1. Introduction
Candida is a type of fungus present in the oral flora of healthy subjects (Arendorf, 1980); it has a wide distribution that includes the gastrointestinal, urinary, and respiratory tracts (Martins, 2014). Fifteen subspecies of Candida cause different diseases in humans; although, 90% of subspecies are harmless, the most common pathogenic species are Candida albicans, Candida glabrata, Candida tropicalis, Candida parapsilosis, and Candida krusei (Pappas, 2016). Candida is considered an opportunistic organism that causes oral infections; it is mainly found under conditions of malnutrition, changes in pH, and/or high concentrations of glucose in saliva (Madhavan, 2011). The proliferation of the different subspecies of Candida is increased in patients with type 2 diabetes mellitus (T2DM) (De Resende, 2006; Guimarães, 2012; Khosravi, 2008; Tang, 2015) because they present with high levels of glucose (NICE, 2018), immunological changes and immunosuppression (Calvet, 2001), and changes to their general pH levels induced by their chronic metabolic syndrome (Lipsa, 2018). In 2017, according to the WHO and the International Diabetes Federation, the world diabetic population was estimated at 425 million people (King, 1998; Agarwal, 2005). Candida can be found locally or generally (Davenport, 1970), but it is characterized by a reduction in saliva when it is found in the mouth (Kadir, 2002) as well as microvascular degeneration and neutrophil activity, which is associated with the high levels of glucose found in patients with T2DM (Duggan, 2015; Wilson, 1986). In addition, an increase in digestive enzymes has been reported in patients with T2DM and candidiasis (Pinto, 2008; Motta-Silva, 2010; Calderone, 2001), which benefits Candida because patients with diabetes suffer a generalized state of immunosuppression (Balan, 2015; Nowakowska, 2004; Dorko, 2005). Two hundred subspecies of Candida have been characterized, of which C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and C. krusei (Feller, 2014) are most frequently detected in patients with candidiasis. Such species diversity makes Candida difficult to treat (Asmundsdóttir, 2008). Of these subspecies, C. albicans is the most aggressive; it is associated with a 93% increase in the development of stomatitis (Moosazadeh, 2016). The immune response of patients with candidiasis is initiated with pathogens associated with molecular patterns, which are recognized by specific receptors of the innate immune response known as pathogen recognition receptors (PRRs) (Netea, 2015; Netea, 2008). The innate immune response to C. albicans occurs through monocytes, neutrophils, macrophages, and dendritic cells; these express PRRs and establish the first line of defense against C. albicans infection (Netea, 2015). Candida albicans can be recognized by beta-glucans, dectin-1, and lecithin type C receptors, primarily contained in its membrane, as well as by TLR2 and TLR4 expression in the human immune response (Brown, 2001; Cambi, 2003; Netea, 2006). As part of the Candida infection response, a proinflammatory response is triggered by activation of the NLRP3 inflammasome (Jaeger, 2016; Jaeger, 2015), which in turn activates IL-1b (Groslambert, 2018).
Specific diagnoses of oral candidiasis in patients with T2DM are important for improving treatment because resistance to therapeutic procedures is largely due to misdiagnosis. Therefore, the objective of the present study was to identify Candida species associated with oral alterations in patients with T2DM using polymerase chain reaction (PCR). In addition, the results of molecular diagnoses were compared with conventional diagnoses obtained using Papanicolau (Pap) smear. Furthermore, the expression of TLR2 and TLR4 genes was evaluated as potential biomarkers of the immune response.
2. Materials and Methods
2.1. Subjects
This research was approved by the Research Ethics Committee of the IPN School of Medicine (No. ESM.CE-01/01-29-2016). Written informed consent was obtained from all participants. The experiments were performed in accordance with the ethical principles of the Declaration of Helsinki and were consistent with the Good Clinical Practice Guidelines. The participants included 18 adults of both sexes who attended a dental consultation at the Centro Interdisciplinario de Ciencias de la Salud, unidad Santo Tomas (CICS UST) del Instituto Politécnico Nacional; these patients were ≥40 years old, had been diagnosed with T2DM, and met the inclusion criteria. In addition, 18 healthy adults of both sexes aged ≥40 years were used as a control group. Peripheral venous blood was drawn from each subject in the morning and glycated hemoglobin HbA1c was determined. Palatal mucosa smears were performed on patients diagnosed with grade I, II, and III prosthetic stomatitis as well as T2DM. Samples were fixed with absolute ethanol and staining was performed according to the Pap technique.
2.2. Gene Expression Analysis and Genotyping
Total RNA was isolated from smears of the palatal mucosa using TRIzol reagent according to the manufacturer’s instructions (TriPure Isolation Reagent; Roche Applied Science, Indianapolis, IN, USA). The amount and purity of the isolated RNA was quantified nanophotometrically by measuring the optical densities at 260 and 280 nm. The integrity of all samples was confirmed by agarose gel electrophoresis. Before reverse transcription, RNA samples were treated further with amplification-grade DNase I (Invitrogen, Carlsbad, CA, USA) to remove trace amounts of contaminating DNA. All RNA samples were stored in RNA elution solution at −80°C. Subsequently, 0.5–1.0 µg of total RNA was subjected to reverse transcription using a First-Strand cDNA Synthesis Kit (Roche Diagnostics, GmbH Mannheim, Germany) with random hexamer primers in an Eppendorf thermocycler according to the manufacturer’s instructions. Finally, the concentration of the newly generated cDNA was determined nanospectrophotometrically.
Real-time quantitative PCR (qPCR) assays were performed using specific oligonucleotide primers that were generated using online assay design software (
https://qpcr.probefinder.com/organism.jsp). The sequences of the primers used to determine gene expression were as follows:
TLR2 forward: 5′-CCT TTG GAT CCT GCT TGC-3′;
TLR2 reverse: 5′-CGT TCT CTC AGG TGA CTG CTC-3′;
TLR4 forward: 5′-TCC ATG CAT TGA TAA GTA ATA TTA GGA-3′;
TLR4 reverse: 5′-CTC TCC TGC GTG AGA CCA G-3′. The sequence of the primers used to determine the genotype of each
Candida strain with its specific alignment temperature were as follows:
C. albicans: 5′-AGC TGC CGC CAG AGG TCT AA-3′ (583 bp);
C. glabrata: 5′-TTG TCT GAG CTC GGA GAG AG-3′ (929 bp);
C. parapsilosis: 5′-GTC AAC CGA TTA TTT AAT AG-3′ (570 bp);
C. tropicalis: 5′-GAT TTG CTT AAT TGC CCC AC-3′ (583 bp);
Candida dubliniensis: 5′-CTC AAA CCC CTA GGG TTT GG-3′ (591 bp);
C. krusei: 5′-CTG GCC GAG CGA ACT AGA CT-3′ (590 bp);
C. guilliermondii: 5′-TTG GCC TAG AGA TAG GTT GG-3′ (668 bp);
Candida lusitaniae: 5′-TTC GGA GCA ACG CCT AAC CG-3′ (433 bp); and a universal reverse primer: 5′-TTC TTT TCC TCC GCT TAT TG-3′. For gene expression analysis, the reaction mixture, containing 1 µL of standard cDNA at an appropriate dilution, was prepared according to the manufacturer’s instructions (Roche Diagnostics, GmbH). A Light Cycler Nano Real-Time PCR System (Roche Diagnostics, Mannheim, Germany) was used for all amplifications, with the following settings used for Universal Probe Library-based assays: an initial denaturation step for 10 min at 95°C and then 45 cycles of 10 s at 94°C, 20 s at 60°C, and 5 s at 72°C. Each qPCR assay included a standard curve of four serial dilution points for each gene. The mRNA levels were normalized to the expression of the endogenous control, 18S mRNA. All qPCR experiments require standardization of the reaction efficiency curves for gene expression. We calculated the mRNA levels using the comparative parameter threshold cycle (Ct) method. In the standardization formula, 2
-ΔΔCt, the value 2 corresponds to the qPCR efficiency reaction (dynamic range curve or dilutions of the constitutive gene).
2.3. Statistical Analysis
Descriptive statistics were performed and the expression data are expressed as means ± standard deviations (SD). Data were analyzed using either the unpaired Student’s t-test or a one-way analysis of variance. GraphPad Prism version 8.00 for Windows and SPSS were used to conduct the statistical analyses and graph the data.
3. Results
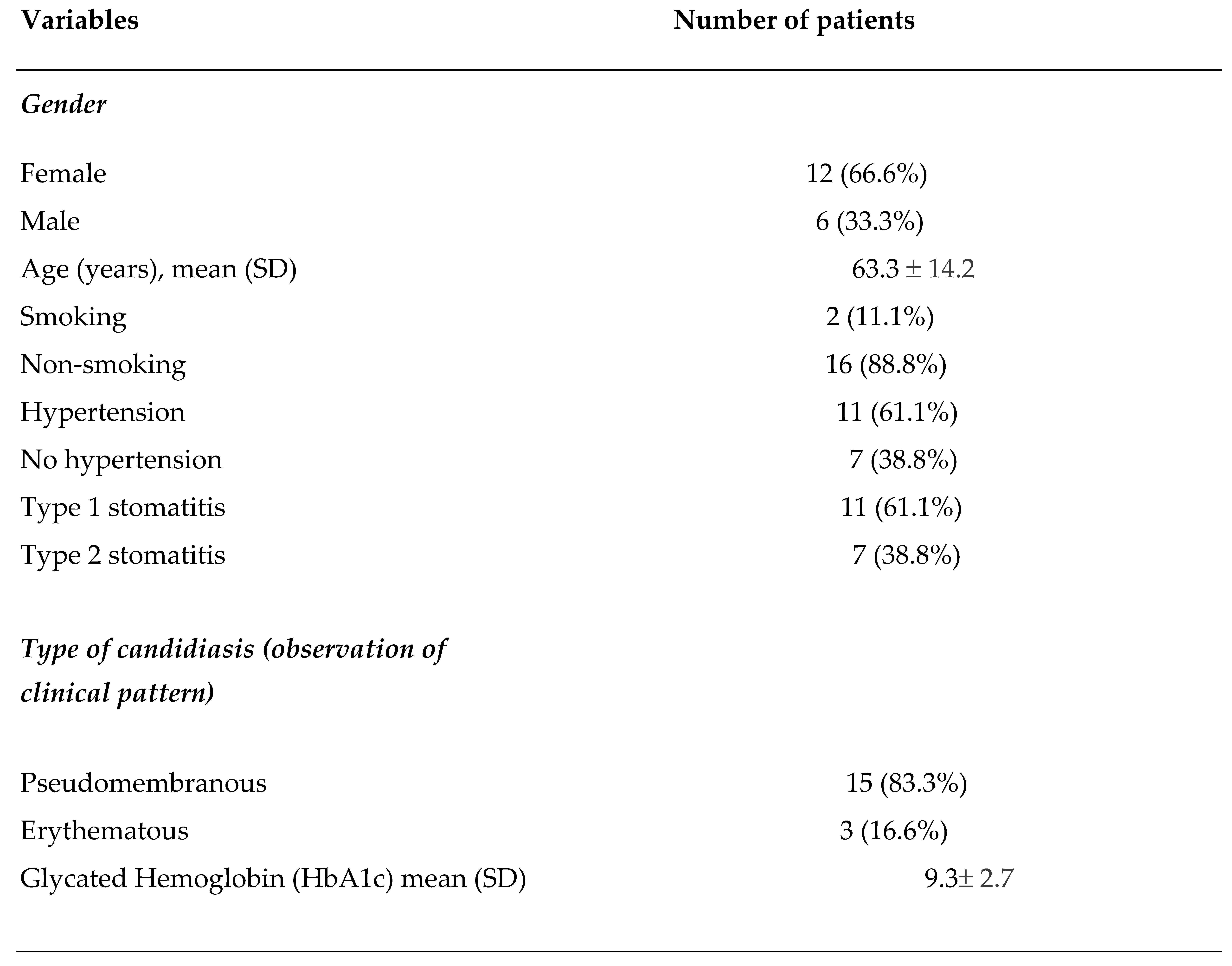
The clinical characteristics of the 18 patients included in this study are shown in
Table 1. In patients with T2DM, the mean age was 63.3 ± 14.2 years. Most patients were female (66.6%), most were nonsmokers, and a high percentage were hypertensive (61.1%). Type I stomatitis was predominant (61.1%) compared with Type II stomatitis (38.8%). When reviewing the clinical patterns, an increase in pseudomembranous candidiasis (83.3%) and a high percentage of glycosylated hemoglobin were observed.
Exfoliative cytology of the samples from patients with T2DM confirmed the presence of
Candida.
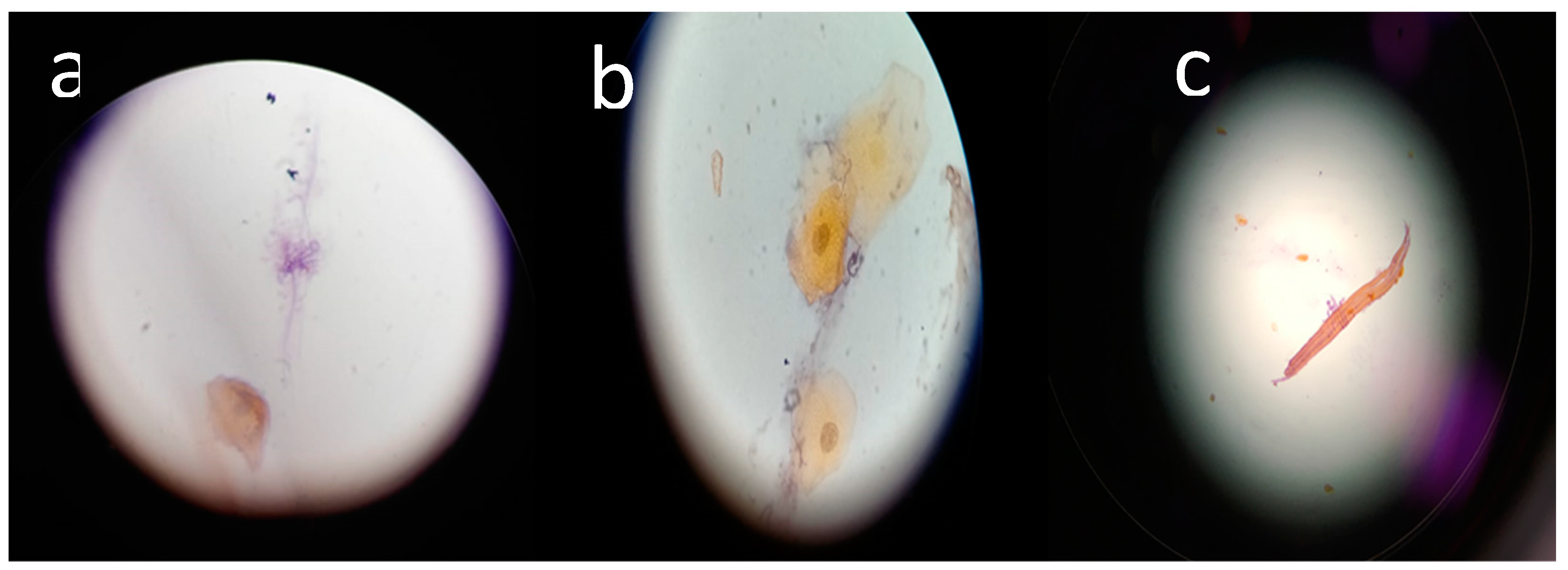
Figure 1 shows the hyphae of the
Candida types; tangled tubular hyphae were observed (
Figure 1a), hyphae related to desquamated epithelial cells were also observed (
Figure 1b), and
Candida was shown to be completely developed (
Figure 1c).
Identification by end-point PCR results and the use of specific primers for each studied species of
Candida is shown in
Table 2. Interestingly, species coexpression was found among the study subjects. The species with the highest coexpression were C.
guilliermondii and
C. krusei. In four patients,
Candida species were not found.
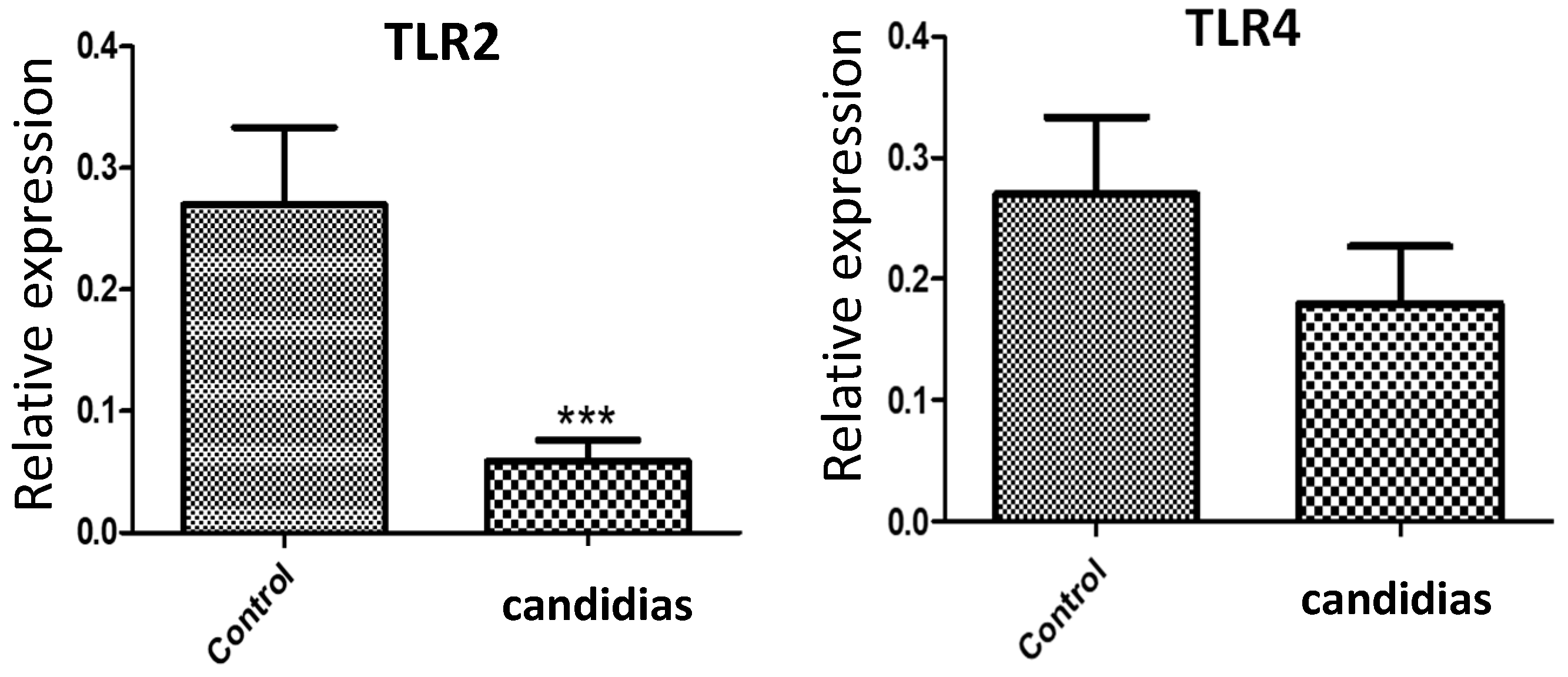
In addition, qPCR analysis revealed a significant decrease (
p < 0.0001) in the mRNA expression of
TLR2 in the candidiasis group relative to that in the control group (
Figure 2). In contrast, there was no significant difference in
TLR4 expression between the groups.
4. Discussion
Here, the most common variants of Candida were identified in the oral cavity of patients with diabetes and candidiasis. When using Pap smears, a conventional method for the diagnosis of Candida, it was possible to detect 3 of 19 patients with the clinical characteristics of this pathology when the patients had dental prosthesis. Oral candidiasis is usually diagnosed using physical examination, clinical history, and exfoliative cytology with SBP or Pap staining (Picciani, 2013; Miller, 2002; Hellstein, 2019). However, the guide of the European Society of Clinical Microbiology and Infectious Diseases recommends cultivating the fungus in a specific medium for the diagnosis of Candida (Cuenca-Estrella, 2012). The recommendations made by the American Society for Infectious Diseases for the treatment of oropharyngeal candidiasis is to provide clotrimazole or nystatin as the first treatment, especially in patients who have not had the disease for a long period (Pappas, 2009).
As well as the Pap technique, molecular techniques were used to determine the presence of Candida subtypes in the patients studied. Consequently, seven species of Candida were identified in patients with diabetes and oral prostheses, unlike in Pap staining, which could only identify three patients with Candida. Nevertheless, the Pap technique is still used preferentially in dental consultations for the diagnosis of Candida in the Mexican population. A similar study was previously conducted in Monterrey, Mexico, in which Candida species were identified using culture media after collecting samples from the microbiology departments of each hospital participating in the study; C. albicans, C. parapsilosis, C. tropicalis, C. glabrata, C. krusei, and C. guilliermondii were mainly identified in a population studied from 2004 to 2007 (González, 2008).
Recent studies indicate an alarming increase in global Candida infections and drug resistance. For example, there are extensive reports of intrinsic and developed resistance to azole antifungals among several Candida species including C. albicans, C. parapsilosis, C. tropicalis, C. krusei, and C. glabrata. Azole antifungals, such as fluconazole, are often the preferred treatment for many Candida infections (Srivastava, 2018; Whaley, 2016). Therefore, there is a need to seek new strategies for Candida diagnosis and treatment. Immunity or resistance involves the participation of the innate immune response through the TLR, DC-SIGN, galectin, and dectin receptors (Cheng, 2012; Richardson, 2015). In addition, the pharmacological resistance of Candida has been shown to initially depend on the concentration and type of drug used as well as the immunological capacity of the infected individual (Wuyts, 2018). Moreover, recent studies indicate that Candida has mutagenic capabilities that allow it to develop pharmacological resistance (Lockhart, 2017). The development of resistance to treatments for candidiasis is mainly caused by the low pharmacological efficiency of such treatment to combat all Candida species observed during infection (Pristov, 2019; Berman, 2020). Given such resistance, the standard clinical approach to treating patients with candidiasis has been questioned, particularly given that the conventional diagnostic method, i.e., by means of stains, does not confirm the Candida species and therefore the treatment offered is not species-specific and may be ineffective. In contrast, molecular diagnosis allows for rapid identification of Candida species and the implementation of an appropriate treatment that can help avoid the systemic damage that is caused by advanced contagion of species that typically develop resistance.
The damage caused by Candida in the oral epithelium provokes an inflammatory response that causes the release of inflammatory mediators and the production of recognition patterns of molecules such as TLRs. Jianwei et al. (2018) demonstrated the activation of TLR2 by the presence of C. albicans; consistent with these findings, Shaoru (2004) found that both TLR2 and TLR4 mRNA expression was low in patients diagnosed with candidiasis that did not receive pharmacological treatment. Similarly, no significant differences in TLR4 expression were found between healthy subjects and those infected with Candida. In addition, Pie et al. (2019) found that subjects infected with Candida showed downregulation of IL-1β (Pei, 2019). Similar to these previous results, we found a significant decrease in the gene expression of TLR2 in the candidiasis group, unlike the expression of TLR4 that did not differ between the candidiasis and control groups. Because diabetes mellitus can be associated with the presence of more predominant types of Candida, the analysis of additional molecules involved in the immune response to each type of Candida will facilitate the development of more effective pharmacological treatments.
Currently, the strategy applied for the clinical care of patients infected by Candida in Mexico lacks the specificity to identify all species that may be present in patients with candidiasis. Additionally, the general treatment provided to these patients leads to the development of pharmacological resistance by some Candida species. This favors the development of resistant species, which contributes to increasing the clinical complications that can compromise the lives of patients.
5. Conclusions
In conclusion, it is advisable to use molecular diagnostic tools to accurately determine the species of Candida present in each patient, which will allow the administration of treatment that guarantees the elimination of all Candida species present. Such changes will contribute to advancing individualized medicine in Mexico.
Author Contributions
Nadia Mabel Pérez-Vielma: Conceptualization, Writing - Original Draft, Methodology, Data curation, Modesto Gómez-López: Methodology, Data curation María de los Ángeles Martínez-Godínez: Conceptualization, Formal analysis, Data curation Ana Laura Luna-Torres: Formal analysis, Conceptualization , Aarón Dominguez López: Methodology, Data curation Ángel Miliar-García: Supervision, Writing - Review & Editing
Funding
Please add: “The present study was performed with the support of the Instituto Politécnico Nacional. This study was supported by a SIP 20180429 grant to AMG. AMG, NMPV, MGL are SNI fellows.
Institutional Review Board Statement
This research was approved by the Research Ethics Committee of the IPN School of Medicine (No. ESM.CE-01/01-29-2016). Written informed consent was obtained from all participants. The experiments were performed in accordance with the ethical principles of the Declaration of Helsinki and were consistent with the Good Clinical Practice Guidelines.
Data Availability Statement
data will be available upon request.
Acknowledgments
We thank teacher Alejandra Sosa Gómez and teacher Martha Miranda Ruiz for the contribution to the development of this project.
Conflicts of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Agarwal, S. , Raman, R., Paul, P. G., Rani, P. K., Uthra, S., Gayathree, R., McCarty, C., Kumaramanickavel, G., Sharma, T., & Nethralaya, S. Diabetic retinopathy epidemiology and molecular genetic study (SN–DREAMS 1): Study design and research methodology. Ophthalmic Epidemiology 2005, 12, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Arendorf, T. M. , & Walker, D. M. The prevalence and intra-oral distribution of Candida albicans in man. Archives of Oral Biology 1980, 25, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Asmundsdóttir, L. R. , Erlendsdóttir, H., Haraldsson, G., Guo, H., Xu, J., & Gottfredsson, M. Molecular epidemiology of candidemia: Evidence of clusters of smoldering nosocomial infections. Clinical Infectious Diseases 2008, 47, e17–e24. [Google Scholar] [CrossRef] [PubMed]
- alan, P., B Gogineni, S., Kumari N, S., Shetty, V., Lakshman Rangare, A., Castelino, R. L., & Areekat, K, F. Candida carriage rate and growth characteristics of saliva in diabetes mellitus patients: A case-control study. Journal of Dental Research, Dental Clinics, Dental Prospects 2015, 9, 274–279. [CrossRef]
- Berman, J. , & Krysan, D. J. Drug resistance and tolerance in fungi. Nature Reviews. Microbiology 2020, 18, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Brown, G. D. , & Gordon, S. Immune recognition. A new receptor for β-glucans. Nature 2001, 413, 36–37. [Google Scholar] [CrossRef]
- Calderone, R. A. , & Fonzi, W. A. Virulence factors of Candida albicans. Trends in Microbiology 2001, 9, 327–335. [Google Scholar] [CrossRef]
- Calvet, H. M. , & Yoshikawa, T. T. Infections in diabetes. Infectious Disease Clinics of North America 2001, 15, 407–21. [Google Scholar] [CrossRef]
- Calvet, H. M. , & Yoshikawa, T. T. Infections in diabetes. Infectious Disease Clinics of North America 2001, 15, 407–21. [Google Scholar] [CrossRef]
- Cambi, A. , Gijzen, K., de Vries, l. J., Torensma, R., Joosten, B., Adema, G. J., Netea, M. G., Kullberg, B. J., Romani, L., & Figdor, C. G. The C-type lectin DC-SIGN (CD209) is an antigen-uptake receptor for Candida albicans on dendritic cells. European Journal of Immunology 2003, 33, 532–538. [Google Scholar] [CrossRef]
- Cheng, S. C. , Joosten, L. A., Kullberg, B. J., & Netea, M. G. Interplay between Candida albicans and the mammalian innate host defense. Infection and Immunity 2012, 80, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Cuenca-Estrella, M. , Verweij, P. E., Arendrup, M. C., Arikan-Akdagli, S., Bille, J., Donnelly, J. P., Jensen, H. E., Lass-Flörl, C., Richardson, M. D., Akova, M., Bassetti, M., Calandra, T., Castagnola, E., Cornely, O. A., Garbino, J., Groll, A. H., Herbrecht, R., Hope, W. W., Kullberg, B. J., Lortholary, O., Meersseman, W., Petrikkos, G., Roilides, E., Viscoli, C., & Ullmann, A. J. ESCMID* guideline for the diagnosis and management of Candida diseases 2012: Diagnostic procedures. Clinical Microbiology and Infection 2012, 18, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Davenport, J. C. The oral distribution of candida in denture stomatitis. British Dental Journal 1970, 129, 151–156. [Google Scholar] [CrossRef] [PubMed]
- De Resende, M. A. , de Sousa, L. V. N. F., de Oliveira, R. C. B. W., Koga-Ito, C. Y., & Lyon, J. P. Prevalence and antifungal susceptibility of yeasts obtained from the oral cavity of elderly individuals. Mycopathologia 2006, 162, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Dorko, E. , Baranová, Z., Jenča, A., Kizek, P., Pilipčinec, E., & Tkáčiková, L. Diabetes mellitus and candidiases. Folia Microbiologica 2005, 50, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Duggan, S. , Essig, F., Hünniger, K., Mokhtari, Z., Bauer, L., Lehnert, T., Brandes, S., Häder, A., Jacobsen, I. D., Martin, R., Figge, M. T., & Kurzai, O. Neutrophil activation by Candida glabrata but not Candida albicans promotes fungal uptake by monocytes. Cellular Microbiology 2015, 17, 1259–1276. [Google Scholar] [CrossRef]
- Feller, L. , Khammissa, R. A. G., Chandran, R., Altini, M., & Lemmer, J. Oral candidosis in relation to oral immunity. Journal of Oral Pathology and Medicine 2014, 43, 563–569. [Google Scholar] [CrossRef]
- González G., M. , Elizondo, M., & Ayala, J. Trends in species distribution and susceptibility of bloodstream isolates of candida collected in Monterrey, Mexico, to seven antifungal agents: Results of a 3-year (2004 to 2007) surveillance study. Journal of Clinical Microbiology 2008, 46, 2902–2905. [Google Scholar] [CrossRef]
- Groslambert, M. , & Py, B. F. Spotlight on the NLRP3 inflammasome pathway. Journal of Inflammation Research 2018, 11, 359–374. [Google Scholar] [CrossRef]
- Guimarães, T. , Nucci, M., Mendonça, J. S., Martinez, R., Brito, L. R., Silva, N., Moretti, M. L., Salomão, R., & Colombo, A. L. Epidemiology and predictors of a poor outcome in elderly patients with candidemia. International Journal of Infectious Diseases 2012, 16, e442–e447. [Google Scholar] [CrossRef]
- Hellstein, J. W. , & Marek, C. L. Candidiasis, red and white manifestations in the oral cavity. Head and Neck Pathology 2019, 13, 25–32. [Google Scholar] [CrossRef]
- Jaeger, M. , Carvalho, A., Cunha, C., Plantinga, T. S., van de Veerdonk, F., Puccetti, M., Galosi, C., Joosten, L. A., Dupont, B., Kullberg, B. J., Sobel, J. D., Romani, L., & Netea, M. G. Association of a variable number tandem repeat in the NLRP3 gene in women with susceptibility to RVVC. European Journal of Clinical Microbiology and Infectious Diseases 2016, 35, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, M. , van der Lee, R., Cheng, S. C., Johnson, M. D., Kumar, V., Ng, A., Plantinga, T. S., Smeekens, S. P., Oosting, M., Wang, X., Barchet, W., Fitzgerald, K., Joosten, L. A. B., Perfect, J. R., Wijmenga, C., van de Veerdonk, F. L., Huynen, M. A., Xavier, R. J., Kullberg, B. J., & Netea, M. G. The RIG-I-like helicase receptor MDA5 (IFIH1) is involved in the host defense against Candida infections. European Journal of Clinical Microbiology and Infectious Diseases 2015, 34, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. , Geng, F., Sun, H., Wang, X., Zhang, H., Yang, Q., & Zhang, J. Candida albicans induces TLR2/MyD88/NF-κB signaling and inflammation in oral lichen planus-derived keratinocytes. Journal of Infection in Developing Countries 2018, 12, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Kadir, T. , Pisiriciler, R., Akyüz, S., Yarat, A., Emekli, N., & Ipbüker, A. Mycological and cytological examination of oral candidal carriage in diabetic patients and non-diabetic control subjects: Thorough analysis of local aetiologic and systemic factors. Journal of Oral Rehabilitation 2002, 29, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, A. R. , Yarahmadi, S., Baiat, M., Shokri, H., & Pourkabireh, M. Factors affecting the prevalence of yeasts in the oral cavity of patients with diabetes mellitus. Journal de Mycologie Médicale 2008, 18, 83–88. [Google Scholar] [CrossRef]
- King, H. , Aubert, R. E., & Herman, W. H. Global burden of diabetes, 1995–2025: Prevalence, numerical estimates, and projections. Diabetes Care 1998, 21, 1414–1431. [Google Scholar] [CrossRef] [PubMed]
- Bhuyan, L. , Hassan, S., Dash, K. C., Panda, A., Behura, S. S., & Ramachandra, S. Candida species diversity in oral cavity of Type 2 diabetic patients and their in vitro antifungal susceptibility. Contemporary Clinical Dentistry 2018, 9, S83–S88. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S. R. , Ghannoum, M. A., & Alexander, B. D. Establishment and use of epidemiological cutoff values for molds and yeasts by use of the Clinical and Laboratory Standards Institute M57 standard. Journal of Clinical Microbiology 2017, 55, 1262–1268. [Google Scholar] [CrossRef]
- Madhavan, P. , Jamal, F., & Chong, P. P. Laboratory isolation and identification of Candida species. Journal of Applied Sciences 2011, 11, 2870–2877. [Google Scholar] [CrossRef]
- Martins, N. , Ferreira, I. C. F. R., Barros, L., Silva, S., & Henriques, M. Candidiasis: Predisposing factors, prevention, diagnosis and alternative treatment. Mycopathologia 2014, 177, 223–240. [Google Scholar] [CrossRef] [PubMed]
- Miller, D. J. Diagnosis and management of Candida and other fungal infections of the head and neck. Current Infectious Disease Reports 2002, 4, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Moosazadeh, M. , Akbari, M., Tabrizi, R., Ghorbani, A., Golkari, A., Banakar, M., Sekhavati, E., Kavari, S.H., & Lankarani, K.B. Denture stomatitis and Candida albicans in Iraninan population: A systemic review and meta-analysis. Journal of Dentistry 2016, 17, 283–292. [Google Scholar] [PubMed]
- Motta-Silva, A. C. , Aleva, N. A., Chavasco, J. K., Armond, M. C., França, J. P., & Pereira, L. J. Erythematous oral candidiasis in patients with controlled type II diabetes mellitus and complete dentures. Mycopathologia 2010, 169, 215–223. [Google Scholar] [CrossRef]
- Netea, M. G. , Brown, G. D., Kullberg, B. J., & Gow, N. A. An integrated model of the recognition of Candida albicans by the innate immune system. Nature Reviews. Microbiology 2008, 6, 67–78. [Google Scholar] [CrossRef]
- Netea, M. G. , Gow, N. A., Munro, C. A., Bates, S., Collins, C., Ferwerda, G., Hobson, R. P., Bertram, G., Hughes, H. B., Jansen, T., Jacobs, L., Buurman, E. T., Gijzen, K., Williams, D. L., Torensma, R., McKinnon, A., MacCallum, D. M., Odds, F. C., Van der Meer, J. W., Brown, A. J., & Kullberg, B. J. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and toll-like receptors. Journal of Clinical Investigation 2006, 116, 1642–1650. [Google Scholar] [CrossRef]
- Netea, M. G. , Joosten, L. A., van der Meer, J. W., Kullberg, B. J., & van de Veerdonk, F. L. Immune defence against Candida fungal infections. Nature Reviews. Immunology 2015, 15, 630–642. [Google Scholar] [CrossRef]
- 38. NICE Type 2 diabetes: Prevention in people at high. (2018). Risk.
- Nowakowska, D. , Kurnatowska, A., Stray-Pedersen, B., & Wilczyński, J. Species distribution and influence of glycemic control on fungal infections in pregnant women with diabetes. Journal of Infection 2004, 48, 339–346. [Google Scholar] [CrossRef]
- Pappas, P. G. , Kauffman, C. A., Andes, D., Benjamin Jr, D. K., Calandra, T. F., Edwards Jr, J. E., Filler, S. G., Fisher, J. F., Kullberg, B. J., Ostrosky-Zeichner, L., & Reboli, A. C. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America 2016, 62, e1–e50. [Google Scholar]
- Pappas, P. G. , Kauffman, C. A., Andes, D., Benjamin, D. K., Jr., Calandra, T. F., Edwards, J. E., Jr., Filler, S. G., Fisher, J. F., Kullberg, B. J., Ostrosky-Zeichner, L., Reboli, A. C., Rex, J. H., Walsh, T. J., Sobel, J. D., & Infectious Diseases Society of America. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clinical Infectious Diseases 2009, 48, 503–535. [Google Scholar] [CrossRef] [PubMed]
- Ye, P. , Wang, X., Ge, S., Chen, W., Wang, W., & Han, X. Long-term cigarette smoking suppresses NLRP3 inflammasome activation in oral mucosal epithelium and attenuates host defense against Candida albicans in a rat model. Biomedicine and Pharmacotherapy 2019, 113, 108597. [Google Scholar] [CrossRef]
- Picciani, B. L. , Michalski-Santos, B., Carneiro, S., Sampaio, A. L., Avelleira, J. C., Azulay, D. R., Pinto, J. M., & Dias, E. P. Oral candidiasis in patients with psoriasis: Correlation of oral examination and cytopathological evaluation with psoriasis disease severity and treatment. Journal of the American Academy of Dermatology 2013, 68, 986–991. [Google Scholar] [CrossRef]
- Pinto, E. , Ribeiro, I. C., Ferreira, N. J., Fortes, C. E., Fonseca, P. A., & Figueiral, M. H. Correlation between enzyme production, germ tube formation and susceptibility to fluconazole in Candida species isolated from patients with denture-related stomatitis and control individuals. Journal of Oral Pathology and Medicine 2008, 37, 587–592. [Google Scholar] [CrossRef]
- Pristov, K. E. , & Ghannoum, M. A. Resistance of Candida to azoles and echinocandins worldwide. Clinical Microbiology and Infection 2019, 25, 792–798. [Google Scholar] [CrossRef]
- Richardson, J. P. , & Moyes, D. L. Adaptive immune responses to Candida albicans infection. Virulence 2015, 6, 327–337. [Google Scholar] [CrossRef]
- Zhang, S. , Li, J., Jia, X., & Wu, Y. The expression of toll-like receptor 2 and 4 mRNA in local tissues of model of oropharyngeal candidiasis in mice. Journal of Huazhong University of Science and Technology (Medical Science) 2004, 24, 639–641. [Google Scholar] [CrossRef]
- Srivastava, V. , Singla, R. K., & Dubey, A. K. Emerging virulence, drug resistance and future anti-fungal drugs for candida pathogens. Current Topics in Medicinal Chemistry 2018, 18, 759–778. [Google Scholar] [CrossRef]
- Tang, H. J. , Liu, W. L., Lin, H. L., & Lai, C. C. Epidemiology and prognostic factors of candidemia in elderly patients. Geriatrics and Gerontology International 2015, 15, 688–693. [Google Scholar] [CrossRef]
- Whaley, S. G. , Berkow, E. L., Rybak, J. M., Nishimoto, A. T., Barker, K. S., & Rogers, P. D. Azole antifungal resistance in candida albicans and emerging non-albicans candida species. Frontiers in Microbiology 2016, 7, 2173. [Google Scholar] [CrossRef]
- Wilson, R. M. , & Reeves, W. G. Neutrophil phagocytosis and killing in insulin-dependent diabetes. Clinical and Experimental Immunology 1986, 63, 478–484. [Google Scholar] [PubMed]
- Wuyts, J. , Van Dijck, P., & Holtappels, M. Fungal persister cells: The basis for recalcitrant infections? PLOS Pathogens 2018, 14, e1007301. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).