Submitted:
31 January 2024
Posted:
01 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Collection of isolates
Identification of isolates by PCR-RFLP of the ribosomal ITS region
Identification of Candida auris
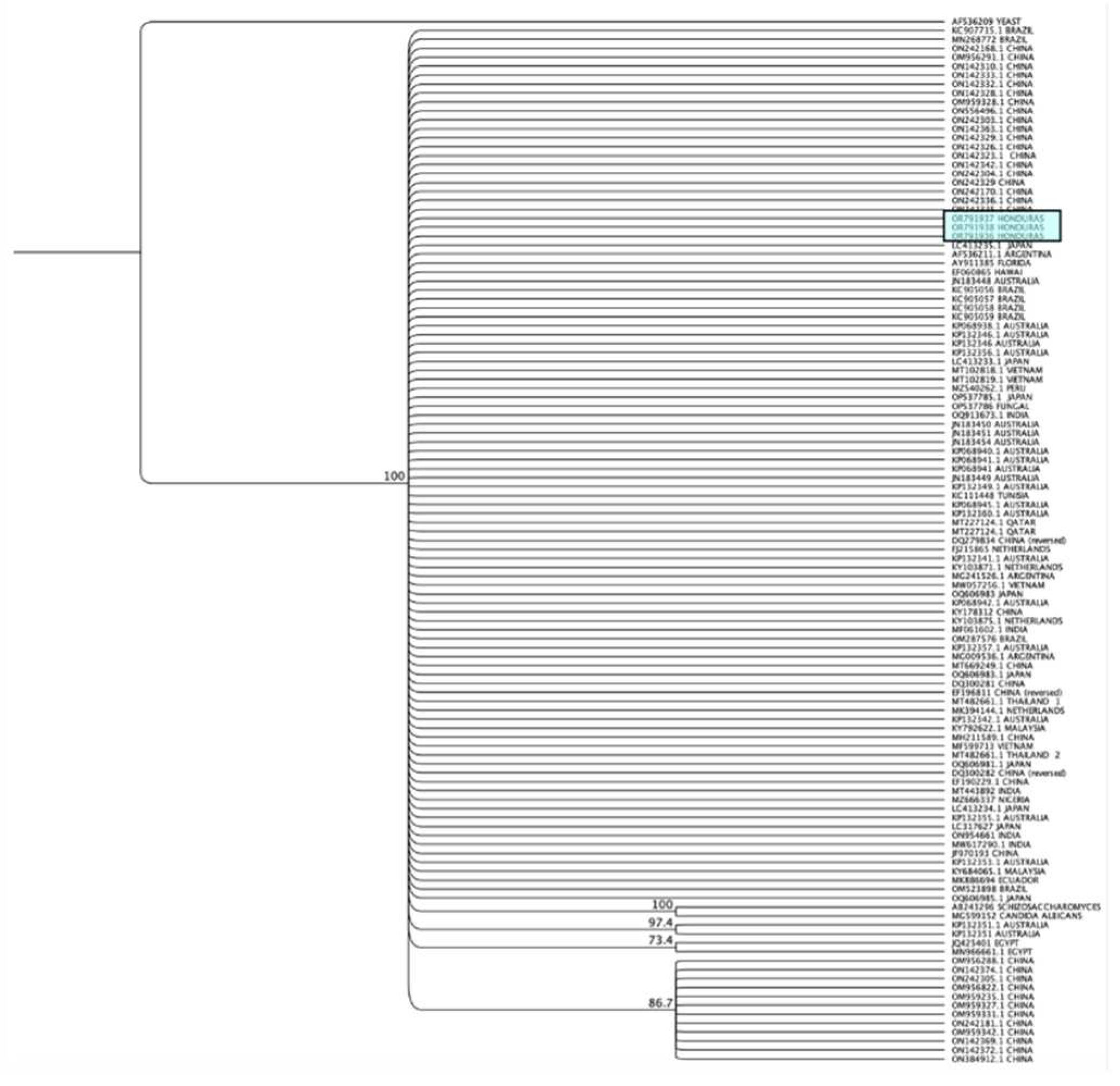
Sequence analysis and construction of cladograms
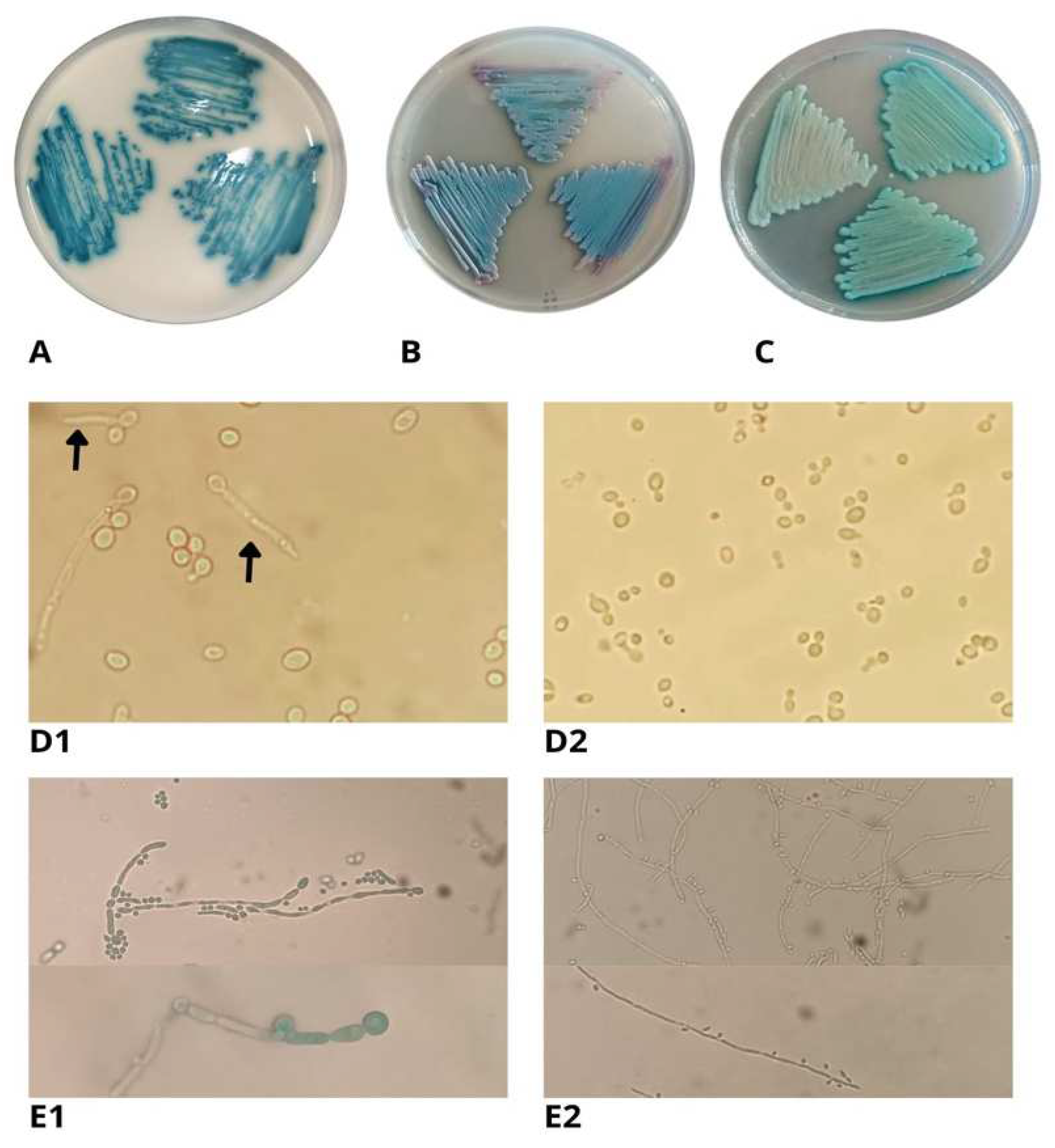
Phenotypic identification
Microbiological identification and chromogenic culture media
Antifungal sensitivity testing
Assays for hydrolytic enzyme activity of Kodamaea ohmeri
Assessment of Kodamaea ohmeri’s ability to produce biofilms
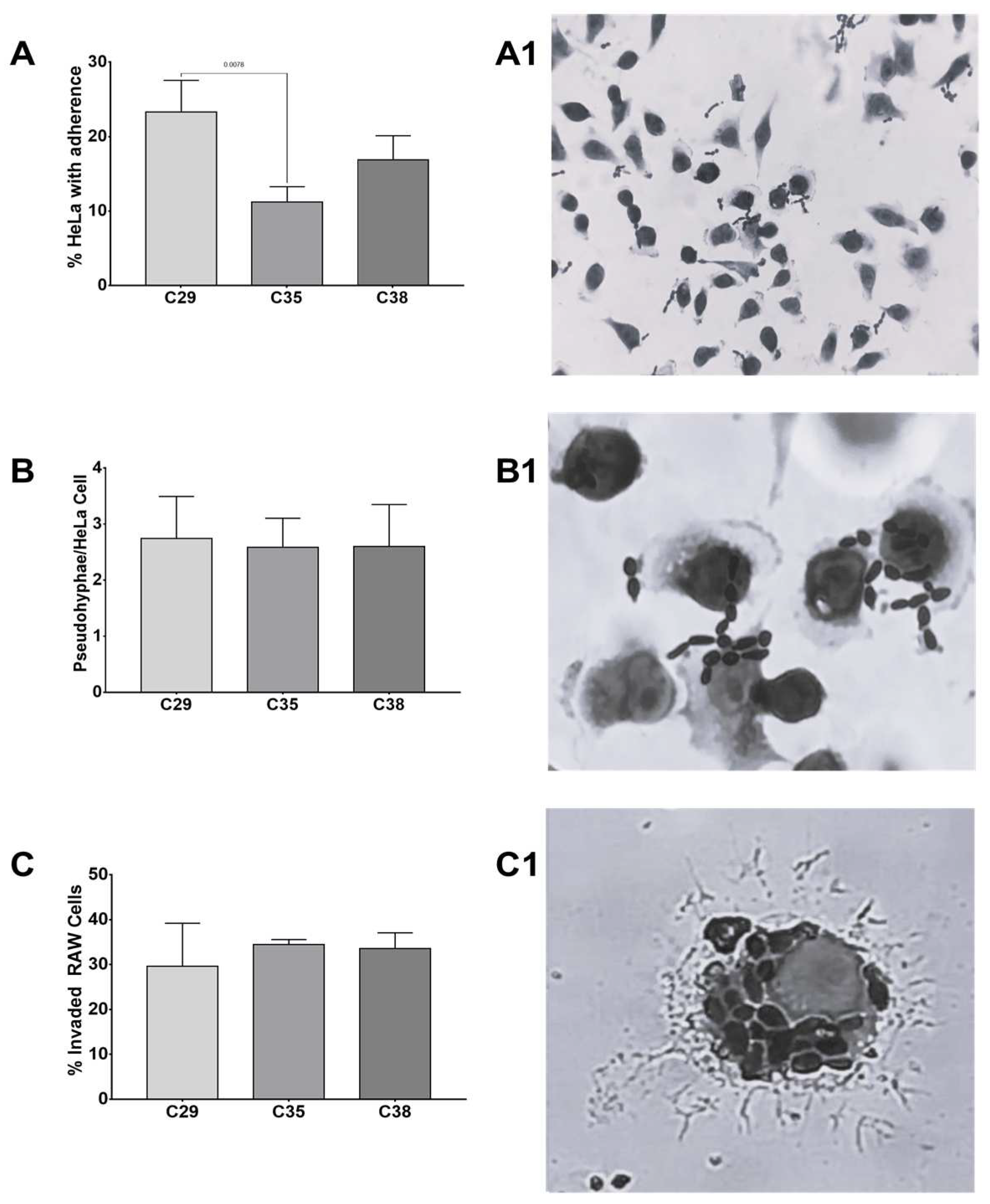
Invasion and adherence assays
3. Results
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choi, J.; Kim, S.-H. A genome Tree of Life for the Fungi kingdom. Proceedings of the National Academy of Sciences 2017, 114, 9391–9396. [Google Scholar] [CrossRef] [PubMed]
- Wickes, B.L.; Wiederhold, N.P. Molecular diagnostics in medical mycology. Nature communications 2018, 9, 5135. [Google Scholar] [CrossRef] [PubMed]
- Wiederhold, N.P. Emerging fungal infections: New species, new names, and antifungal resistance. Clinical Chemistry 2022, 68, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Nnadi, N.E.; Carter, D.A. Climate change and the emergence of fungal pathogens. PLoS Pathogens 2021, 17, e1009503. [Google Scholar] [CrossRef] [PubMed]
- van Rhijn, N.; Bromley, M. The consequences of our changing environment on life threatening and debilitating fungal diseases in humans. Journal of Fungi 2021, 7, 367. [Google Scholar] [CrossRef] [PubMed]
- Bastos, R.W.; Rossato, L.; Goldman, G.H.; Santos, D.A. Fungicide effects on human fungal pathogens: Cross-resistance to medical drugs and beyond. PLoS Pathogens 2021, 17, e1010073. [Google Scholar] [CrossRef]
- Casadevall, A.; Kontoyiannis, D.P.; Robert, V. On the emergence of Candida auris: climate change, azoles, swamps, and birds. MBio 2019, 10. [Google Scholar] [CrossRef]
- Kurtzman, C.; Fell, J.W.; Boekhout, T. The yeasts: a taxonomic study; Elsevier: 2011.
- Jin, S.; Jin, Z. A case of septicemia caused by Kodamaea ohmeri. Chin J Clin Lab Sci 1994, 3, 167. [Google Scholar]
- Ioannou, P.; Papakitsou, I. Kodamaea ohmeri infections in humans: A systematic review. Mycoses 2020, 63, 636–643. [Google Scholar] [CrossRef]
- Bergman, M.; Gagnon, D.; Doern, G.V. Pichia ohmeri fungemia. Diagnostic microbiology and infectious disease 1998, 30, 229–231. [Google Scholar] [CrossRef]
- Zhou, M.; Li, Y.; Kudinha, T.; Xu, Y.; Liu, Z. Kodamaea ohmeri as an emerging human pathogen: a review and update. Frontiers in Microbiology 2021, 12, 736582. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Rudramurthy, S.; Kale, P.; Hariprasath, P.; Dhaliwal, M.; Singhi, S.; Rao, K. Epidemiological study of a large cluster of fungaemia cases due to Kodamaea ohmeri in an Indian tertiary care centre. Clinical Microbiology and Infection 2014, 20, O83–O89. [Google Scholar] [CrossRef] [PubMed]
- Otag, F.; Kuyucu, N.; Erturan, Z.; Sen, S.; Emekdas, G.; Sugita, T. An outbreak of Pichia ohmeri infection in the paediatric intensive care unit: case reports and review of the literature. Mycoses 2005, 48, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, J.; Dong, L.; Mai, J.; Zhang, L.; Zhu, J. Clinical features and homological analysis of Pichia ohmeri-caused hospital-acquired fungemia in premature infants. Zhonghua yi xue za zhi 2013, 93, 285–288. [Google Scholar] [PubMed]
- Ortiz, B.; Aguilar, K.; Galindo, C.; Molina, L.; Fontecha, G. Candida species isolated from clinical samples in a tertiary hospital in Honduras: Where is Candida auris? Current Medical Mycology 2022, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Fontecha, G.; Montes, K.; Ortiz, B.; Galindo, C.; Braham, S. Identification of cryptic species of four Candida complexes in a culture collection. Journal of Fungi 2019, 5, 117. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, M.; Bartolomé Álvarez, J.; Lockhart, S.R.; Valentín, E.; Ruiz-Gaitán, A.C.; Eraso, E.; de Groot, P.W.J. Identification of Candida auris and related species by multiplex PCR based on unique GPI protein-encoding genes. Mycoses 2021, 64, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Giusiano, G.; Piontelli, E. Hongos oportunistas levaduriformes y filamentosos comunes en clínica. 2016.
- Neji, S.; Hadrich, I.; Trabelsi, H.; Abbes, S.; Cheikhrouhou, F.; Sellami, H.; Makni, F.; Ayadi, A. Virulence factors, antifungal susceptibility and molecular mechanisms of azole resistance among Candida parapsilosis complex isolates recovered from clinical specimens. Journal of biomedical science 2017, 24, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Price, M.F.; Wilkinson, I.D.; Gentry, L.O. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia: Journal of Medical and Veterinary Mycology 1982, 20, 7–14. [Google Scholar] [CrossRef]
- Saiprom, N.; Wongsuk, T.; Oonanant, W.; Sukphopetch, P.; Chantratita, N.; Boonsilp, S. Characterization of Virulence Factors in Candida Species Causing Candidemia in a Tertiary Care Hospital in Bangkok, Thailand. Journal of Fungi 2023, 9, 353. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Faria-Gonçalves, P.; Oliveira, A.S.; Gaspar, C.; Rodrigues, L.; Palmeira-de-Oliveira, R.; Martinez-de-Oliveira, J.; Gonçalves, T.; Palmeira-de-Oliveira, A.; Rolo, J. Vulvovaginal Candida albicans Clinical Isolates’ Resistance to Phagocytosis In-Vitro. Life 2022, 12, 838. [Google Scholar] [CrossRef]
- Farooq, H.; Monowar, T.; Chinni, S.V.; Latt, S.S.; Zainol, N.H.; Sabesan, G.S. Epidemiology and molecular identification of mixed yeast isolates in Malaysia: A way forward. Current medical mycology 2022, 8, 35. [Google Scholar] [CrossRef]
- Mohammadi, R.; Mirhendi, H.; Rezaei-Matehkolaei, A.; Ghahri, M.; Shidfar, M.R.; Jalalizand, N.; Makimura, K. Molecular identification and distribution profile of Candida species isolated from Iranian patients. Medical Mycology 2013, 51, 657–663. [Google Scholar] [CrossRef]
- Kord, M.; Elmimoghaddam, A.; Hashemi, S.J.; Reziae, S.; Daie Ghazvini, R.; Salehi, M.; Abdollahi, A.; Ahmadi, A.; Getso, M.I.; Boekhout, T.; et al. Comparison of PCR-RFLP with 21-plex PCR and rDNA Sequencing for Identification of Clinical Yeast Isolates. Mycopathologia 2021, 186, 213–220. [Google Scholar] [CrossRef]
- Montes, K.; Ortiz, B.; Galindo, C.; Figueroa, I.; Braham, S.; Fontecha, G. Identification of Candida species from clinical samples in a Honduran tertiary hospital. Pathogens 2019, 8, 237. [Google Scholar] [CrossRef]
- Almeida, F.; Rodrigues, M.L.; Coelho, C. The still underestimated problem of fungal diseases worldwide. Frontiers in microbiology 2019, 10, 214. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A. Fungal diseases in the 21st century: the near and far horizons. Pathogens & immunity 2018, 3, 183. [Google Scholar]
- Johnson, E.M. Rare and emerging Candida species. Current Fungal Infection Reports 2009, 3, 152–159. [Google Scholar] [CrossRef]
- Ellwanger, J.H.; Chies, J.A.B. Candida auris emergence as a consequence of climate change: Impacts on Americas and the need to contain greenhouse gas emissions. The Lancet Regional Health–Americas 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Akinbobola, A.B.; Kean, R.; Hanifi, S.M.A.; Quilliam, R.S. Environmental reservoirs of the drug-resistant pathogenic yeast Candida auris. PLoS pathogens 2023, 19, e1011268. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Trubl, G.; Taş, N.; Jansson, J.K. Permafrost as a potential pathogen reservoir. One Earth 2022, 5, 351–360. [Google Scholar] [CrossRef]
- Morales-López, S.E.; Garcia-Effron, G. Infections due to rare Cryptococcus species. A literature review. Journal of fungi 2021, 7, 279. [Google Scholar] [CrossRef] [PubMed]
- Costa, G.L.d.; Negri, M.; Miranda, R.P.R.d.; Corrêa-Moreira, D.; Pinto, T.C.A.; Ramos, L.d.S.; Ferreira, D.G.; Salomão, B.; Fumian, T.M.; Mannarino, C.F. Candida palmioleophila: A New Emerging Threat in Brazil? Journal of Fungi 2023, 9, 770. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Bing, J.; Xu, X.; Zheng, Q.; Hu, T.; Hao, Y.; Li, S.; Nobile, C.J.; Zhan, P.; Huang, G. Candida vulturna outbreak caused by cluster of multidrug-resistant strains, China. Emerging Infectious Diseases 2023, 29, 1425. [Google Scholar] [CrossRef] [PubMed]
- Kabtani, J.; Boulanouar, F.; Militello, M.; Cassagne, C.; Ranque, S. Candida massiliensis sp. nov. Isolated from a Clinical Sample. Mycopathologia 2023, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kano, R.; Kimura, U.; Kakurai, M.; Hiruma, J.; Kamata, H.; Suga, Y.; Harada, K. Trichophyton indotineae sp. nov.: a new highly terbinafine-resistant anthropophilic dermatophyte species. Mycopathologia 2020, 185, 947–958. [Google Scholar] [CrossRef]
- Rosa, C.A.; Lachance, M.-A.; Starmer, W.T.; Barker, J.S.F.; Bowles, J.M.; Schlag-Edler, B. Kodamaea nitidulidarum, Candida restingae and Kodamaea anthophila, three new related yeast species from ephemeral flowers. International Journal of Systematic and Evolutionary Microbiology 1999, 49, 309–318. [Google Scholar] [CrossRef]
- Mtibaa, L.; Souid, H.; Jemli, B.; Hajjej, Z.; Halweni, C. Kodamaea ohmeri. An Emerging Yeast in Tunisia: First Identification in Three Case Reports and Literature Review. J Med Microb Diagn 2019, 8, 1000299. [Google Scholar]
- Agrawal, V.; Bhagwat, A.; Vishalakshi, V.; Gode, V.; Sawant, C. Exploring the potential of chromogenic medium for the identification of medically important yeast species other than Candida. Int J Pharm Pharm Sci 2014, 6, 291–294. [Google Scholar]
- Lee, J.S.; Shin, J.H.; Kim, M.-N.; Jung, S.-I.; Park, K.H.; Cho, D.; Kee, S.J.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Kodamaea ohmeri isolates from patients in a university hospital: identification, antifungal susceptibility, and pulsed-field gel electrophoresis analysis. Journal of clinical microbiology 2007, 45, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Yan, J.; Gao, Z.; Yang, H.; Tang, Y.; Yang, L. Subcutaneous granuloma caused by Kodamaea ohmeri in an immunocompromised patient in China. Australasian Journal of Dermatology 2020, 61, e213–e216. [Google Scholar] [CrossRef] [PubMed]
- Biswal, D.; Sahu, M.; Mahajan, A.; Advani, S.H.; Shah, S. Kodameae ohmeri–An emerging yeast: two cases and literature review. Journal of Clinical and Diagnostic Research: JCDR 2015, 9, DD01. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Yu, S.; Kudinha, T.; Xiao, M.; Wang, H.; Xu, Y.; Zhao, H. Identification and antifungal susceptibility profiles of Kodamaea ohmeri based on a seven-year multicenter surveillance study. Infection and drug resistance 2019, 1657–1664. [Google Scholar] [CrossRef] [PubMed]
- Sathi, F.A.; Aung, M.S.; Paul, S.K.; Nasreen, S.A.; Haque, N.; Roy, S.; Ahmed, S.; Alam, M.M.; Khan, S.; Rabbany, M.A. Clonal Diversity of Candida auris, Candida blankii, and Kodamaea ohmeri Isolated from Septicemia and Otomycosis in Bangladesh as Determined by Multilocus Sequence Typing. Journal of Fungi 2023, 9, 658. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Overview of antifungal ECOFFs and clinical breakpoints for yeasts, m.a.d.u.t.E.E.D., E.Def 9.4 and E.Def 11.0 procedures. Version 4.0, 2023. http://www.eucast.org.
- Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibility testing of yeasts 4th ed CLSI standard M27. Wayne, P. 2017.
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiology and immunology 2009, 53, 41–44. [Google Scholar] [CrossRef]
- Cendejas-Bueno, E.; Gomez-Lopez, A.; Mellado, E.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Identification of pathogenic rare yeast species in clinical samples: comparison between phenotypical and molecular methods. Journal of clinical microbiology 2010, 48, 1895–1899. [Google Scholar] [CrossRef]
- Sipiczki, M.; Tap, R.M. Candida vulturna pro tempore sp. nov., a dimorphic yeast species related to the Candida haemulonis species complex isolated from flowers and clinical sample. International Journal of Systematic and Evolutionary Microbiology 2016, 66, 4009–4015. [Google Scholar] [CrossRef]
- Schaller, M.; Borelli, C.; Korting, H.C.; Hube, B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 2005, 48, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Brunke, S.; Mogavero, S.; Kasper, L.; Hube, B. Virulence factors in fungal pathogens of man. Current opinion in microbiology 2016, 32, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Giacobino, J.; Montelli, A.C.; Barretti, P.; Bruder-Nascimento, A.; Caramori, J.T.; Barbosa, L.; Bagagli, E. Fungal peritonitis in patients undergoing peritoneal dialysis (PD) in Brazil: molecular identification, biofilm production and antifungal susceptibility of the agents. Medical Mycology 2016, 54, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Maciel, N.O.; Johann, S.; Brandão, L.R.; Kucharíková, S.; Morais, C.G.; Oliveira, A.P.; Freitas, G.J.; Borelli, B.M.; Pellizzari, F.M.; Santos, D.A. Occurrence, antifungal susceptibility, and virulence factors of opportunistic yeasts isolated from Brazilian beaches. Memórias do Instituto Oswaldo Cruz 2019, 114. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Johnson, A.D. Candida albicans biofilms and human disease. Annual review of microbiology 2015, 69, 71–92. [Google Scholar] [CrossRef]
- Malinovská, Z.; Čonková, E.; Váczi, P. Biofilm Formation in medically important Candida species. Journal of Fungi 2023, 9, 955. [Google Scholar] [CrossRef]
- Ramage, G.; Borghi, E.; Rodrigues, C.F.; Kean, R.; Williams, C.; Lopez-Ribot, J. Our current clinical understanding of Candida biofilms: where are we two decades on? APMIS 2023. [Google Scholar] [CrossRef] [PubMed]
- Wächtler, B.; Citiulo, F.; Jablonowski, N.; Förster, S.; Dalle, F.; Schaller, M.; Wilson, D.; Hube, B. Candida albicans-epithelial interactions: dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PloS one 2012, 7, e36952. [Google Scholar] [CrossRef]
- de Souza, C.; Perini, H.; Caloni, C.; Furlaneto-Maia, L.; Furlaneto, M. Adhesion of Candida tropicalis to polystyrene and epithelial cell lines: Insights of correlation of the extent of adherent yeast cells among distinct surfaces. Journal de Mycologie Médicale 2020, 30, 101043. [Google Scholar] [CrossRef]
- de Souza, C.M.; Perini, H.F.; Verri, W.A.; Zaninelli, T.H.; Furlaneto-Maia, L.; Furlaneto, M.C. Changes in adhesion of Candida tropicalis clinical isolates exhibiting switch phenotypes to polystyrene and HeLa cells. Mycopathologia 2021, 186, 81–91. [Google Scholar] [CrossRef]
- Pote, S.T.; Sonawane, M.S.; Rahi, P.; Shah, S.R.; Shouche, Y.S.; Patole, M.S.; Thakar, M.R.; Sharma, R. Distribution of Pathogenic Yeasts in Different Clinical Samples: Their Identification, Antifungal Susceptibility Pattern, and Cell Invasion Assays. Infection and Drug Resistance 2020, 13, 1133–1145. [Google Scholar] [CrossRef]
- Dalle, F.; Wächtler, B.; L’Ollivier, C.; Holland, G.; Bannert, N.; Wilson, D.; Labruère, C.; Bonnin, A.; Hube, B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cellular microbiology 2010, 12, 248–271. [Google Scholar] [CrossRef]
- Erwig, L.P.; Gow, N.A.R. Interactions of fungal pathogens with phagocytes. Nature Reviews Microbiology 2016, 14, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Romani, L. Immunity to fungal infections. Nature Reviews Immunology 2011, 11, 275–288. [Google Scholar] [CrossRef]
- Trovato, L.; Calvo, M.; Scalia, G.; Oliveri, S. A Comparative Prospective Study in Evaluating Candida spp. In Vitro Susceptibility through Micronaut-AM and Sensititre Yeast-One. Microbiology Research 2023, 14, 1077–1088. [Google Scholar] [CrossRef]
- Altinbaş, R.; BARIŞ, A.; Şen, S.; Öztürk, R.; Kiraz, N. Comparison of the Sensititre YeastOne antifungal method with the CLSI M27-A3 reference method to determine the activity of antifungal agents against clinical isolatesof Candida spp. Turkish Journal of Medical Sciences 2020, 50, 2024–2031. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Turnidge, J.; Alastruey-Izquierdo, A.; Botterel, F.; Canton, E.; Castro, C.; Chen, Y.-C.; Chen, Y.; Chryssanthou, E.; Dannaoui, E. Method-dependent epidemiological cutoff values for detection of triazole resistance in Candida and Aspergillus species for the Sensititre YeastOne colorimetric broth and Etest agar diffusion methods. Antimicrobial agents and chemotherapy 2019, 63. [Google Scholar] [CrossRef]
| Antifungal drug | Isolate (MIC) | ||
|---|---|---|---|
| C-29 (mg/L) | C-35 (mg/L) | C-38 (mg/L) | |
| Amphotericin B | 0.5 | 0.25 | 0.5 |
| Anidulafungin | 0.25 | 0.12 | 0.12 |
| Caspofungin | 2.0 | 1.0 | 0.25 |
| Micafungin | 0.12 | 0.25 | 0.12 |
| Fluconazole | 4.0 | 4.0 | 4.0 |
| Itraconazole | 0.12 | 0.12 | 0.12 |
| Posaconazole | 0.06 | 0.12 | 0.06 |
| Voriconazole | 0.03 | 0.03 | 0.03 |
| 5-Flucytosine | ≤ 0.06 | ≤ 0.06 | ≤ 0.06 |
| (Pz)** | ||||||
|---|---|---|---|---|---|---|
| Isolate | Biofilm production* | Gelatin | BSA | Casein | Phospholipase | Hemolisin |
| C-29 | 0.831 ± 0.12 Strong producer |
0.57 ± 0.04 | 0.48 ± 0.02 | 0.83 ± 0.06 | 0.67 ± 0.03 | 0.30 ± 0.02 |
| C-35 | 1.245 ± 0.16 Strong producer |
0.51 ± 0.05 | 0.38 ± 0.05 | 0.89 ± 0.01 | 0.62 ± 0.03 | 0.30 ± 0.04 |
| C-38 | 0.727 ± 0.15 Strong producer |
0.56 ± 0.02 | 0.46 ± 0.04 | 0.89 ± 0.02 | 0.67 ± 0.05 | 0.30 ± 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





