies RPE pathology in cell culture and whole animal models of Smith-Lemli-Opitz Syndrome. Autophagy 2018, 14, 1796–1817. [Google Scholar] [CrossRef]
- Mauch, D.H.; Nagler, K.; Schumacher, S.; Goritz, C.; Muller, E.C.; Otto, A.; Pfrieger, F.W. CNS synaptogenesis promoted by glia-derived cholesterol. Science 2001, 294, 1354–1357. [Google Scholar] [CrossRef]
- Mercer, A.J.; Szalewski, R.J.; Jackman, S.L.; Van Hook, M.J.; Thoreson, W.B. Regulation of presynaptic strength by controlling Ca2+ channel mobility: effects of cholesterol depletion on release at the cone ribbon synapse. J Neurophysiol 2012, 107, 3468–3478. [Google Scholar] [CrossRef]
- Thoreson, W.B.; Mercer, A.J.; Cork, K.M.; Szalewski, R.J. Lateral mobility of L-type calcium channels in synaptic terminals of retinal bipolar cells. Mol Vis 2013, 19, 16–24. [Google Scholar]
- Amaratunga, A.; Abraham, C.R.; Edwards, R.B.; Sandell, J.H.; Schreiber, B.M.; Fine, R.E. Apolipoprotein E is synthesized in the retina by Muller glial cells, secreted into the vitreous, and rapidly transported into the optic nerve by retinal ganglion cells. J Biol Chem 1996, 271, 5628–5632. [Google Scholar] [CrossRef]
- Shanmugaratnam, J.; Berg, E.; Kimerer, L.; Johnson, R.J.; Amaratunga, A.; Schreiber, B.M.; Fine, R.E. Retinal Muller glia secrete apolipoproteins E and J which are efficiently assembled into lipoprotein particles. Brain Res Mol Brain Res 1997, 50, 113–120. [Google Scholar] [CrossRef]
- Fliesler, S.J.; Keller, R.K. Isoprenoid metabolism in the vertebrate retina. Int J Biochem Cell Biol 1997, 29, 877–894. [Google Scholar] [CrossRef]
- Fliesler, S.J.; Schroepfer, G.J., Jr. Metabolism of mevalonic acid in cell-free homogenates of bovine retinas. Formation of novel isoprenoid acids. J Biol Chem 1983, 258, 15062–15070. [Google Scholar] [CrossRef]
- Fliesler, S.J.; Schroepfer, G.J., Jr. In vitro metabolism of mevalonic acid in the bovine retina. J Neurochem 1986, 46, 448–460. [Google Scholar] [CrossRef]
- Fliesler, S.J.; Florman, R.; Rapp, L.M.; Pittler, S.J.; Keller, R.K. In vivo biosynthesis of cholesterol in the rat retina. FEBS Lett 1993, 335, 234–238. [Google Scholar] [CrossRef]
- Fliesler, S.J.; Florman, R.; Keller, R.K. Isoprenoid lipid metabolism in the retina: dynamics of squalene and cholesterol incorporation and turnover in frog rod outer segment membranes. Exp Eye Res 1995, 60, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Fliesler, S.J.; Keller, R.K. Metabolism of [3H]farnesol to cholesterol and cholesterogenic intermediates in the living rat eye. Biochem Biophys Res Commun 1995, 210, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Keller, R.K.; Fliesler, S.J.; Nellis, S.W. Isoprenoid biosynthesis in the retina. Quantitation of the sterol and dolichol biosynthetic pathways. J Biol Chem 1988, 263, 2250–2254. [Google Scholar] [CrossRef] [PubMed]
- Bjorkhem, I.; Meaney, S. Brain cholesterol: long secret life behind a barrier. Arterioscler Thromb Vasc Biol 2004, 24, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.B.; Mast, N.; Bederman, I.R.; Li, Y.; Brunengraber, H.; Bjorkhem, I.; Pikuleva, I.A. Cholesterol in mouse retina originates primarily from in situ de novo biosynthesis. J Lipid Res 2016, 57, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Tserentsoodol, N.; Sztein, J.; Campos, M.; Gordiyenko, N.V.; Fariss, R.N.; Lee, J.W.; Fliesler, S.J.; Rodriguez, I.R. Uptake of cholesterol by the retina occurs primarily via a low density lipoprotein receptor-mediated process. Mol Vis 2006, 12, 1306–1318. [Google Scholar] [PubMed]
- Clark, B.S.; Stein-O'Brien, G.L.; Shiau, F.; Cannon, G.H.; Davis-Marcisak, E.; Sherman, T.; Santiago, C.P.; Hoang, T.V.; Rajaii, F.; James-Esposito, R.E.; et al. Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron 2019, 102, 1111–1126. [Google Scholar] [CrossRef] [PubMed]
- Voigt, A.P.; Whitmore, S.S.; Lessing, N.D.; DeLuca, A.P.; Tucker, B.A.; Stone, E.M.; Mullins, R.F.; Scheetz, T.E. Spectacle: An interactive resource for ocular single-cell RNA sequencing data analysis. Exp Eye Res 2020, 200, 108204. [Google Scholar] [CrossRef] [PubMed]
- Castro-Perez, J.; Previs, S.F.; McLaren, D.G.; Shah, V.; Herath, K.; Bhat, G.; Johns, D.G.; Wang, S.P.; Mitnaul, L.; Jensen, K.; et al. In vivo D2O labeling to quantify static and dynamic changes in cholesterol and cholesterol esters by high resolution LC/MS. J Lipid Res 2011, 52, 159–169. [Google Scholar] [CrossRef]
- Kim, T.Y.; Wang, D.; Kim, A.K.; Lau, E.; Lin, A.J.; Liem, D.A.; Zhang, J.; Zong, N.C.; Lam, M.P.; Ping, P. Metabolic labeling reveals proteome dynamics of mouse mitochondria. Mol Cell Proteomics 2012, 11, 1586–1594. [Google Scholar] [CrossRef]
- Lee, W.N.; Bassilian, S.; Ajie, H.O.; Schoeller, D.A.; Edmond, J.; Bergner, E.A.; Byerley, L.O. In vivo measurement of fatty acids and cholesterol synthesis using D2O and mass isotopomer analysis. Am J Physiol 1994, 266, E699–E708. [Google Scholar] [CrossRef]
- Diraison, F.; Pachiaudi, C.; Beylot, M. In vivo measurement of plasma cholesterol and fatty acid synthesis with deuterated water: determination of the average number of deuterium atoms incorporated. Metabolism 1996, 45, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Di Buono, M.; Jones, P.J.; Beaumier, L.; Wykes, L.J. Comparison of deuterium incorporation and mass isotopomer distribution analysis for measurement of human cholesterol biosynthesis. J Lipid Res 2000, 41, 1516–1523. [Google Scholar] [CrossRef]
- Keller, R.K.; Small, M.; Fliesler, S.J. Enzyme blockade: a nonradioactive method to determine the absolute rate of cholesterol synthesis in the brain. J Lipid Res 2004, 45, 1952–1957. [Google Scholar] [CrossRef]
- Pittler, S.J.; Fliesler, S.J.; Rapp, L.M. Novel morphological changes in rat retina induced by intravitreal injection of lovastatin. Exp Eye Res 1992, 54, 149–152. [Google Scholar] [CrossRef]
- Pittler, S.J.; Fliesler, S.J.; Fisher, P.L.; Keller, P.K.; Rapp, L.M. In vivo requirement of protein prenylation for maintenance of retinal cytoarchitecture and photoreceptor structure. J Cell Biol 1995, 130, 431–439. [Google Scholar] [CrossRef]
- Fitzky, B.U.; Moebius, F.F.; Asaoka, H.; Waage-Baudet, H.; Xu, L.; Xu, G.; Maeda, N.; Kluckman, K.; Hiller, S.; Yu, H.; et al. 7-Dehydrocholesterol-dependent proteolysis of HMG-CoA reductase suppresses sterol biosynthesis in a mouse model of Smith-Lemli-Opitz/RSH syndrome. J Clin Invest 2001, 108, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Kanuri, B.; Fong, V.; Ponny, S.R.; Tallman, K.A.; Rao, S.R.; Porter, N.; Fliesler, S.J.; Patel, S.B. Generation and validation of a conditional knockout mouse model for the study of the Smith-Lemli-Opitz syndrome. J Lipid Res 2021, 62, 100002. [Google Scholar] [CrossRef] [PubMed]
- Wassif, C.A.; Brownson, K.E.; Sterner, A.L.; Forlino, A.; Zerfas, P.M.; Wilson, W.K.; Starost, M.F.; Porter, F.D. HEM dysplasia and ichthyosis are likely laminopathies and not due to 3beta-hydroxysterol Delta14-reductase deficiency. Hum Mol Genet 2007, 16, 1176–1187. [Google Scholar] [CrossRef]
- Li, X.; Roberti, R.; Blobel, G. Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum. Nature 2015, 517, 104–107. [Google Scholar] [CrossRef]
- Gc, J.B.; Chen, J.; Pokharel, S.M.; Mohanty, I.; Mariasoosai, C.; Obi, P.; Panipinto, P.; Bandyopadhyay, S.; Bose, S.; Natesan, S. Molecular basis for the recognition of 24-(S)-hydroxycholesterol by integrin alphavbeta3. Sci Rep 2023, 13, 9166. [Google Scholar] [CrossRef]
- Cocciadiferro, D.; Mazza, T.; Vecchio, D.; Biagini, T.; Petrizzelli, F.; Agolini, E.; Villani, A.; Minervino, D.; Martinelli, D.; Rizzo, C.; et al. Exploiting in silico structural analysis to introduce emerging genotype-phenotype correlations in DHCR24-related sterol biosynthesis disorder: a case study. Front Genet 2023, 14, 1307934. [Google Scholar] [CrossRef] [PubMed]
- Korade, Z.; Kenworthy, A.K.; Mirnics, K. Molecular consequences of altered neuronal cholesterol biosynthesis. J Neurosci Res 2009, 87, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Song, A.J.; Palmiter, R.D. Detecting and Avoiding Problems When Using the Cre-lox System. Trends Genet 2018, 34, 333–340. [Google Scholar] [CrossRef]
- St-Onge, L.; Furth, P.A.; Gruss, P. Temporal control of the Cre recombinase in transgenic mice by a tetracycline responsive promoter. Nucleic Acids Res 1996, 24, 3875–3877. [Google Scholar] [CrossRef]
- Luo, L.; Ambrozkiewicz, M.C.; Benseler, F.; Chen, C.; Dumontier, E.; Falkner, S.; Furlanis, E.; Gomez, A.M.; Hoshina, N.; Huang, W.H.; et al. Optimizing Nervous System-Specific Gene Targeting with Cre Driver Lines: Prevalence of Germline Recombination and Influencing Factors. Neuron 2020, 106, 37–65. [Google Scholar] [CrossRef] [PubMed]
- Madisen, L.; Zwingman, T.A.; Sunkin, S.M.; Oh, S.W.; Zariwala, H.A.; Gu, H.; Ng, L.L.; Palmiter, R.D.; Hawrylycz, M.J.; Jones, A.R.; et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010, 13, 133–140. [Google Scholar] [CrossRef]
- Ramachandra Rao, S.; Fliesler, S.J. A simple, rapid fluorescent reporter-based method for detection of ectopic cre recombinase expression in presumed retinal cell type-targeted mouse lines. Exp Eye Res 2023, 235, 109637. [Google Scholar] [CrossRef]
- Choi, E.H.; Suh, S.; Einstein, D.E.; Leinonen, H.; Dong, Z.; Rao, S.R.; Fliesler, S.J.; Blackshaw, S.; Yu, M.; Peachey, N.S.; et al. An inducible Cre mouse for studying roles of the RPE in retinal physiology and disease. JCI Insight 2021, 6. [Google Scholar] [CrossRef]
- Prasov, L.; Glaser, T. Pushing the envelope of retinal ganglion cell genesis: context dependent function of Math5 (Atoh7). Dev Biol 2012, 368, 214–230. [Google Scholar] [CrossRef]
- Gregorian, C.; Nakashima, J.; Le Belle, J.; Ohab, J.; Kim, R.; Liu, A.; Smith, K.B.; Groszer, M.; Garcia, A.D.; Sofroniew, M.V.; et al. Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. J Neurosci 2009, 29, 1874–1886. [Google Scholar] [CrossRef]
- Ramachandra Rao, S.; Skelton, L.A.; Wu, F.; Onysk, A.; Spolnik, G.; Danikiewicz, W.; Butler, M.C.; Stacks, D.A.; Surmacz, L.; Mu, X.; et al. Retinal Degeneration Caused by Rod-Specific Dhdds Ablation Occurs without Concomitant Inhibition of Protein N-Glycosylation. iScience 2020, 23, 101198. [Google Scholar] [CrossRef] [PubMed]
- Essner, E.; Gordon, S.R. Observations on the permeability of the choriocapillaris of the eye. Cell Tissue Res 1983, 231, 571–577. [Google Scholar] [CrossRef]
- Yang, R.; Liu, Q.; Zhang, M. The Past and Present Lives of the Intraocular Transmembrane Protein CD36. Cells 2022, 12, 171. [Google Scholar] [CrossRef]
- Noske, U.M.; Schmidt-Erfurth, U.; Meyer, C.; Diddens, H. Lipid metabolism in retinal pigment epithelium. Possible significance of lipoprotein receptors. Ophthalmologe 1998, 95, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.G.; Bailey, K.R.; Kane, J.P.; Schwartz, D.M. Human retinal pigment epithelial cells express scavenger receptors BI and BII. Biochem Biophys Res Commun 2002, 292, 1017–1022. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Jiang, A.; Liang, J.; Meng, H.; Chang, B.; Gao, H.; Qiao, X. Expression of VLDLR in the retina and evolution of subretinal neovascularization in the knockout mouse model's retinal angiomatous proliferation. Invest Ophthalmol Vis Sci 2008, 49, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Hayes, K.C.; Lindsey, S.; Stephan, Z.F.; Brecker, D. Retinal pigment epithelium possesses both LDL and scavenger receptor activity. Invest Ophthalmol Vis Sci 1989, 30, 225–232. [Google Scholar] [PubMed]
- Gordiyenko, N.; Campos, M.; Lee, J.W.; Fariss, R.N.; Sztein, J.; Rodriguez, I.R. RPE cells internalize low-density lipoprotein (LDL) and oxidized LDL (oxLDL) in large quantities in vitro and in vivo. Invest Ophthalmol Vis Sci 2004, 45, 2822–2829. [Google Scholar] [CrossRef]
- Fliesler, S.J.; Peachey, N.S.; Herron, J.; Hines, K.M.; Weinstock, N.I.; Ramachandra Rao, S.; Xu, L. Prevention of Retinal Degeneration in a Rat Model of Smith-Lemli-Opitz Syndrome. Sci Rep 2018, 8, 1286. [Google Scholar] [CrossRef]
- Mast, N.; Bederman, I.R.; Pikuleva, I.A. Retinal Cholesterol Content Is Reduced in Simvastatin-Treated Mice Due to Inhibited Local Biosynthesis Albeit Increased Uptake of Serum Cholesterol. Drug Metab Dispos 2018, 46, 1528–1537. [Google Scholar] [CrossRef]
- Mast, N.; El-Darzi, N.; Li, Y.; Pikuleva, I.A. Quantitative characterizations of the cholesterol-related pathways in the retina and brain of hamsters. J Lipid Res 2023, 64, 100401. [Google Scholar] [CrossRef]
- El-Darzi, N.; Mast, N.; Dailey, B.; Denker, J.; Li, Y.; Vance, J.; Pikuleva, I.A. Characterizations of Hamster Retina as a Model for Studies of Retinal Cholesterol Homeostasis. Biology (Basel) 2021, 10, 1003. [Google Scholar] [CrossRef] [PubMed]
- Tserentsoodol, N.; Gordiyenko, N.V.; Pascual, I.; Lee, J.W.; Fliesler, S.J.; Rodriguez, I.R. Intraretinal lipid transport is dependent on high density lipoprotein-like particles and class B scavenger receptors. Mol Vis 2006, 12, 1319–1333. [Google Scholar] [PubMed]
- Pikuleva, I.A. Cholesterol-metabolizing cytochromes P450. Drug Metab Dispos 2006, 34, 513–520. [Google Scholar] [CrossRef]
- Farhan, F.; Almarhoun, M.; Wong, A.; Findlay, A.S.; Bartholomew, C.; Williams, M.T.S.; Hurd, T.W.; Shu, X. Deletion of TSPO Causes Dysregulation of Cholesterol Metabolism in Mouse Retina. Cells 2021, 10, 3066. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.C.; Johnson, K.M. Cholesterol, reactive oxygen species, and the formation of biologically active mediators. J Biol Chem 2008, 283, 15521–15525. [Google Scholar] [CrossRef]
- Ban, N.; Lee, T.J.; Sene, A.; Dong, Z.; Santeford, A.; Lin, J.B.; Ory, D.S.; Apte, R.S. Disrupted cholesterol metabolism promotes age-related photoreceptor neurodegeneration. J Lipid Res 2018, 59, 1414–1423. [Google Scholar] [CrossRef]
- Storti, F.; Klee, K.; Todorova, V.; Steiner, R.; Othman, A.; van der Velde-Visser, S.; Samardzija, M.; Meneau, I.; Barben, M.; Karademir, D.; et al. Impaired ABCA1/ABCG1-mediated lipid efflux in the mouse retinal pigment epithelium (RPE) leads to retinal degeneration. Elife 2019, 8. [Google Scholar] [CrossRef]
- Saadane, A.; Mast, N.; Charvet, C.D.; Omarova, S.; Zheng, W.; Huang, S.S.; Kern, T.S.; Peachey, N.S.; Pikuleva, I.A. Retinal and nonocular abnormalities in Cyp27a1(-/-)Cyp46a1(-/-) mice with dysfunctional metabolism of cholesterol. Am J Pathol 2014, 184, 2403–2419. [Google Scholar] [CrossRef]
- Fliesler, S.J.; Vaughan, D.K.; Jenewein, E.C.; Richards, M.J.; Nagel, B.A.; Peachey, N.S. Partial rescue of retinal function and sterol steady-state in a rat model of Smith-Lemli-Opitz syndrome. Pediatr Res 2007, 61, 273–278. [Google Scholar] [CrossRef]
- Jira, P. Cholesterol metabolism deficiency. Handb Clin Neurol 2013, 113, 1845–1850. [Google Scholar] [CrossRef] [PubMed]
- Porter, F.D. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet 2008, 16, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Horvat, S.; McWhir, J.; Rozman, D. Defects in cholesterol synthesis genes in mouse and in humans: lessons for drug development and safer treatments. Drug Metab Rev 2011, 43, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, M.D.; Christie, J.M.; Eroglu, Y.; Freeman, K.A.; Steiner, R.D. Treatment of Smith-Lemli-Opitz syndrome and other sterol disorders. Am J Med Genet C Semin Med Genet 2012, 160C, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Ballout, R.A.; Livinski, A.; Fu, Y.P.; Steiner, R.D.; Remaley, A.T. Statins for Smith-Lemli-Opitz syndrome. Cochrane Database Syst Rev 2022, 11, CD013521. [Google Scholar] [CrossRef] [PubMed]
- Curcio, C.A.; Johnson, M.; Rudolf, M.; Huang, J.D. The oil spill in ageing Bruch membrane. Br J Ophthalmol 2011, 95, 1638–1645. [Google Scholar] [CrossRef]
- Curcio, C.A. Soft Drusen in Age-Related Macular Degeneration: Biology and Targeting Via the Oil Spill Strategies. Invest Ophthalmol Vis Sci 2018, 59, AMD160–AMD181. [Google Scholar] [CrossRef]
- Klein, R.; Myers, C.E.; Buitendijk, G.H.; Rochtchina, E.; Gao, X.; de Jong, P.T.; Sivakumaran, T.A.; Burlutsky, G.; McKean-Cowdin, R.; Hofman, A.; et al. Lipids, lipid genes, and incident age-related macular degeneration: the three continent age-related macular degeneration consortium. Am J Ophthalmol 2014, 158, 513–524. [Google Scholar] [CrossRef]
- Memarzadeh, E.; Heidari-Soureshjani, S. The Relationship between Statin and Risk of Age-Related Macular Degeneration: A Systematic Review and Meta-Analysis. J Ophthalmol 2022, 2022, 8564818. [Google Scholar] [CrossRef]
- Lymperopoulou, C.; Kandarakis, S.A.; Tzanaki, I.; Mylona, I.; Xanthos, T.; Agouridis, A.P. The Effect of Statins on Ocular Disorders: A Systematic Review of Randomized Controlled Trials. Pharmaceuticals (Basel) 2023, 16, 711. [Google Scholar] [CrossRef]
- Smith, R.T.; Olsen, T.W.; Chong, V.; Kim, J.; Hammer, M.; Lema, G.; Deobhakta, A.; Tan, A.; Tong, Y.; Tai, K.; et al. Subretinal drusenoid deposits, Age related macular degeneration, and Cardiovascular disease. Asia Pac J Ophthalmol (Phila) 2024, 100036. [Google Scholar] [CrossRef] [PubMed]
- Moir, J.; Aggarwal, S.; Skondra, D. Repurposing medications for treatment of age-related macular degeneration: Insights from novel approaches to data mining. Exp Biol Med (Maywood) 2023, 248, 798–810. [Google Scholar] [CrossRef]
- Zheng, E.; Madura, P.; Grandos, J.; Broncel, M.; Pawlos, A.; Wozniak, E.; Gorzelak-Pabis, P. When the same treatment has different response: The role of pharmacogenomics in statin therapy. Biomed Pharmacother 2024, 170, 115966. [Google Scholar] [CrossRef]
- Agnello, F.; Mauro, M.S.; Rochira, C.; Landolina, D.; Finocchiaro, S.; Greco, A.; Ammirabile, N.; Raffo, C.; Mazzone, P.M.; Spagnolo, M.; et al. PCSK9 inhibitors: current status and emerging frontiers in lipid control. Expert Rev Cardiovasc Ther 2023, 1–18. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




