1. Introduction
Inflammation constitutes a crucial component of the body's immune reaction. It may manifest as an acute response, rapidly emerging (such as following trauma or during an infection), or it can persist over an extended period (chronic) in association with conditions like autoimmune diseases or cancer [
1,
2,
3]. Inflammation indices are released into the bloodstream as inflammation indices in response to significant inflammation in a particular organ or tissue, resulting from injury, ischemia, or infection. Assessing these indicators proves valuable in identifying the presence of diseases involving inflammation and tracking their progression, particularly post-initiation of therapy[
4,
5]. Moreover, these markers can aid in assessing the risk of developing specific pathologies[
4].
Typically, tests for inflammatory markers are prescribed alongside other diagnostic examinations when a doctor suspects an acute or chronic disease linked to tissue or cell damage in the patient[
4,
5]. Inflammation often leads to the production of inflammatory cytokines, vital proteins within the immune system that play a pivotal role in the inflammatory response. These proteins are essential for activating and coordinating immune cells involved in defending the body against injury and infection. Nevertheless, an excessive amount of inflammatory cytokines can be detrimental, emphasizing the significance of maintaining a proper balance in their production and activation[
6,
7].
There are also anti-inflammatory cytokines. These cytokines play a role in balancing the immune system and modulating the inflammatory response. While inflammatory cytokines promote inflammation to defend the body from injury and infection, anti-inflammatory cytokines act to limit and control this process, contributing to the resolution of inflammation [
8,
9].
Some notable anti-inflammatory cytokines include interleukin (IL-10). These play a key role in maintaining homeostasis and preventing an excessive inflammatory response that could damage tissues. The balance between inflammatory and anti-inflammatory cytokines is critical for adequate regulation of the immune response[
6,
7,
8,
9,
10].
The current study employed molecular docking studies [
11,
12] to investigate various drugs and natural substances in their interaction with TNF-alpha.
The objective was to understand which compounds exhibit a strong binding affinity within the ligand binding site of this cytokine.
Tumor necrosis factor (TNF), a vital cytokine produced by white blood cells, plays a crucial role in regulating the immune response, particularly in the context of tumor-related pathologies. TNF functions by promoting inflammation, facilitating the production of cells involved in the inflammatory response, and aiding in cellular healing[
13,
14]. Often denoted as TNF-alpha, it holds significance in inflammatory bowel diseases (IBD), including Crohn's disease and ulcerative colitis[
15].
3. Results and Discussion
Inflammation plays a pivotal role in the body's immune response, presenting as either an acute reaction that rapidly arises (e.g., post-trauma or during an infection) or as a chronic state that persists over an extended duration, commonly associated with conditions such as autoimmune diseases or cancer [
1,
2,
3].
The present investigation utilized molecular docking studies [
11,
12] to explore the interactions of various drugs and natural substances with TNF-alpha. The primary goal was to discern which compounds demonstrate a robust binding affinity within the ligand binding site of this cytokine.
Tumor necrosis factor (TNF), an essential cytokine produced by white blood cells, plays a pivotal role in regulating the immune response, particularly in the context of tumor-related pathologies. TNF operates by promoting inflammation, facilitating the production of cells involved in the inflammatory response, and aiding in cellular healing[
13,
14]. Commonly referred to as TNF-alpha, it carries significance in inflammatory bowel diseases (IBD), including Crohn's disease and ulcerative colitis[
15].Developing effective TNF-alpha inhibitors is a promising avenue for intervention in various health conditions and underscores the importance of targeted therapies in the field of medicine. Molecular docking methods [
16,
17,
18] are useful for this goal.
Indeed, the present work presents for the first time, some potential drugs TNF-alpha inhibitors as Dactolisib with binding energy of -10.1 kcal/mol, Nilotinib, with binding energy of -9.6 kcal/mol, Eltrombopag with binding energy of -9.7 kcal/mol, Radotinib with binding energy of -9.6 kcal/mol and Nilotinib with binding energy of -9.7. Additonally Hypericin with binding energy of -9.9 kcal/mol could be considered natural TNF-alpha inhibitor. These computational results are preliminary and necessary for further biological studies to confirm the results of these data are very important for the scientific community, potentially applicable in the field of infiammatory malatties.
Molecular docking methods [
16,
17,
18] are proved valuable in the pursuit of identifying TNF-alpha inhibitors. In this study, novel potential drugs with inhibitory effects on TNF-alpha are introduced for the first time. Dactolisib exhibited a promising binding energy of -10.1 kcal/mol, followed by Nilotinib at -9.6 kcal/mol, Eltrombopag at -9.7 kcal/mol, Radotinib at -9.6 kcal/mol, and Nilotinib again at -9.7 kcal/mol. Additionally, Hypericin demonstrated a noteworthy binding energy of -9.9 kcal/mol, positioning it as a potential natural TNF-alpha inhibitor.
These computational findings, though preliminary, lay the foundation for further biological studies that are crucial to confirming their efficacy. The implications of these results extend to the scientific community, holding potential applications in the realm of inflammatory diseases.
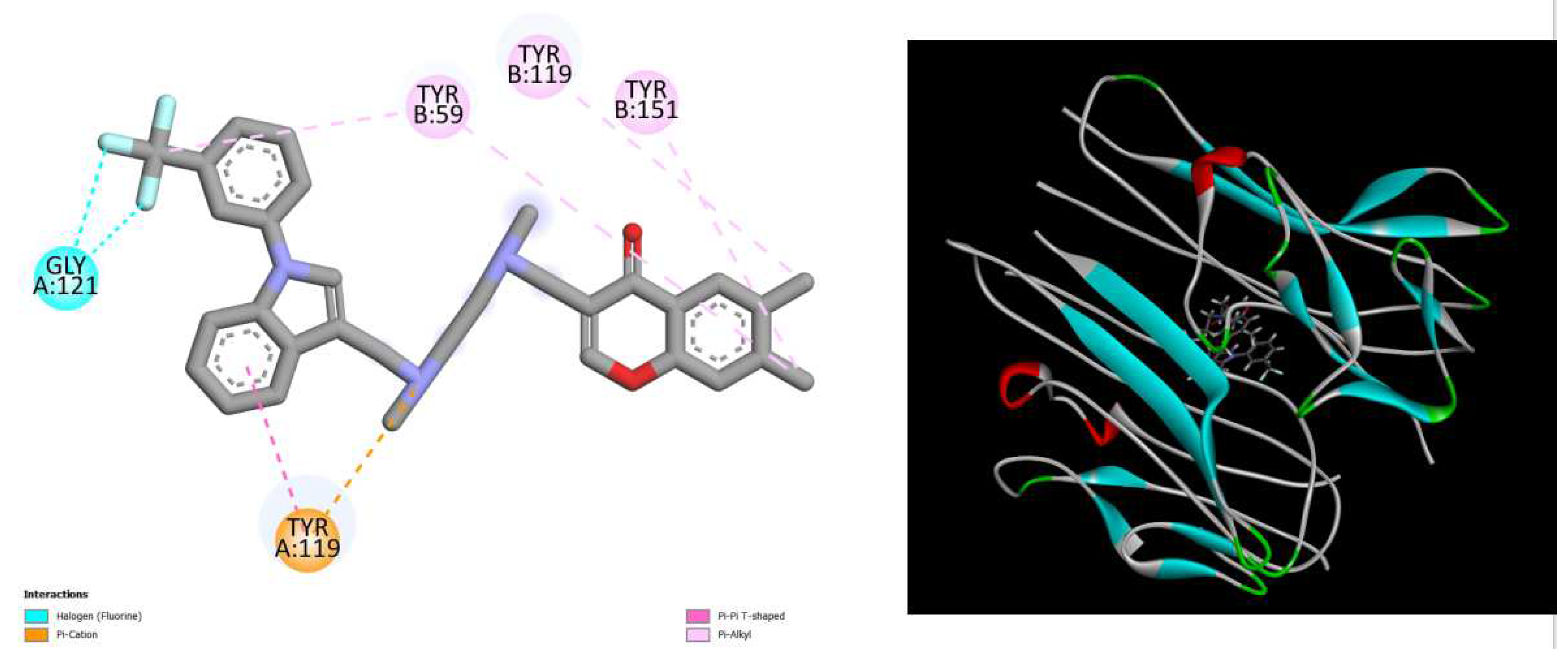
Figure 1.
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with crystal ligand 307(6,7-DIMETHYL-3-[(METHYL{2-[METHYL({1-[3-(TRIFLUOROMETHYL)PHENYL]-1H-INDOL-3-YL}METHYL)AMINO]ETHYL}AMINO)METHYL]-4H-CHROMEN-4-ONE) with binding energy of -8.8 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and 307. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of 307.
Figure 1.
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with crystal ligand 307(6,7-DIMETHYL-3-[(METHYL{2-[METHYL({1-[3-(TRIFLUOROMETHYL)PHENYL]-1H-INDOL-3-YL}METHYL)AMINO]ETHYL}AMINO)METHYL]-4H-CHROMEN-4-ONE) with binding energy of -8.8 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and 307. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of 307.
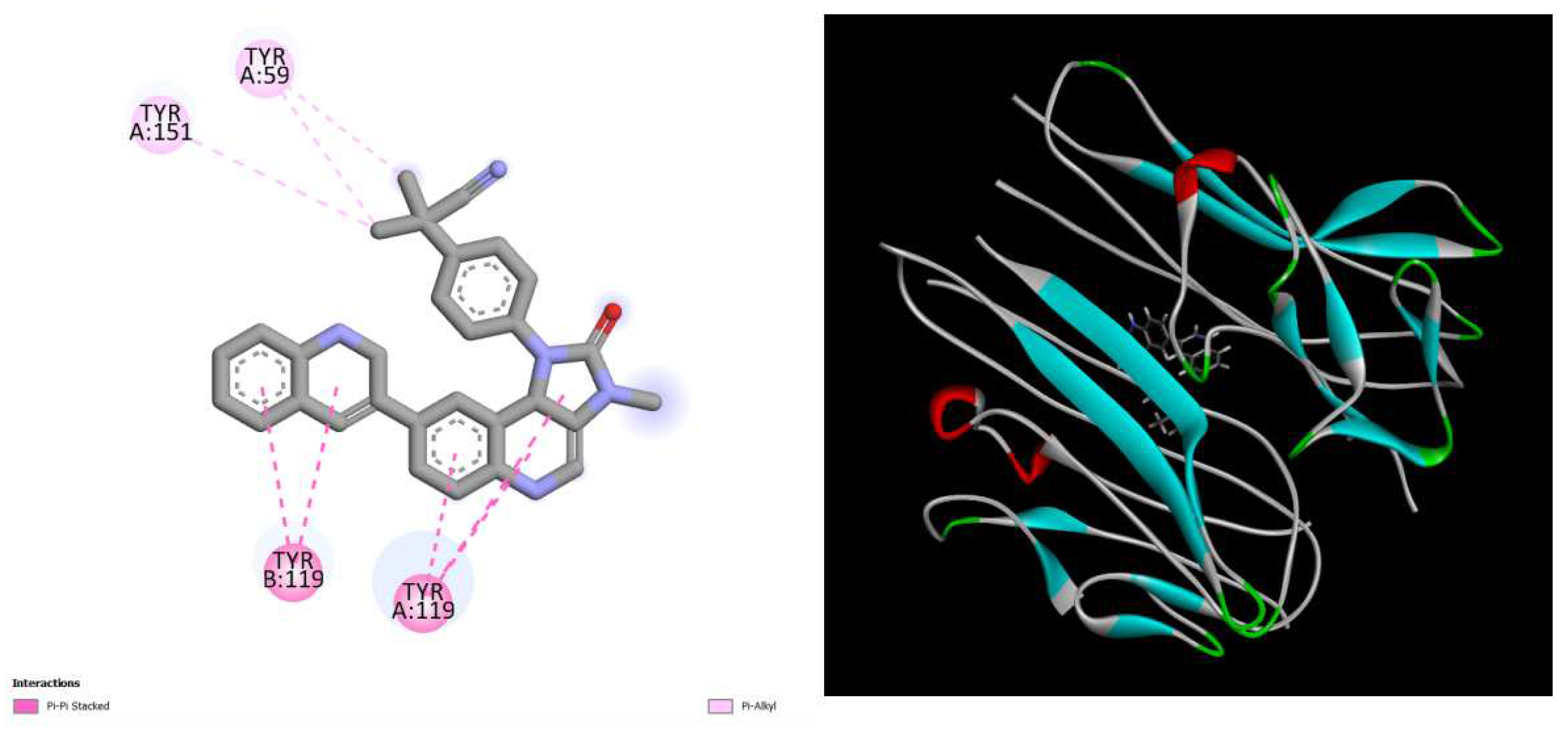
Figure 2.
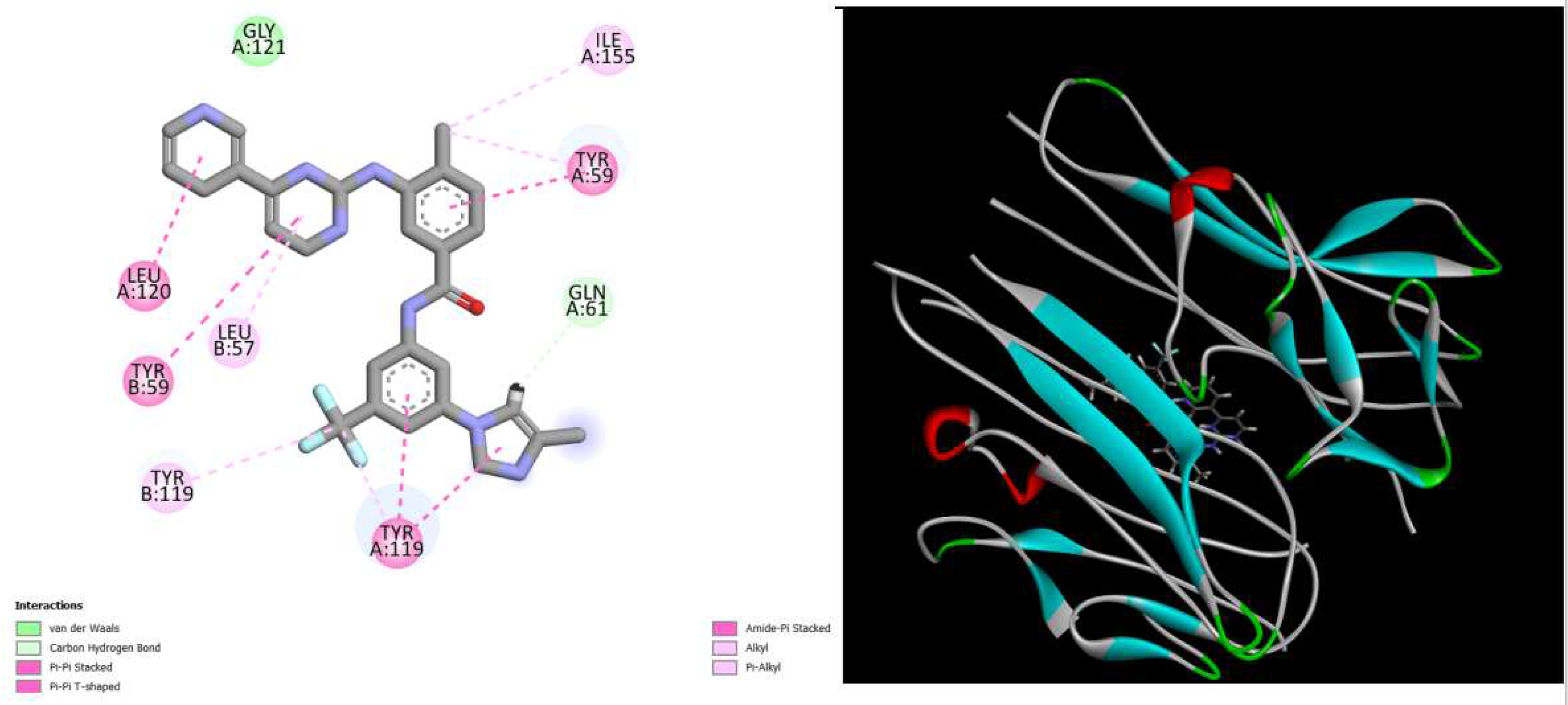
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with docked dactolisib with binding energy of -10.1 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and dactolisib. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of dactolisib.
Figure 2.
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with docked dactolisib with binding energy of -10.1 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and dactolisib. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of dactolisib.
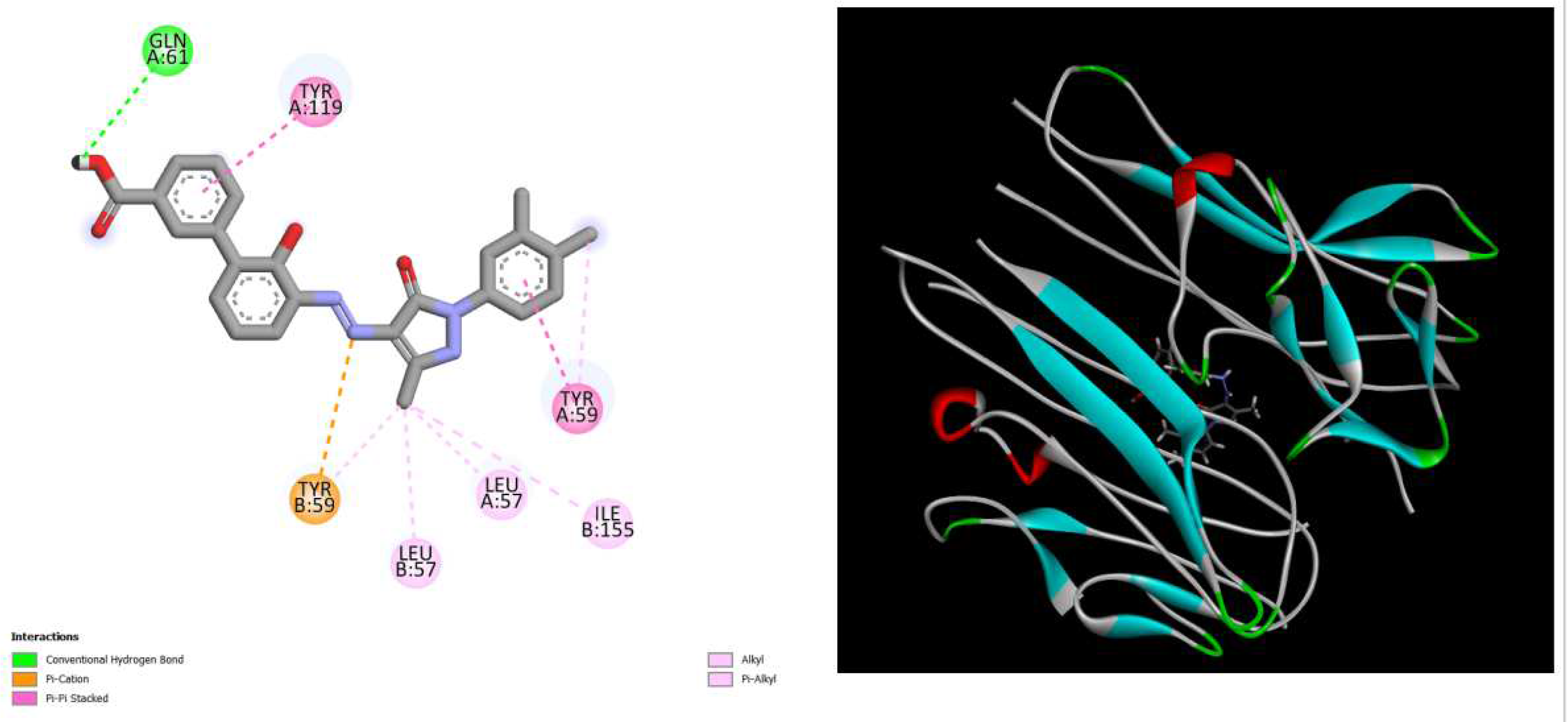
Figure 3.
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with docked Eltrombopag with binding energy of -9.7 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Eltrombopag. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Eltrombopag.
Figure 3.
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with docked Eltrombopag with binding energy of -9.7 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and Eltrombopag. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Eltrombopag.
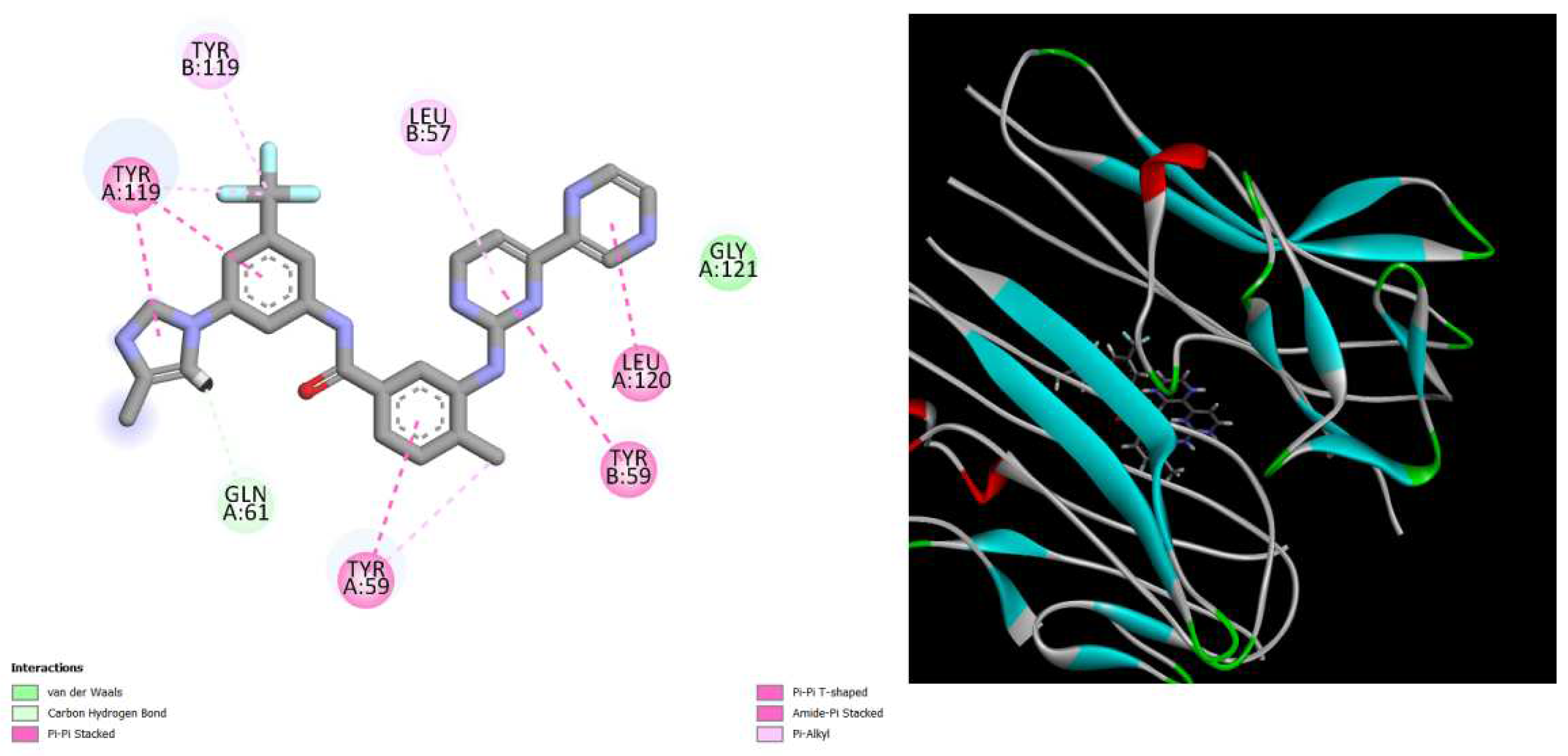
Figure 4.
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with docked Radotinib with binding energy of -9.6 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and dRadotinib.
Figure 4.
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with docked Radotinib with binding energy of -9.6 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and dRadotinib.
Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Radotinib.
Figure 5.
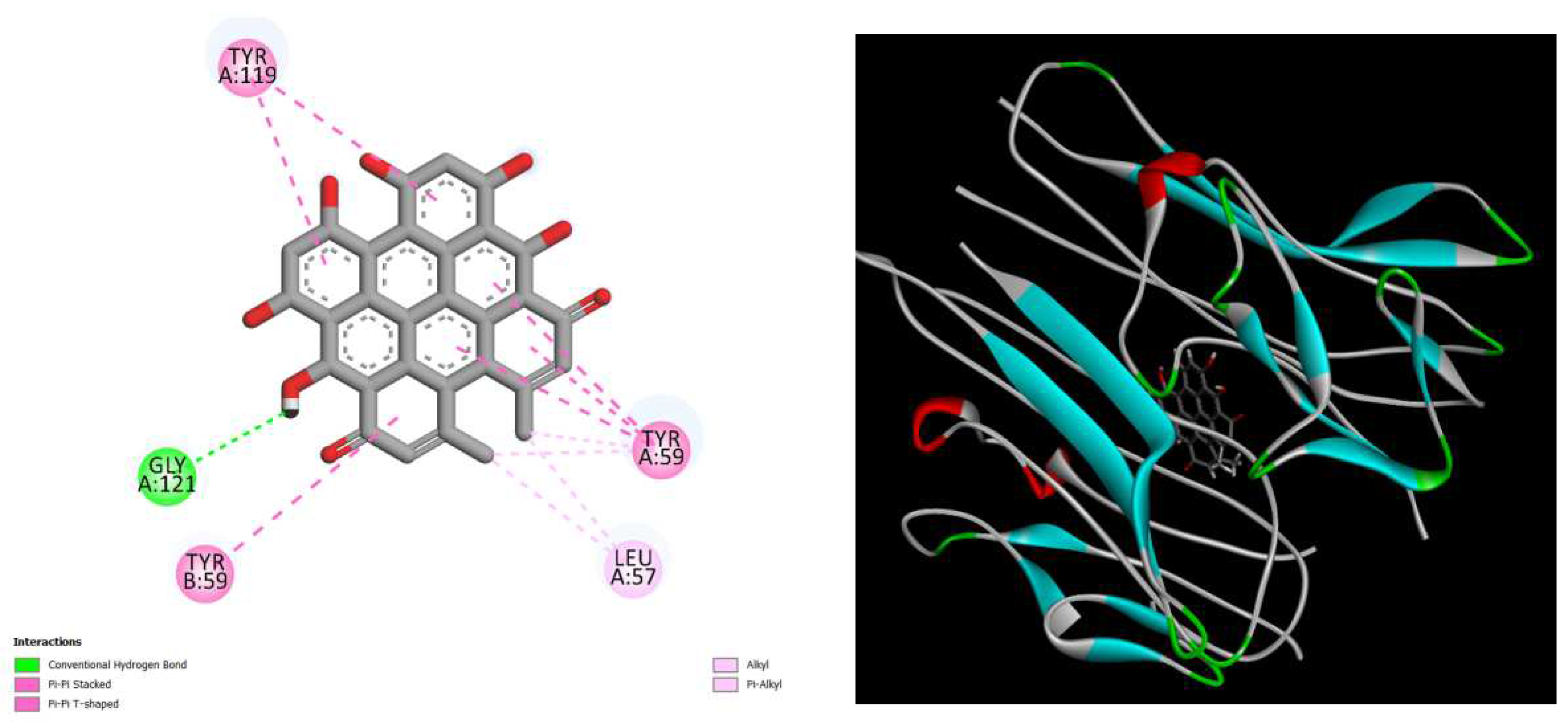
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with docked hypericin with binding energy of -9.9 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and hypericin Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of hypericin.
Figure 5.
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with docked hypericin with binding energy of -9.9 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and hypericin Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of hypericin.
Figure 6.
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with docked nilotinib with binding energy of -9.6 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and nilotinib. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of nilotinib.
Figure 6.
displays the docking outcomes of TNF-alpha Crystal Structure of TNF-alpha in conjunction with docked nilotinib with binding energy of -9.6 kcal/mol within the Ligand Binding Site, as analyzed by Autodock Vina with pyrx program. On the left side, 2D diagrams illustrate the residue interactions between the protein and nilotinib. Meanwhile, the right side exhibits the Ligand Binding Site of the protein, highlighting the specific location of nilotinib.