2. Results
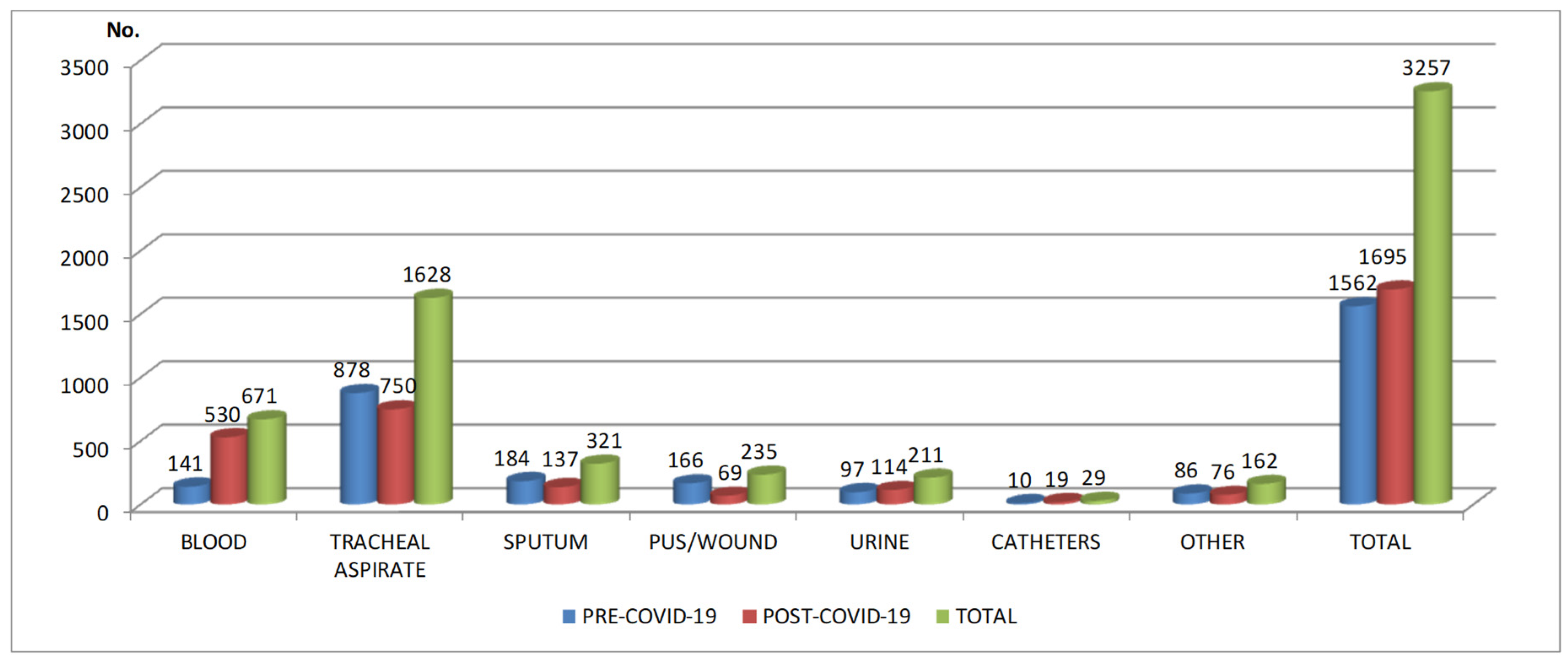
In this research were collected 3257 samples. 1562 samples and 1940 bacterial isolates (
Figure 1) were obtained in pre-COVID-19 period from 1267 patients admitted in ICU (797 males - 62.90%; 470 females – 37.10%; average age: 64±18.60) (
Table 1). 1695 samples and 2281 bacterial isolates were obtained from 1354 patients in post-COVID-19 period (841 males – 62.11%; 513 females – 37.89%; average age: 64±17,51).
In pre-COVID-19 period, most samples came from respiratory tract (1062 – 68%), pus/wound swabs (166-10.63%) and blood (141- 9.02%). In post-COVID-19 period, almost half of the samples originated from respiratory tract (887 – 52.33%), while blood samples were more than three times higher compared with pre-COVID-19 period (530- 31.27%). The number of pus/wound swabs from which pathogens were isolated was less than half in post-COVID-19 period (69-4.07%), compared with pre-COVID-19 period (166-10.63%) (
Table 2).
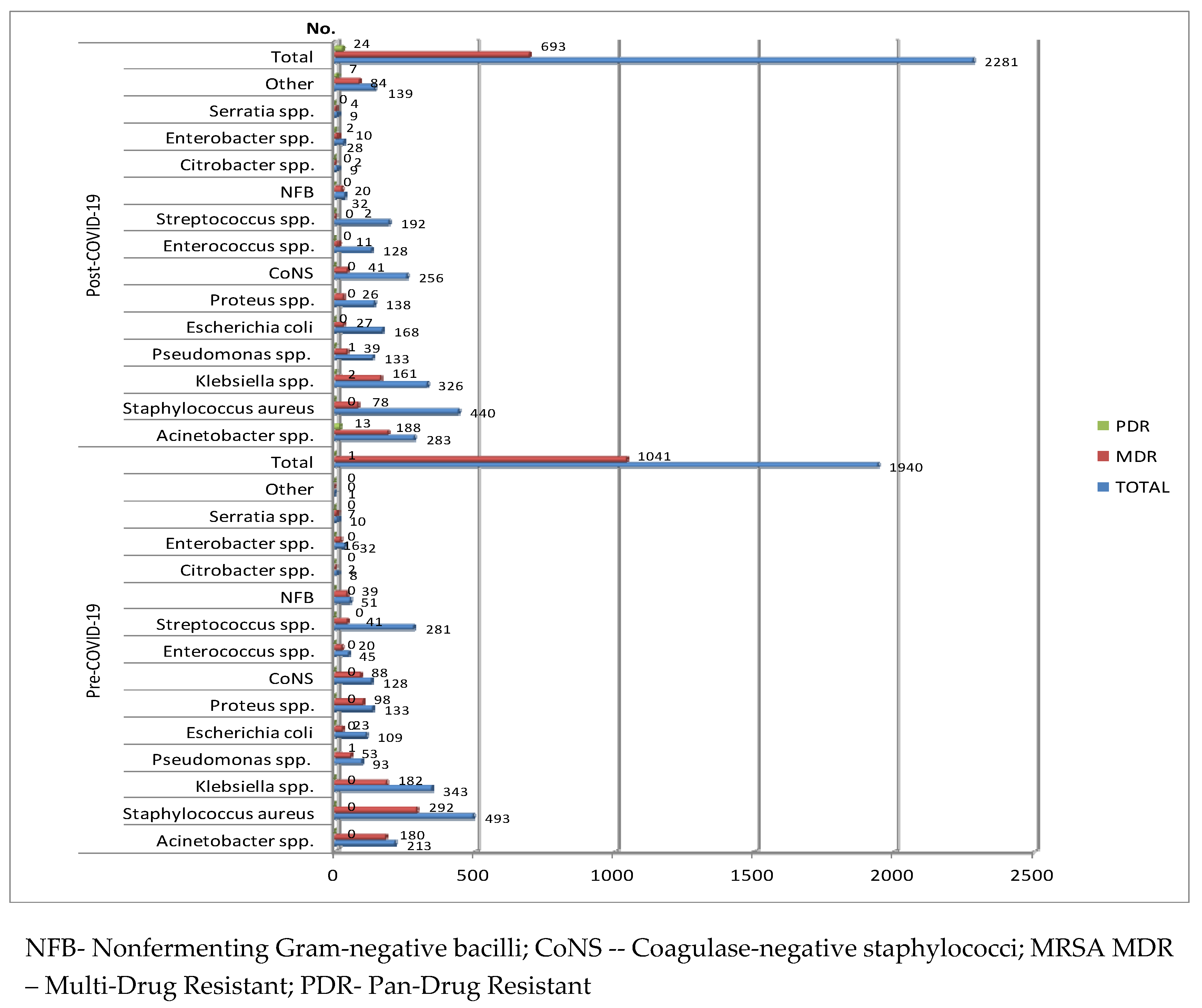
The most frequently isolated of all micro-organisms identified in the harvested samples in both the pre-COVID-19 and post-COVID-19 period were Staphylococcus aureus and Klebsiella spp. (
Figure 2).
In the pre-COVID-19 period, the most commonly reported Gram-negative bacterial pathogens registered was Klebsiella spp. (343-17.68%), followed by Acinetobacter spp. (213-10.98%), Proteus (133—6.86%), E. coli (109-5.62%) and Pseudomonas spp. (93-4.80%). More than half of the strains (1041-53.66%) were multidrug-resistant (MDR). (fig.2). The most prevalent Gram-positive bacterial pathogens were Staphylococcus aureus (S. aureus) (493-25.41%), followed by Streptococcus spp. (281-14.48%). 315 strains of S. aureus were Methicillin-Resistant (MRSA - Methicillin-Resistant Staphylococcus Aureus).
In the post-COVID-19 period, the hierarchy is preserved in the case of Gram-negative bacterial pathogens, but the percentage of strains of Acinetobacter spp. (12.41%-283), E. coli (168-7.36%) and Pseudomonas spp. (133-5.83%) has increased, while that of Klebsiella spp. has decreased (14.29%-326). In the case of Gram-positive pathogens, there has been an important increase in the number and percentage of the strains of Coagulase-negative staphylococci (CoNS) (256-11.22%) and Enterococcus spp. (128-5.61%), while those of Staphylococcus aureus and Streptococcus spp. were lower.
Almost 70% from all the pathogens identified in the samples harvested in the pre-COVID-19 period and almost 60% in the post-COVID-19 period were found in tracheal aspirate/sputum (
Figure 1) and Staphylococcus aureus and Klebsiella spp. were the most common isolated pathogens from these samples (
Figure 1).
More than half of the isolated pathogens in blood samples were CoNS (55.19%) in the pre-COVID-19 period, followed by Klebsiella spp. (11.03%). In the post-COVID-19, CoNS were the most prevalent (40.13%), followed by Acinetobacter spp. (8.15%) and Klebsiella spp.(5.7%). The most prevalent pathogen in tracheal aspirate/sputum was S. aureus, both in the pre-COVID-19 (30.03%) and in the post-COVID-19 period (28.13%). In urine specimens Klebsiella spp. (30.55%) was the most predominant pathogen in the pre-COVID-19 period, while E coli occupied the first place in the post-COVID-19 period (33.04%).
S. aureus held, in the pre-COVID-19 period, the highest percentage of isolated pathogens from pus/wound swabs among isolated pathogens (22.70%), while Klebsiella spp. was the most prevalent in the post-COVID-19 period, followed by Acinetobacter spp., Proteus spp. and S. aureus (12.90%). A small number of pathogens was isolated from intravascular catheters, S. aureus being the most prevalent in both periods (
Table 2).
More than half of all the pathogens were MDR in the pre-COVID-19 period and almost a third in post-COVID-19 period era. Among the MDR Gram-positive pathogens, almost 60% of the S. aureus strains (292/493), 70% of the CoNS strains (88/128) and 45% (20/45) of the Enterococcus spp. strains were MDR in the pre-COVID-19 period. In the post COVID-19 period, only 17.72% of the S. aureus strains (78/440) were MDR. Although the number of CoNS strains was almost double (256) in the same period, only 16% were MDR. The same thing was found in the case of strains of Enterococcus spp. (128 strains-11 MDR) (
Figure 2).
In the case of the most frequently isolated Gram-negative bacteria, a very high percentage of MDR isolates was found in the pre-COVID-19 period in the case of Acinetobacter spp. (84.5%), Nonfermenting Gram-negative bacilli (NFB) (76.47%), Klebsiella spp. (53.06%), Proteus spp. (73.68%), Pseudomonas spp. (56.98%), (figure....). Also, a pandrug-resistant (PDR) Pseudomonas strain was isolated. One fifth of the E. coli strains (23/109) were MDR. Although in the post COVID-19 period, there has been a decrease in the number and percentage of Gram-negative strains, 16 strains were PDR, of which 13 were Acinetobacter spp. (
Figure 2)
The antibiotic resistance rates of the Gram-positive and Gram-negative isolates are summarized in
Table 3 and
Table 4.
Table 3.
Distribution of pathogens isolated from different specimen types in ICU, County Emergency Clinical Hospital Craiova, Romania, pre- and post-COVID-19 periods.
Table 3.
Distribution of pathogens isolated from different specimen types in ICU, County Emergency Clinical Hospital Craiova, Romania, pre- and post-COVID-19 periods.
| |
Klebsiella spp. |
Escherichia coli |
Pseudomonasspp.
|
|
Antimicrobialagent
|
Pre-COVID-19(n= 343)
|
Post-COVID-19(n=326)
|
p value |
Pre-COVID-19(n=109)
|
Post-COVID-19(n=168)
|
pvalue
|
Pre-COVID-19(n=93)
|
Post-COVID-19(n=133)
|
pvalue
|
| Amoxicillin/clavulanic acid |
217(66.36%) |
194 (62.98%) |
0.15 |
31(29.52%) |
60 (36.58%) |
0.23 |
18(100%) |
7 (87.5%) |
0.12 |
| Ceftazidime |
209(65.31%) |
223 (69.68%) |
0.23 |
32(31.68%) |
64 (38.55%) |
0.25 |
53(60.92%) |
86 (67.72%) |
0.30 |
| Ceftriaxone |
213(65.34%) |
212 (67.30%) |
0.59 |
37(35.58%) |
55 (33.95%) |
0.78 |
14(82.35%) |
7 (70%) |
0.45 |
| Cefotaxime |
99(65.56%) |
137 (65.55%) |
<0.001* |
7(26.92%) |
4033.78% |
0.31 |
9(75%) |
9 (81.82%) |
0.69 |
| Cefazolin |
152(77.16%) |
211 (79.32%) |
0.57 |
32(57.14%) |
62 (51.24%) |
0.46 |
5(100%) |
6 (100%) |
- |
| Cefepime |
132(61.68%) |
172 (55.30%) |
0.14 |
21(33.33%) |
40 (25.97%) |
0.27 |
52(76.47%) |
55 (58.51%) |
0.01* |
| Imipenem |
108(40.60%) |
113 (45.56%) |
0.25 |
16(16.49%) |
7 (6.14%) |
0.01* |
45(58.44%) |
35 (47.30%) |
0.17 |
| Meropenem |
111(45.68%) |
106 (44.35%) |
0.76 |
2(2.60%) |
9(9.57%) |
0.06 |
52(65%) |
47 (57.32%) |
0.31 |
| Ciprofloxacin |
199(59.76%) |
179 (61.30%) |
0.69 |
43(39.82%) |
64 (43.54%) |
0.74 |
49(55.68%) |
76 (62.29%) |
0.33 |
| Levofloxacin |
40(55.56%) |
82 (59%) |
0.63 |
13(52%) |
21 (28.76%) |
0.03 |
31(60.78%) |
49 (56.98%) |
0.21 |
| Piperacillin/tazobactam |
35(71.43%) |
119 (61.34%) |
0.19 |
0(0%) |
24(24%) |
0.14 |
28(36.84%) |
39 (47.56%) |
0.36 |
| Colistin |
1(0.38%) |
40 (20.51%) |
<0.001* |
3(4.69%) |
25 (32.46%) |
<0.001* |
0(0%) |
4 (4.88%) |
0.03 |
| Gentamicin |
112(44.62%) |
203 (64.85%) |
<0.001* |
21(36.94%) |
60 (37.73%) |
0.90 |
38(61.29%) |
77 (60.63%) |
0.93 |
| Aztreonam |
182(56.35%) |
180 (73.60%) |
<0.001* |
27(27%) |
31 (27.43%) |
0.94 |
39(50%) |
34 (39.08%) |
0.15 |
Table 3.
(continued) Distribution of pathogens isolated from different specimen types in ICU, County Emergency Clinical Hospital Craiova, Romania, pre- and post-COVID-19 periods.
Table 3.
(continued) Distribution of pathogens isolated from different specimen types in ICU, County Emergency Clinical Hospital Craiova, Romania, pre- and post-COVID-19 periods.
| |
Acinetobacter spp. |
Proteus spp. |
|
Antimicrobialagent
|
Pre-COVID-19(n=213)
|
Post-COVID-19(n=283)
|
p value |
Pre-COVID-19(n=133)
|
Post-COVID-19(n=138)
|
p value |
| Amoxicillin/clavulanic acid |
87(94.57%) |
47 (100%) |
0.10 |
100(78.13%) |
96 (73.85%) |
0.42 |
| Ceftazidime |
165(93.75%) |
265 (96.01%) |
0.27 |
89(72.95%) |
100 (73.53%) |
0.91 |
| Ceftriaxone |
189(97.73%) |
270 (97.12%) |
0.84 |
99(76.15%) |
91 (67.41%) |
0.11 |
| Cefotaxime |
159(95.78%) |
230 (95.43%) |
0.86 |
45(80.36%) |
71 (72.45%) |
0.27 |
| Cefazolin |
52(100%) |
49 (100%) |
- |
80(94.12%) |
103 (84.43%) |
0.03* |
| Cefepime |
132(95.65%) |
141 (84.43%) |
<0.001* |
40(54.80%) |
28 (21.37%) |
<0.001* |
| Imipenem |
77(90.58%) |
156 (90.17%) |
0.91 |
49(46.67%) |
33 (36.67%) |
0.15 |
| Meropenem |
150(88.76%) |
192 (88.48%) |
0.93 |
19(19.79%) |
10 (22.73%) |
0.69 |
| Ciprofloxacin |
190(92.23%) |
165 (94.83%) |
0.30 |
82(68.33%) |
93 (75%) |
0.24 |
| Levofloxacin |
47(88.68%) |
104 (88.88%) |
0.96 |
20(86.96%) |
41 (75.93%) |
0.27 |
| Piperacillin/tazobactam |
118(86.76%) |
186 (91.62%) |
0.14 |
0(0%) |
20 (26.32%) |
0.06 |
| Colistin |
7(3.37%) |
36 (18.09%) |
<0.001* |
95(100%) |
56 (100%) |
- |
| Gentamicin |
70(86.42%) |
236 (85.19%) |
0.78 |
72(76.59%) |
75 (55.97%) |
0.001* |
| Aztreonam |
77(96.25%) |
47 (81.03%) |
0.003* |
34(27.2%) |
24 (26.97%) |
0.96 |
Table 4.
Antimicrobial resistance pattern of Gram-positive bacteria isolated from patients hospitalized in ICU, County Emergency Clinical Hospital Craiova, Romania, pre- and post-COVID-19 periods.
Table 4.
Antimicrobial resistance pattern of Gram-positive bacteria isolated from patients hospitalized in ICU, County Emergency Clinical Hospital Craiova, Romania, pre- and post-COVID-19 periods.
| |
Staphylococcus aureus |
CoNS |
|
Antimicrobialagent
|
Pre-COVID-19(n=493)
|
Post-COVID-19(n=440)
|
pvalue
|
Pre-COVID-19(n=128)
|
Post-COVID-19(n=256)
|
p value |
| Ciprofloxacin |
296(61.67%) |
259 (59.27%) |
0.45 |
90(70.87%) |
149 (59.36%) |
0.18 |
| Clindamycin |
366(76.25%) |
259 (77.72%) |
0.59 |
85(66.93%) |
163 (68.20%) |
0.80 |
| Clarithromycin |
219(56.74%) |
243 (60.90%) |
0.23 |
73(67.60%) |
54 (73.97%) |
0.35 |
| Doxycycline |
190(40.08%) |
135 (51.72%) |
0.002* |
52(54.74%) |
7 (43.75%) |
0.41 |
| Erythromycin |
359(74.17%) |
239 (55.58%) |
<0.001* |
100(80.65%) |
99 (68.27%) |
0.02* |
| Linezolid |
1(0.22%) |
8 (3.13%) |
<0.001* |
16(23.88%) |
11(9.65%) |
0.009 |
| Penicillin |
474(98.34%) |
382 (87.01%) |
<0.001* |
112(93.33%) |
231 (93.52%) |
0.94 |
| Rifampicin |
252(53.16%) |
263 (64.93%) |
<0.001* |
57(45.24%) |
157 (62.55%) |
0.001* |
| Tetracycline |
224(62.57%) |
25 (78.12%) |
0.07 |
85(71.43%) |
81 (72.32%) |
0.88 |
| Oxacillin |
354(72.99%) |
229 (73.40%) |
0.89 |
104(81.89%) |
119 (76.28%) |
0.25 |
| Vancomycin |
2(16.67%) |
1 (3.58%) |
0.14 |
12(15.79%) |
1 (0.91%) |
<0.001* |
| Teicoplanin |
5(26.31%) |
55 (53.40%) |
0.03* |
16(26.67%) |
96 (63.57%) |
<0.001* |
Table 4.
(continued) Antimicrobial resistance pattern of Gram-positive bacteria isolated from patients hospitalized in ICU, County Emergency Clinical Hospital Craiova, Romania, pre- and post-COVID-19 periods.
Table 4.
(continued) Antimicrobial resistance pattern of Gram-positive bacteria isolated from patients hospitalized in ICU, County Emergency Clinical Hospital Craiova, Romania, pre- and post-COVID-19 periods.
| |
Streptococcusspp.
|
Enterococcus spp. |
|
Antimicrobialagent
|
Pre-COVID-19(n=281)
|
Post-COVID-19(n=192)
|
p value |
Pre-COVID-19(n=45)
|
Post-COVID-19(n=128)
|
pvalue
|
| Ciprofloxacin |
5(100%) |
3 (100%) |
- |
37(86.05%) |
91 (72.8%) |
0.07 |
| Clindamycin |
80 (29.31%) |
50 (27.03%) |
0.59 |
1(100%) |
1 (50%) |
- |
| Clarithromycin |
84(49.12%) |
53 (27.03%) |
<0.001* |
4(100%) |
5 (71.43%) |
0.23 |
| Doxycycline |
24(10.57%) |
16 (19.75%) |
0.03* |
25(80.65%) |
19 (33.33%) |
<0.001* |
| Erythromycin |
144(52.55%) |
46 (29.11%) |
<0.001* |
6(100%) |
42 (75%) |
0.16 |
| Linezolid |
0(0%) |
0 (0%) |
- |
2(4.54%) |
1 (1.20%) |
0.23 |
| Penicillin |
200(85.11%) |
74 (62.18%) |
<0.001* |
16(38.09%) |
60 (61.22%) |
0.01* |
| Rifampicin |
4(20%) |
3 (3.61%) |
0.008 |
- |
19 (51.35%) |
- |
| Tetracycline |
41(35.04%) |
34 (32.69%) |
0.71 |
6(75%) |
13 (41.93%) |
0.09 |
| Oxacillin |
170(91.89%) |
37(87.70%) |
0.73 |
- |
24 (96%) |
- |
| Vancomycin |
3(1.11%) |
0 (0%) |
0.26 |
7(29.17%) |
4 (5.33%) |
0.001* |
| Teicoplanin |
- |
- |
- |
12(29.27%) |
27 (36.98%) |
0.40 |
In the case of antimicrobial resistance rates of the Gram-negative bacteria, almost 80% from the Klebsiella spp. strains isolated in both periods were resistant to first generation cephalosporins, around 65% to third-generation and 55-60% to fourth-generation cephalosporins (
Table 3). Overall, the number of isolated Klebsiella spp. strains decreased in the post COVID-19 period, but the number of resistant strains were higher, with statistically significant difference (p<0.001) for Cefotaxime. Around 45% of the strains were resistant to carbapenems (imipenem and meropenem) and around 55% to fluoroquinolones, with no statistically significant difference between the two periods. Klebsiella spp. had a statistically significant increase in resistance rates (p<0.001) in the post COVID-19 period compared with to the pre-COVID-19 period, against colistin (0.38% to 20.51%), gentamicin (44.62% to 64.85%) and aztreonam (56.35% to 3.60%). For piperacillin-tazobactam, it was registered a decrease in the resistance rates, but without statistical significance. Two PDR Klebsiella strains were found (
Figure 2).
In the case of E. coli strains, it was registered a significant increase in the resistance rate against colistin (4.69% to 32.46%, p<0.001) and a significant decrease against levofloxacin (52% to 28.76%, p=0.03) and imipenem (16.49% to 6.14%, p=0.01). The number of the isolated strains was higher in the post-COVID-19 period, with an increased resistance rate against amoxicillin/clavulanic acid, ceftazidime, cefotaxime and meropenem, but without statistical significance (
Table 3). The highest resistance rate was against cefazolin, over 50%. The percentage of MDR strains decreased in the post-COVID-19 period (16.07%).
Antimicrobial resistance in Acinetobacter spp. isolates revealed a very high level of resistance against the third generation cephalosporins (over 95%). High resistance was found also to the carbapenems (around 90%), piperacillin-tazobactam (from 88% to 91%), fluoroquinolones (from 88% to 95%) and aminoglycosides (around 85%) (
Table 3). In the post-COVID-19 period the number of Acinetobacter sp. strains were higher, with a significant increase of the antimicrobial resistance rate against colistin (3.37% to 18.09%, p<0.001) and a significant decrease against cefepime (95.65% to 84.43%, p<0.001) and aztreonam (96.25% to 81.03%, p=0.003). All the isolated Acinetobacter spp. strains were found resistant to amoxicillin/clavulanic acid and cefazolin. Also, almost 5% of the Acinetobacter spp. strains were PDR.
In the case of Pseudomonas spp. strains, the number of the isolated strains was higher in the post-COVID-19 period and the highest resistance rate in both periods was against cefazolin and amoxicillin/clavulanic acid (100%). An increase in the antimicrobial resistance rate against third generation cephalosporins was found for ceftazidime (from 60.92% to 67.72%), cefotaxime (from 75% to 81.82%) and a decrease for ceftriaxone (from 82.35% to 70%), without statistical significance. Antibiotic resistance rate against fourth generation cephalosporins (cefepime) was significantly lower in the post-COVID-19 period (from 76.47% to 58.51%, p=0.01). Carbapenem resistance rate decreased also, but without statistical significance. The lower resistance rate was against colistin, with significant increase (p=0.03) in the post-COVID-19 period (4.88%), compared to the pre-COVID19 period (0%) (
Table 3). Although the percentage of the MDR Pseudomonas spp. strains was almost halved in the post-COVID-19 period, a PDR strain was isolated in both periods (
Figure 2).
The research revealed in the case of Proteus spp. strains a high resistance rate against amoxicillin/clavulanic acid, first and third-generation cephalosporins, fluoroquinolones and gentamicin, but the rate was lower in the post-COVID-19 period, the difference being statistically significant for cefazolin (from 94.12% to 84.43%, p=0.03), cefepime (from 54.80% to 21.37%, p<0.001) and gentamicin (from 76.59% to 55.97%, p<0.001). This correlates with the decrease of the percentage of MDR strains, from 73.68% to 18.04%. The resistance rate against piperacillin/tazobactam was higher, but without statistical significance. All the tested strains were resistant to colistin.
Among the MDR Gram-positive pathogens, more than 50% of the Staphylococcus aureus strains were MRSA (315-63.89% in the pre-COVID-19 period and 223-50.68% in the post-COVID-19 period). Almost 60% were MDR in the pre-COVID-19 period, but around 20% in the post-COVID-19 period. There were statistically significant increases in antimicrobial resistance rate in the post-COVID-19 period against doxycycline (40.08% to 51.72%, p=0.002), linezolid (0.22% to 3.13%, p<0.001), rifampicin (53.16% to 64.93%, p<0.001) and teicoplanin (26.31% to 53.40%, p=0.03%). Significant decreases were found against erythromycin (74.17% to 55.58%, p<0.001) and penicillin (98.34% to 87.01%, p<0.001) (
Table 4).
The number of isolated strains of CoNS was almost double in the post-COVID-19 period, but only 15% were MDR, compared to almost 70% in the pre-COVID-19 period (
Figure 2). Over 90% of the strains were resistant to penicillin and 70% to tetracycline. High and increasing resistance rates were registered against clindamycin and clarithromycin, with no statistical significance. Significant increases were recorded in the post-COVID-19 period against rifampicin (45.24% to 62.55%, p=0.001) and teicoplanin (26.67% to 63.57%, p<0.001). In the same period, significantly decreased the antibiotic resistance rate against erythromycin (from 80.65% to 68.27%, p=0.02), vancomycin (from 15.79% to 0.91%, p<0.001) and rifampicin (from 45.24% to 62.55%, p=0.001).
All the Streptococcus spp. strains were resistant to ciprofloxacin in both periods. Half of the strains were MDR in the pre-COVID-19 period, reaching 60% in the post-COVID-19 period. A high resistance was observed against penicillin (85.11%), but significantly decreased in the post-COVID-19 period (62.18%, p<0.001). A significant decrease was registered also against oxacillin (from 91.89% to 34.15%, p<0.001), erythromycin (from 52.55% to 29.11%, p<0.001) and clarithromycin (from 49.12% to 27.03 %, p<0.001). The resistance rate of the Streptococcus spp. strains against doxycycline increased significantly, from 10.57% to 19.75%, p<0.001.
Enterococcus spp. resistance rate against penicillin was significantly higher in the post-COVID-19 period (61.22% compared to 34.15%, p=0.01) and lower against doxycycline (80.65% to 33.33%, p<0.001) and vancomycin (29.17% to 5.33%, p<0.001) (
Table 4). The percentage of MDR strains was five times higher in the pre-COVID-19 period (44.44%), compared to the post-COVID-19 period (8.6%) (
Figure 2).
3. Discussion
According to the European Antimicrobial Resistance Surveillance Network (EARS-Net) Report for 2022, AMR remains a concern in the EU/EEA, especially regarding the continuous increase in carbapenem-resistant K. pneumoniae and vancomycin- resistant E. faecium [
4].
The COVID-19 pandemic has challenged health systems in all countries, with the reorganization of hospitals, with limited access for the patients with other conditions than COVID-19. Due to the lack of specific treatment, antibiotics have been empirically administered mainly to patients with severe forms of disease, and those hospitalized in ICU. Inappropriate use of antibiotics in viral illness and in the absence of bacterial co-infection may be an enabling factor for the selection of multidrug-resistant strains. The use of non-invasive diagnostic techniques can lead to the elimination of the risk of contracting an infection with multidrug resistant pathogens [
6].
Our research aims to determine whether the COVID-19 pandemic significantly changed the resistance of pathogens involved in cases of infection in hospitalized patients in ICU - Emergency Clinical County Hospital of Craiova, in selected periods before and after COVID-19. To our knowledge, no similar studies have been published examining the possible changes of antimicrobial resistance in Romania, pre- and post-COVID-19 period. Only one study investigated possible changes in uropathogen resistance in female patients before and during the COVID-19 pandemic [
7,
8]. The study found the greatest increase in resistance in Klebsiella spp. and Pseudomonas against quinolones, consistent with other research [
9] (and a significant increase in resistance to carbapenems. A decrease in resistance to Penicillin was observed in Enterococcus spp.
There are several other studies who have found a relationship between COVID-19 and AMR, suggesting that some conditions, often including increased antibiotic usage, may be contributing to the rise of AMR, but most of them included for comparison the period before the pandemic and what happened during the pandemic.
A study conducted in India revealed an overall increase in carbapenem resistance rates between pre-COVID-19 period and COVID-19 period for E. coli, Klebsiella pneumoniae, Acinetobacter baumanii and Pseudomonas aeruginosa [
10].
Another study carried on in a University Hospital in Egypt showed significant increase for XDR species during the COVID-19 era and, in the case of Gram-negative pathogens, a statistically significant increase in the resistance for ampicillin, ciprofloxacin, aztreonam, cefazolin, during the COVID-19 period compared with before. The susceptibility pattern was not different from Gram-positive pathogens [
11].
A significant decrease in resistance for Klebsiella pneumoniae, Pseudomonas and Acinetobacter baumanii carbapenem-resistant, during pandemic compared with pre-COVID-19 period, and a significant increase in the prevalence of E. faecium vancomycin resistant was found in a research conducted in Columbia [
12].
According to a study conducted in Pakistan, S. aureus showed a decrease against oxacillin and erythromycin after the COVID-19 pandemic, with an increasing pattern of resistance in the case of Enterococcus spp. for ampicillin, gentamicin and ciprofloxacin [
13].
In our research, the most frequently isolated pathogens were S. aureus and Klebsiella spp. in both studied periods. CoNS were the most common pathogens involved in blood stream infections during both studied periods, while S. aureus was most commonly isolated from tracheal aspirate/sputum and intravascular catheters. S. aureus was most frequently identified also from pus/wound swabs in the pre-COVID-19 period, while in the post-COVID-19 period have prevailed Gram-negative pathogens. An increasing percentage of isolated Gram-negative strains was observed in the post-COVID-19 period (except for Klebsiella spp.), and of Gram-positive CoNS and Enterococcus spp. One-third of urinary tract infections were caused by Klebsiella spp. in the pre-COVID-19 period and by E. coli in the post-COVID-19 period. These findings are consistent with those from other studies [
14]. Both Enterococcus spp. and other negative pathogens are frequently implicated in the etiology of healthcare-associated infections, in European acute care hospitals [
4].
A very high percentage of MDR isolates in the pre-COVID-19 period was found in the case of Acinetobacter spp., NFB, Klebsiella spp., Proteus spp. and Pseudomonas spp. with an important decrease in the post-COVID -19 period, both for Gram-negative and Gram –positive pathogens, but also with a significant increase in AMR to certain antibiotics.
AMR percentages for Klebsiella spp. had a statistically significant increase (p<0.001) in the post COVID-19 period compared with to the pre-COVID-19 period, against cefotaxime, colistin, gentamicin and aztreonam. Around 50% of the strains were resistant to carbapenems (imipenem and meropenem) and to fluoroquinolones. The increasing resistance to backup antibiotics, such as colistin, is an issue of greatest concern, which may be due to the use of these antibiotics in situations where they are not indicated, contributing to the selection of resistant pathogens. One of the few published studies evaluating changes in antimicrobial resistance [
14], revealed a statistically significant decrease in the resistance of Klebsiella spp. against ceftazidime, colistin and doxycycline, in the post-COVID-19 period compared to the pre-COVID-19 period. A review conducted by Khaznadar et al revealed also that Gram-negative pathogens were those with levels of antimicrobial resistance most affected by the overuse of antibiotics during the pandemic [
15], while a retrospective study conducted in Greece showed an increasing trend in the incidence of resistant Gram-negative bacteria, particularly in ICUs, compared to the pre-pandemic period [
16].
Antimicrobial resistance in Acinetobacter spp. isolates revealed a very high level of resistance against the third-generation cephalosporins (over 95%), carbapenems (around 90%), piperacillin-tazobactam (around 90%), fluoroquinolones (around 90%) and aminoglycosides (around 85%). These findings are consistent with those from other studies [
17,
18,
19,
20].
It was observed also a significantly increasing trend of the antimicrobial resistance rate against colistin (p<0.001) and a significant decrease against fourth-generation cephalosporins (p<0.001) and monobactams (p=0.003). In another research [
14], no statistically significant increase or decrease in resistance rates was observed between study periods for Acinetobacter spp.
In the case of Pseudomonas spp. strains, AMR decreased significantly in the post-COVID-19 period against fourth generation cephalosporins (cefepime, p=0.01) and increased against colistin (p=0.03). An increasing trend was revealed against third generation cephalosporins (ceftazidime, cefotaxime). Carbapenem resistance rate decreased, but without statistical significance. The results were different compared with other study [
14], who revealed a significant decrease in the resistance against cefotaxime and meropenem. P. aeruginosa remains one of the major causes of healthcare-associated infection in Europe [
4], and the increasing trend of resistance to certain antibiotics, including colistin, draws attention to the difficulty of treating infected patients, which is also proven by identification of PDR Pseudomonas spp. strains during both studied periods.
A significantly decreasing trend was observed in the case of Proteus spp. strains resistant first (cefazolin, p=0.03) and fourth-generation cephalosporins (cefepime (p<0.001), and aminoglycosides (gentamicin p<0.001), consistent with other findings [
14] An increase, but without statistical significance was found against piperacillin/tazobactam. The resistance against colistin was 100%, Proteus being inherently resistant to this antibiotic.
In the case of S. aureus isolates, statistically significant increases in antimicrobial resistance rate were found in the post-COVID-19 period against doxycycline (p=0.002), linezolid (p<0.001), rifampicin (p<0.001) and teicoplanin (p=0.03%), while significant decreases were found against erythromycin (p<0.001) and penicillin (p<0.001), a different trend compared to other researchers [
14].
The number of the isolated strains of CoNS was almost double in the post-COVID-19 period Significant increases were recorded in the post-COVID-19 period against rifampicin (p=0.001) and teicoplanin (p<0.001). In the same period, significantly decreased the antibiotic resistance rate against erythromycin (p=0.02), vancomycin (p<0.001) and rifampicin (p=0.001). In another study [
14], in the post- COVID-19 period, a significant decrease was found only against vancomycin.
The number of positive blood samples was more than three times higher in the post-COVID-19 period. Over 60% of S. aureus strains identified in blood samples were MRSA (19/34), less than from the pre-COVID-19 period (almost 80%, but higher than the percentage of invasive isolates resistant to methicillin reported in Romania (25-50%)[
4]. The same declining trend was seen in the EU/EEA population-weighted mean percentage of MRSA[
4].
Almost 90% of Klebsiella spp. strains were carbapenem-resistant in the pre-COVID-19 period, compared to approximately 65% in the post-COVID-19 period. Although decreasing, the percentage is higher than that reported across the country (between 25% and 50%) [
4].
Almost 25% of strains of E. coli responsible for blood stream infections in the post-COVID-19 period were resistant to third-generation cephalosporins, compared to 50% of those isolated from blood samples in the pre-COVID-19 period.
According to the Council of the EU Recommendation regarding the three AMR targets to be achieved by the EU by 2030, these include reducing the total incidence of bloodstream infections with MRSA, third-generation cephalosporin-resistant E. coli and carbapenem-resistant K. pneumoniae, by 15%, 10% and 5%, respectively, by 2030 against the baseline year 2019 [
4].The results highlighted in our study show that these targets were achieved in the incidence rates registered in our hospital. Another important issue in Romania, which is necessary to pay attention to, is that there is the possibility of increasing the number of MDR-TB cases in the post-COVID period, due to the limitation of TB patients' access to diagnosis and treatment during the pandemic. The number of XDR-TB cases already increased before COVID-19 era [
21], including the cases of extrapulmonary tuberculosis which originates from the hematogenous metastatic affects developed during the prime TB infection period [
22], which are diagnosed in almost 20% of all TB cases in high endemic areas[
23].
Our study has several limitations, related to the small number of strains tested in certain situations and the retrospective nature of the study.
4. Methods
The research is a retrospective study of all the isolated bacterial pathogens collected from patients, admitted to the intensive care unit (ICU) of Emergency Clinical County Hospital of Craiova, Romania, in pre- and post-COVID-19 pandemic periods. The hospital provides specialized healthcare to patients from Dolj county and South-West Region of Romania. The selected study periods were from April 1, 2019, to March 31, 2020 (pre-COVID-19 period) and July 1, 2021 to June 30, 2022 (post-COVID-19 period). We chose two periods of one year each, in the second being registered a global decline in cases of SARS-CoV2 infection. Since March 2022 the epidemiological alert period in Romania ended.
Data were collected from the clinical pathology databases of the hospital. The specimens from all patients were sent to the Hospital’s Laboratory of Microbiology. Samples included blood, urine, sputum/tracheal aspirate (respiratory secretion), pus/wound swabs, exudates, intravascular catheters, cerebrospinal fluid, sterile fluids. Cases where it was more than one isolate of the same pathogen from the same patient and the same site of infection were excluded.
All positive bacterial cultures from patients admitted in ICU in the studied period were included. The percentage of MDR strains among the clinical isolates from ICU, was analyzed by taking into consideration resistance to at least three different antibiotic groups: aminoglycosides, cephalosporins, carbapenems, tetracyclines and fluoroquinolones[
24].
Multidrug-resistant (MDR) pathogen is defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories[
24].
Pan-drug-resistant (PDR) is defined as non-susceptibility to all agents tested in the hospital[
25].
The identification of the isolated strains and the analysis of the resistance patterns for the action of the appropriate antibiotics were performed using Vitek 2 Compact system[
26].
The antimicrobial susceptibility test was carried out according to Clinical Laboratory Standard Institute (CLSI) guidelines [
27].
Information about patients’ age, sex, hospital department, sample type, site of infection and antimicrobial resistance pattern have been stored in the in the Hospital`s Information System. Data were entered and analyzed using Microsoft Excel. Continuous variables like age are expressed as mean ± STDEV (standard deviation). The pattern of micro-organisms was analyzed and expressed as percentages. The χ 2 test was used to evaluate the changes in antimicrobial resistance in pre- and post- COVID-19 period, using Epi Info software. If p values were ≤ 0.05, the differences were considered statistically significant.