Submitted:
05 February 2024
Posted:
06 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. The immune response to SARS-CoV-2 infection and vaccination
2.1. The humoral immune response to SARS-CoV-2 infection
2.1.1. COVID-19 and response to vaccination in patients with humoral immunodeficiency or B cell depletion therapy.
2.1.2. Clinical effect of intravenous immunoglobulin (IVIG) administration on outcome of (severe) COVID-19
2.1.3. Passive vaccination with monoclonal antibodies.
2.2. T cell mediated immunity against coronaviruses.
2.2.1. Importance of T-cell immunity
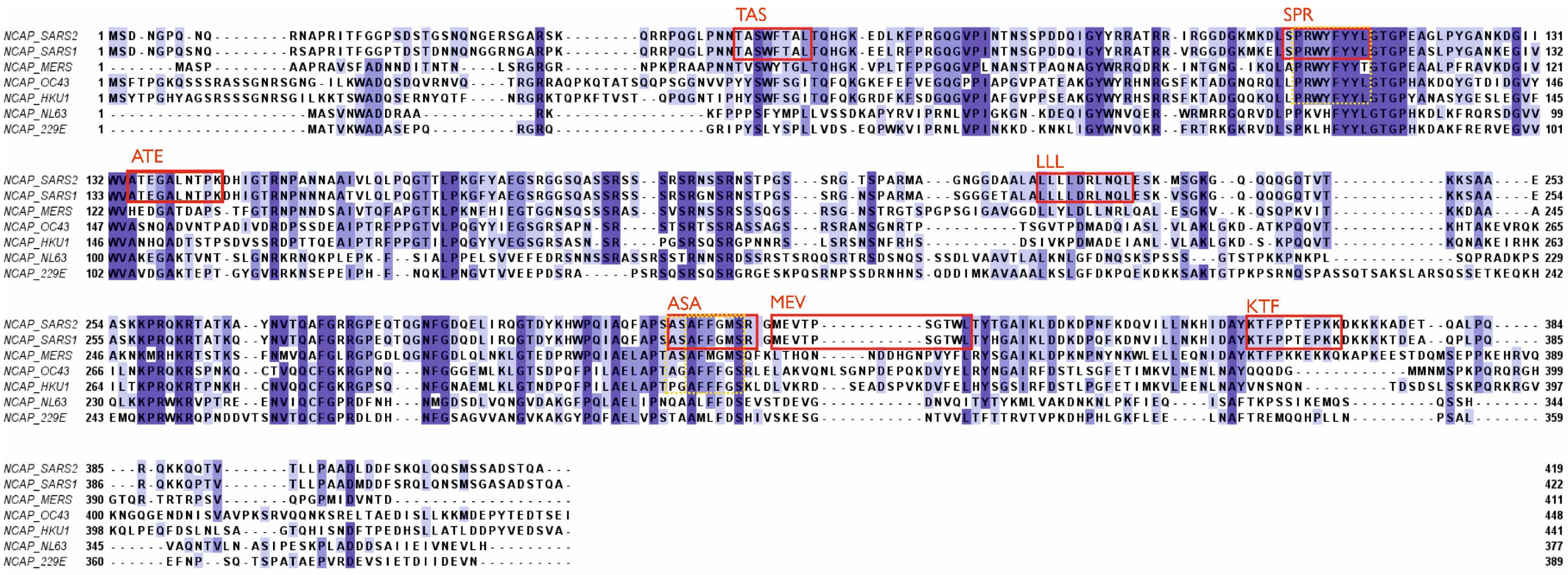
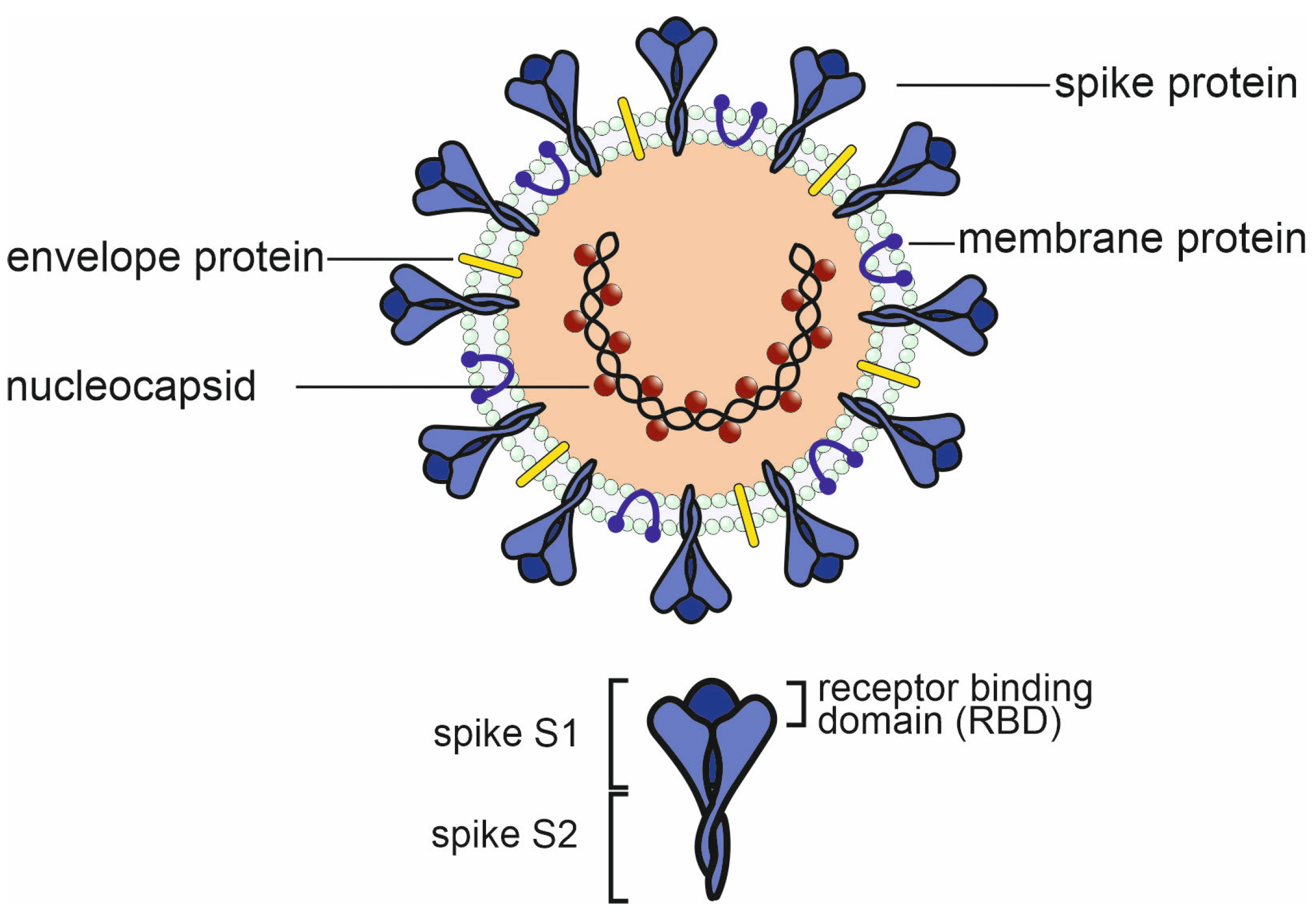
3. Potential negative impact of existing antibodies and memory B cells on future viruses.
3.1. Antibody Dependent Enhancement (ADE)

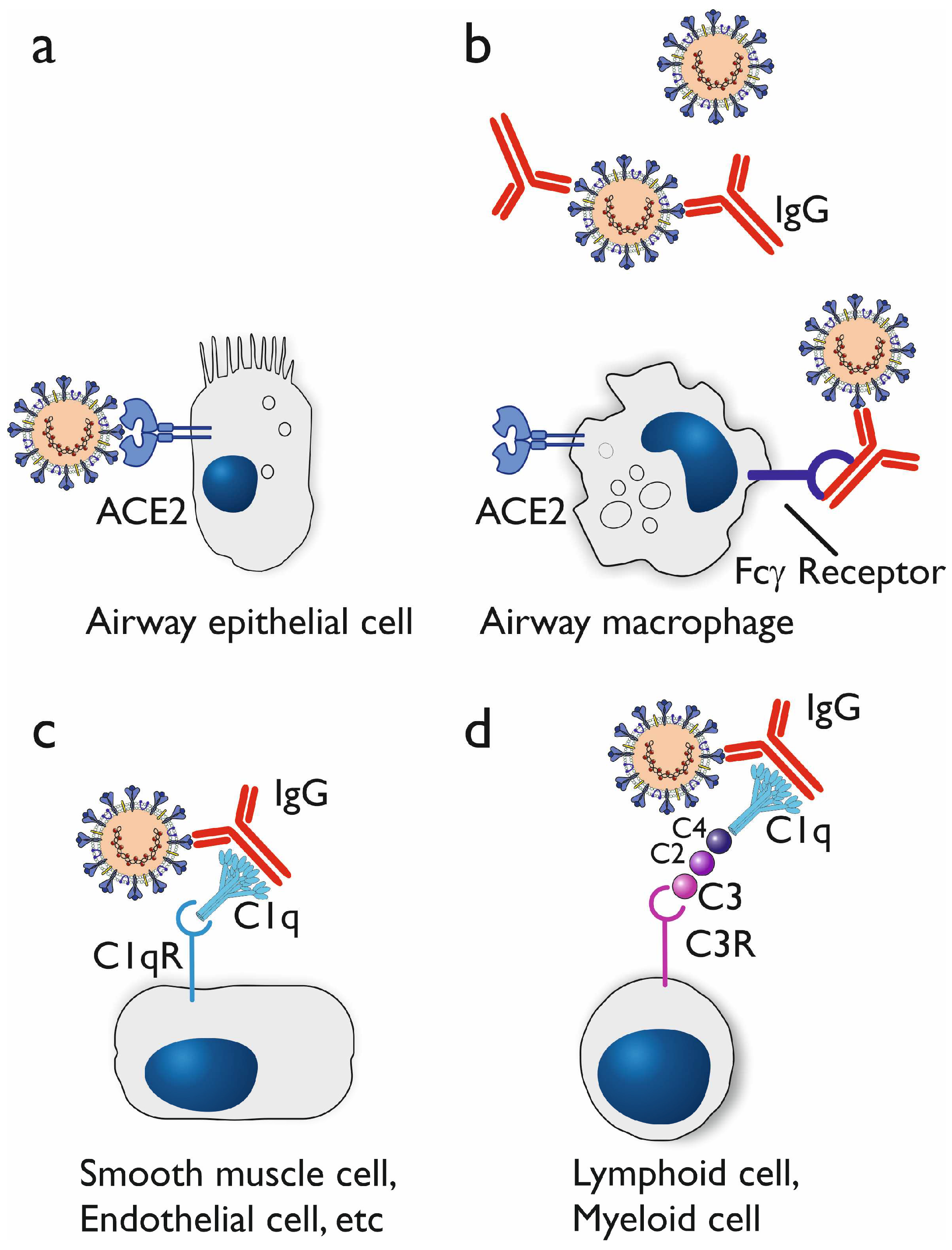
3.1.1. In vitro studies on antibody dependent enhancement
3.1.2. In vivo indications for antibody dependent enhancement
3.1.3. Proposed mechanisms of antibody dependent enhancement
3.2. The original antigenic sin
4. Concluding remarks and outlook for the future
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, et al. China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [CrossRef]
- World Health Organization. Virtual Press Conference on COVID-19 & Other Global Health Emergencies. Available online: https://www.who.int/publications/m/item/virtual-press-conference-on-covid-19---other-global-health-emergencies (accessed on January 14 2024).
- Harris E. WHO Declares End of COVID-19 Global Health Emergency. JAMA. 2023 Jun 6;329(21):1817. [CrossRef]
- Cohen J. COVID’s cold cousins. Four largely ignored coronaviruses circulate in humans without causing great harm and may portend the future for SARS-CoV-2. Science 383, 2024, 141-145. [CrossRef]
- Kaur N, Singh R, Dar Z, Bijarnia RK, Dhingra N, Kaur T. Genetic comparison among various coronavirus strains for the identification of potential vaccine targets of SARS-CoV2. Infect Genet Evol. 2021 Apr;89:104490. [CrossRef]
- Al-Qahtani WS, Alneghery LM, Alqahtani AQS, Al-Kahtani MD, Alkahtani S. A review of comparison study between corona viruses (Sars-cov, mers-cov) and novel corona virus (COVID-19). Revista Mexicana de Ingeniería Química 2020, 19(Sup. 1), 201-212. [CrossRef]
- Yang H, Rao Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat Rev Microbiol. 2021 Nov;19(11):685-700. [CrossRef]
- Tang G, Liu Z, Chen D. Human coronaviruses: Origin, host and receptor. J Clin Virol. 2022 Oct;155:105246. [CrossRef]
- Rajapakse N, Dixit D. Human and novel coronavirus infections in children: a review. Paediatr Int Child Health. 2021 Feb;41(1):36-55. [CrossRef]
- van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. Identification of a new human coronavirus. Nat Med. 2004 Apr;10(4):368-73. [CrossRef]
- Hu B, Huang S, Yin L. The cytokine storm and COVID-19. J Med Virol. 2021 Jan;93(1):250-256. [CrossRef]
- Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med. 2020 Dec 3;383(23):2255-2273. [CrossRef]
- Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020 Jul 30;11(1):3810. [CrossRef]
- Moss P. The T cell immune response against SARS-CoV-2. Nat Immunol. 2022 Feb;23(2):186-193. [CrossRef]
- Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS, Ley AM, Kyu S, Howell JC, Ozturk T, et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol. 2020 Dec;21(12):1506-1516. [CrossRef]
- Zhang Q, Wang Y, Qi C, Shen L, Li J. Clinical trial analysis of 2019-nCoV therapy registered in China. J Med Virol. 2020 Jun;92(6):540-545. [CrossRef]
- Hashem AM, Algaissi A, Almahboub SA, Alfaleh MA, Abujamel TS, Alamri SS, Alluhaybi KA, Hobani HI, AlHarbi RH, Alsulaiman RM, et al. Early Humoral Response Correlates with Disease Severity and Outcomes in COVID-19 Patients. Viruses. 2020 Dec 4;12(12):1390. [CrossRef]
- Liu X, Wang J, Xu X, Liao G, Chen Y, Hu CH. Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg Microbes Infect. 2020 Dec;9(1):1269-1274. [CrossRef]
- Rijkers G, Murk JL, Wintermans B, van Looy B, van den Berge M, Veenemans J, Stohr J, Reusken C, van der Pol P, Reimerink J. Differences in Antibody Kinetics and Functionality Between Severe and Mild Severe Acute Respiratory Syndrome Coronavirus 2 Infections. J Infect Dis. 2020 Sep 14;222(8):1265-1269. [CrossRef]
- Soresina A, Moratto D, Chiarini M, Paolillo C, Baresi G, Focà E, Bezzi M, Baronio B, Giacomelli M, Badolato R. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr Allergy Immunol. 2020 Jul;31(5):565-569. [CrossRef]
- Devassikutty FM, Jain A, Edavazhippurath A, Joseph MC, Peedikayil MMT, Scaria V, Sandhya P, Govindaraj GM. X-Linked Agammaglobulinemia and COVID-19: Two Case Reports and Review of Literature. Pediatr Allergy Immunol Pulmonol. 2021 Sep;34(3):115-118. [CrossRef]
- Quinti I, Locatelli F, Carsetti R. The Immune Response to SARS-CoV-2 Vaccination: Insights Learned From Adult Patients With Common Variable Immune Deficiency. Front Immunol. 2022 Jan 19;12:815404. [CrossRef]
- Vanni A, Salvati L, Mazzoni A, Lamacchia G, Capone M, Francalanci S, Kiros ST, Cosmi L, Puccini B, Ciceri M, et al. Bendamustine impairs humoral but not cellular immunity to SARS-CoV-2 vaccination in rituximab-treated B-cell lymphoma-affected patients. Front Immunol. 2023 Dec 1;14:1322594. [CrossRef]
- Candon S, Lemee V, Leveque E, Etancelin P, Paquin C, Carette M, Contentin N, Bobee V, Alani M, Cardinael N, et al. Dissociated humoral and cellular immune responses after a three-dose schema of BNT162b2 vaccine in patients receiving anti-CD20 monoclonal antibody maintenance treatment for B-cell lymphomas. Haematologica. 2022 Mar 1;107(3):755-758. [CrossRef]
- Ishio T, Tsukamoto S, Yokoyama E, Izumiyama K, Saito M, Muraki H, et al. Anti-CD20 antibodies and bendamustine attenuate humoral immunity to COVID-19 vaccination in patients with B-cell non-Hodgkin lymphoma. Ann Hematol 2023; 12:1–11. [CrossRef]
- Perry C, Luttwak E, Balaban R, Shefer G, Morales MM, Aharon A, Tabib Y, Cohen YC, Benyamini N, Beyar-Katz, et al. Efficacy of the BNT162b2 mRNA COVID-19 vaccine in patients with B-cell non-Hodgkin lymphoma. Blood Adv. 2021 Aug 24;5(16):3053-3061. [CrossRef]
- Lu L, Chan CY, Lim YY, Than M, Teo S, Lau PYW, Ng KH, Yap HK. SARS-CoV-2 Humoral Immunity Persists Following Rituximab Therapy. Vaccines (Basel). 2023 Dec 18;11(12):1864. [CrossRef]
- Bsteh G, Assar H, Hegen H, Heschl B, Leutmezer F, Di Pauli F, Gradl C, Traxler G, Zulehner G, AUT-MuSC investigators. COVID-19 severity and mortality in multiple sclerosis are not associated with immunotherapy: Insights from a nation-wide Austrian registry. PLoS One. 2021 Jul 27;16(7):e0255316. [CrossRef]
- Avouac J, Drumez E, Hachulla E, Seror R, Georgin-Lavialle S, El Mahou S, Pertuiset E, Pham T, Marotte H, FAIR/SFR/SNFMI/SOFREMIP/CRI/IMIDIATE consortium and contributors. COVID-19 outcomes in patients with inflammatory rheumatic and musculoskeletal diseases treated with rituximab: a cohort study. Lancet Rheumatol. 2021 Jun;3(6):e419-e426. [CrossRef]
- Shao Z, Feng Y, Zhong L, Xie Q, Lei M, Liu Z, Wang C, Ji J, Liu H, Gu Z, Hu Z, Su L, Wu M, Liu Z. Clinical efficacy of intravenous immunoglobulin therapy in critical ill patients with COVID-19: a multicenter retrospective cohort study. Clin Transl Immunology. 2020 Oct 14;9(10):e1192. [CrossRef]
- Gharebaghi N, Nejadrahim R, Mousavi SJ, Sadat-Ebrahimi SR, Hajizadeh R. The use of intravenous immunoglobulin gamma for the treatment of severe coronavirus disease 2019: a randomized placebo-controlled double-blind clinical trial. BMC Infect Dis. 2020 Oct 21;20(1):786. [CrossRef]
- Mazeraud A, Jamme M, Mancusi RL, Latroche C, Megarbane B, Siami S, Zarka J, Moneger G, Santoli F, Argaud L, Chillet P, et al. Intravenous immunoglobulins in patients with COVID-19-associated moderate-to-severe acute respiratory distress syndrome (ICAR): multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Respir Med. 2022 Feb;10(2):158-166. [CrossRef]
- Pei L, Zhang S, Huang L, Geng X, Ma L, Jiang W, Li W, Chen D. Antiviral agents, glucocorticoids, antibiotics, and intravenous immunoglobulin in 1142 patients with coronavirus disease 2019: a systematic review and meta-analysis. Pol Arch Intern Med. 2020 Sep 30;130(9):726-733. [CrossRef]
- Kwapisz D, Bogusławska J. Intravenous immunoglobulins (IVIG) in severe/critical COVID-19 adult patients. Biomed Pharmacother. 2023 Jul;163:114851. [CrossRef]
- Piechotta V, Iannizzi C, Chai KL, Valk SJ, Kimber C, Dorando E, Monsef I, Wood EM, Lamikanra AA, Roberts DJ, et al. Convalescent plasma or hyperimmune immunoglobulin for people with COVID-19: a living systematic review. Cochrane Database Syst Rev. 2021 May 20;5(5):CD013600. Update in: Cochrane Database Syst Rev. 2023 Feb 1;2:CD013600. PMID: 34013969; PMCID: PMC8135693. [CrossRef]
- RECOVERY Collaborative Group. Convalescent plasma in patients admitted to hospital with COVID-19 (RECOVERY): a randomised controlled, open-label, platform trial. Lancet. 2021 May 29;397(10289):2049-2059. [CrossRef]
- Writing Committee for the REMAP-CAP Investigators; Estcourt LJ, Turgeon AF, McQuilten ZK, McVerry BJ, Al-Beidh F, Annane D, Arabi YM, Arnold DM, Beane A, Bégin P, et al. Effect of Convalescent Plasma on Organ Support-Free Days in Critically Ill Patients With COVID-19: A Randomized Clinical Trial. JAMA. 2021 Nov 2;326(17):1690-1702. [CrossRef]
- Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, et al., BLAZE-1 Investigators. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N Engl J Med. 2021 Jan 21;384(3):229-237. [CrossRef]
- Dougan M, Nirula A, Azizad M, Mocherla B, Gottlieb RL, Chen P, Hebert C, Perry R, Boscia J, Heller B, et al., BLAZE-1 Investigators. Bamlanivimab plus Etesevimab in Mild or Moderate Covid-19. N Engl J Med. 2021 Oct 7;385(15):1382-1392. [CrossRef]
- Li M, Lou F, Fan H. SARS-CoV-2 variant Omicron: currently the most complete “escapee” from neutralization by antibodies and vaccines. Signal Transduct Target Ther. 2022 Jan 28;7(1):28. [CrossRef]
- Anand P, Puranik A, Aravamudan M, Venkatakrishnan AJ, Soundararajan V. SARS-CoV-2 strategically mimics proteolytic activation of human ENaC. Elife. 2020 May 26;9:e58603. [CrossRef]
- Kotsias F, Cebrian I, Alloatti A. Antigen processing and presentation. Int Rev Cell Mol Biol. 2019;348:69-121. [CrossRef]
- Nagler A, Kalaora S, Barbolin C, Gangaev A, Ketelaars SLC, Alon M, Pai J, Benedek G, Yahalom-Ronen Y, Erez N, Greenberg P, et al. Identification of presented SARS-CoV-2 HLA class I and HLA class II peptides using HLA peptidomics. Cell Rep. 2021 Jun 29;35(13):109305. [CrossRef]
- Ferretti AP, Kula T, Wang Y, Nguyen DMV, Weinheimer A, Dunlap GS, Xu Q, Nabilsi N, Perullo CR, Cristofaro AW, et al. Unbiased Screens Show CD8+ T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity. 2020 Nov 17;53(5):1095-1107.e3. [CrossRef]
- Suardana IBK, Mahardika BK, Pharmawati M, Sudipa PH, Sari TK, Mahendra NB, Mahardika GN. Whole-Genome Comparison of Representatives of All Variants of SARS-CoV-2, Including Subvariant BA.2 and the GKA Clade. Adv Virol. 2023 Mar 9;2023:6476626. [CrossRef]
- Bai Z, Cao Y, Liu W, Li J. The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses. 2021 Jun 10;13(6):1115. [CrossRef]
- Verma J, Kaushal N, Manish M, Subbarao N, Shakirova V, Martynova E, Liu R, Hamza S, Rizvanov AA, Khaiboullina SF, Baranwal M. Identification of conserved immunogenic peptides of SARS-CoV-2 nucleocapsid protein. J Biomol Struct Dyn. 2023 Sep 26:1-17. [CrossRef]
- Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, Chng MHY, Lin M, Tan N, Linster M, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020 Aug;584(7821):457-462. [CrossRef]
- AlKhalifah JM, Seddiq W, Alshehri MA, Alhetheel A, Albarrag A, Meo SA, Al-Tawfiq JA, Barry M. Impact of MERS-CoV and SARS-CoV-2 Viral Infection on Immunoglobulin-IgG Cross-Reactivity. Vaccines (Basel). 2023 Feb 26;11(3):552. [CrossRef]
- Kesheh MM, Hosseini P, Soltani S, Zandi M. An overview on the seven pathogenic human coronaviruses. Rev Med Virol. 2022 Mar;32(2):e2282. [CrossRef]
- Hewitt EW. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology. 2003 Oct;110(2):163-9. [CrossRef]
- Cassioli C, Baldari CT. The Expanding Arsenal of Cytotoxic T Cells. Front Immunol. 2022 Apr 20;13:883010. [CrossRef]
- Tay C, Kanellakis P, Hosseini H, Cao A, Toh BH, Bobik A, Kyaw T. B Cell and CD4 T Cell Interactions Promote Development of Atherosclerosis. Front Immunol. 2020 Jan 10;10:3046. [CrossRef]
- Kared H, Redd AD, Bloch EM, Bonny TS, Sumatoh H, Kairi F, Carbajo D, Abel B, Newell EW, Bettinotti MP, et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J Clin Invest. 2021 Mar 1;131(5):e145476. [CrossRef]
- Vennema H, de Groot RJ, Harbour DA, Dalderup M, Gruffydd-Jones T, Horzinek MC, Spaan WJ. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J Virol. 1990 Mar;64(3):1407-9. [CrossRef]
- Olsen CW, Corapi WV, Ngichabe CK, Baines JD, Scott FW. Monoclonal antibodies to the spike protein of feline infectious peritonitis virus mediate antibody-dependent enhancement of infection of feline macrophages. J Virol. 1992 Feb;66(2):956-65. [CrossRef]
- Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J Virol. 2020 Feb 14;94(5):e02015-19. [CrossRef]
- Takada A, Kawaoka Y. Antibody-dependent enhancement of viral infection: molecular mechanisms and in vivo implications. Rev Med Virol. 2003 Nov-Dec;13(6):387-98. [CrossRef]
- Arvin AM, Fink K, Schmid MA, Cathcart A, Spreafico R, Havenar-Daughton C, Lanzavecchia A, Corti D, Virgin HW. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020 Aug;584(7821):353-363. [CrossRef]
- Pongracz T, Vidarsson G, Wuhrer M. Antibody glycosylation in COVID-19. Glycoconjugate Journal. 2022 Jun;39(3):335-44.
- van Osch, T.L.J., Nouta, J., Derksen, N.I.L., van Mierlo, G., van der Schoot, C.E., Wuhrer, M., Rispens, T., Vidarsson, G.: Fc galactosylation promotes hexamerization of human IgG1, leading to enhanced classical complement activation. J. Immunol. 207(6), 1545–1554 (2021). [CrossRef]
- Weiskopf K, Weissman IL. Macrophages are critical effectors of antibody therapies for cancer. MAbs. 2015;7(2):303-10. [CrossRef]
- Junker F, Gordon J, Qureshi O. Fc Gamma Receptors and Their Role in Antigen Uptake, Presentation, and T Cell Activation. Front Immunol. 2020 Jul 3;11:1393. [CrossRef]
- von Kietzell K, Pozzuto T, Heilbronn R, Grössl T, Fechner H, Weger S. Antibody-mediated enhancement of parvovirus B19 uptake into endothelial cells mediated by a receptor for complement factor C1q. J Virol. 2014 Jul;88(14):8102-15. [CrossRef]
- Eggleton P, Tenner AJ, Reid KB. C1q receptors. Clin Exp Immunol. 2000 Jun;120(3):406-12. [CrossRef]
- Wen J, Cheng Y, Ling R, Dai Y, Huang B, Huang W, Zhang S, Jiang Y. Antibody-dependent enhancement of coronavirus. Int J Infect Dis. 2020 Nov;100:483-489. [CrossRef]
- Thomas S, Smatti MK, Ouhtit A, Cyprian FS, Almaslamani MA, Thani AA, Yassine HM. Antibody-Dependent Enhancement (ADE) and the role of complement system in disease pathogenesis. Mol Immunol. 2022 Dec;152:172-182. [CrossRef]
- Monto AS, Malosh RE, Petrie JG, Martin ET. The Doctrine of Original Antigenic Sin: Separating Good From Evil. J Infect Dis. 2017 Jun 15;215(12):1782-1788. [CrossRef]
- Francis T. On the doctrine of original antigenic sin. Proc Am Philos Soc 1960; 104:572–578.
- Petráš M, Králová Lesná I. SARS-CoV-2 vaccination in the context of original antigenic sin. Hum Vaccin Immunother. 2022 Dec 31;18(1):1949953. [CrossRef]
- Pillai S. SARS-CoV-2 vaccination washes away original antigenic sin. Trends Immunol. 2022 Apr;43(4):271-273. [CrossRef]
- Rijkers GT, van Overveld FJ. The “original antigenic sin” and its relevance for SARS-CoV-2 (COVID-19) vaccination. Clin Immunol Communications. 2021; 1: 13-16. [CrossRef]
- Xia CS, Zhan M, Liu Y, Yue ZH, Song Y, Zhang F, Wang H. SARS-CoV-2 antibody response in SARS survivors with and without the COVID-19 vaccine. Int J Antimicrob Agents. 2023 Oct;62(4):106947. [CrossRef]
- El-Saed A, Othman F, Baffoe-Bonnie H, Almulhem R, Matalqah M, Alshammari L, Alshamrani MM. Symptomatic MERS-CoV infection reduces the risk of future COVID-19 disease; a retrospective cohort study. BMC Infect Dis. 2023 Nov 3;23(1):757. [CrossRef]
- Sette A, Sidney J, Crotty S. T Cell Responses to SARS-CoV-2. Annu Rev Immunol. 2023 Apr 26;41:343-373. Epub 2023 Feb 7. PMID: 36750314. [CrossRef]
- Appelberg S, Ahlén G, Yan J, Nikouyan N, Weber S, Larsson O, Höglund U, Aleman S, Weber F, Perlhamre E, et al. A universal SARS-CoV DNA vaccine inducing highly cross-reactive neutralizing antibodies and T cells. EMBO Mol Med. 2022 Oct 10;14(10):e15821. [CrossRef]
- Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chrétien D, Sanamxay D, et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022 Apr;604(7905):330-336. [CrossRef]
- Shannon CP, Blimkie TM, Ben-Othman R, Gladish N, Amenyogbe N, Drissler S, Edgar RD, Chan Q, Krajden M, Foster LJ, et al. Multi-Omic Data Integration Allows Baseline Immune Signatures to Predict Hepatitis B Vaccine Response in a Small Cohort. Front Immunol. 2020 Nov 30;11:578801. [CrossRef]
- Leslie M. Giant project will chart human immune diversity to improve drugs and vaccines. Science 2024 Jan 5; 383y: 13-14.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





