Submitted:
05 February 2024
Posted:
06 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Intestinal stem cell regulation
2.1. Signaling pathways
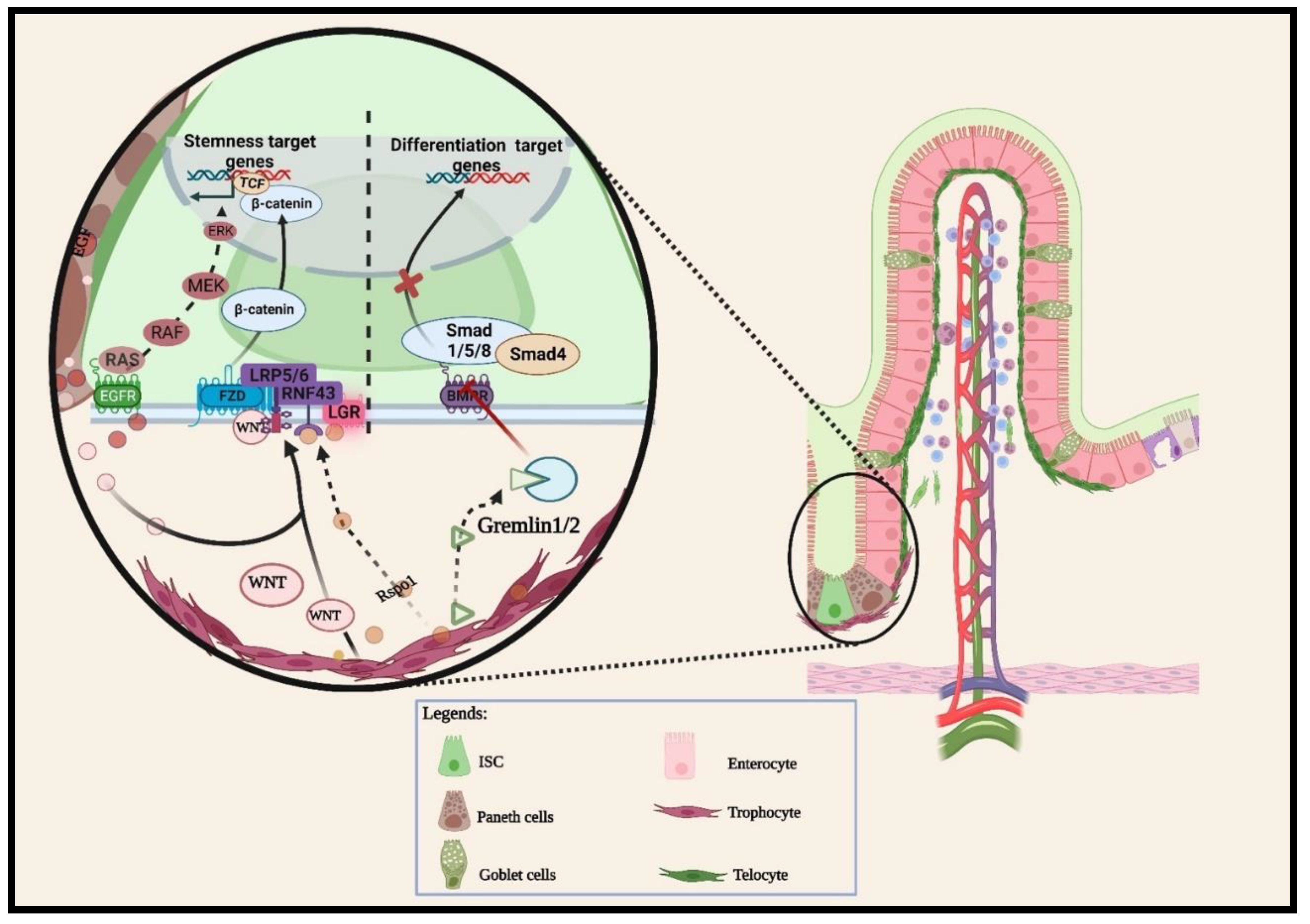
2.1.1. Wnt pathway
2.1.2. BMP pathways
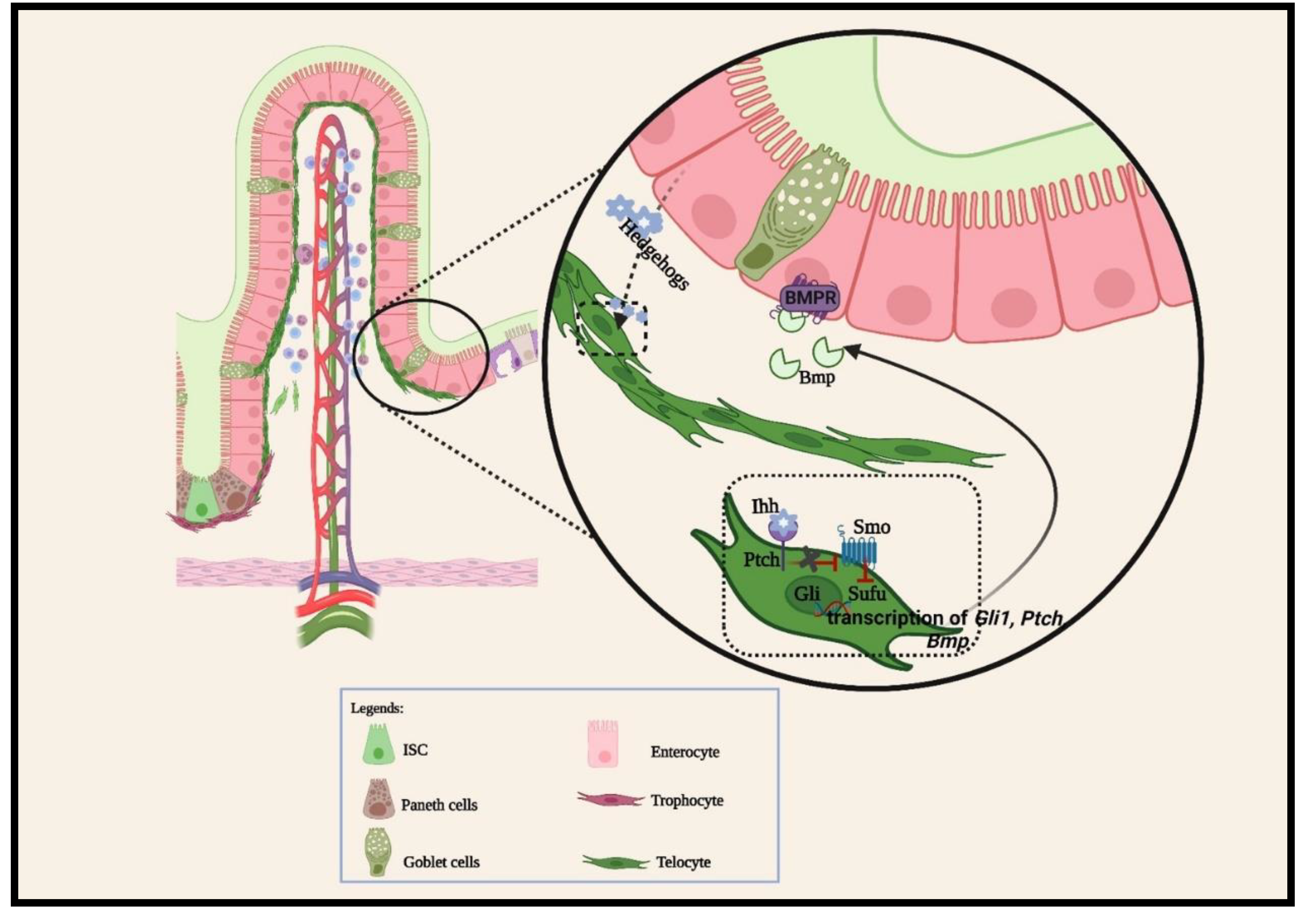
2.1.3. Other cellular signaling pathways
3. Intestinal cell plasticity and regeneration
4. Subepithelial mesenchymal stromal cell characteristics
4.1. Roles of intestinal mesenchymal stromal cells during prenatal intestinal morphogenesis
4.2. Roles of intestinal mesenchymal stromal cells during intestinal homeostasis
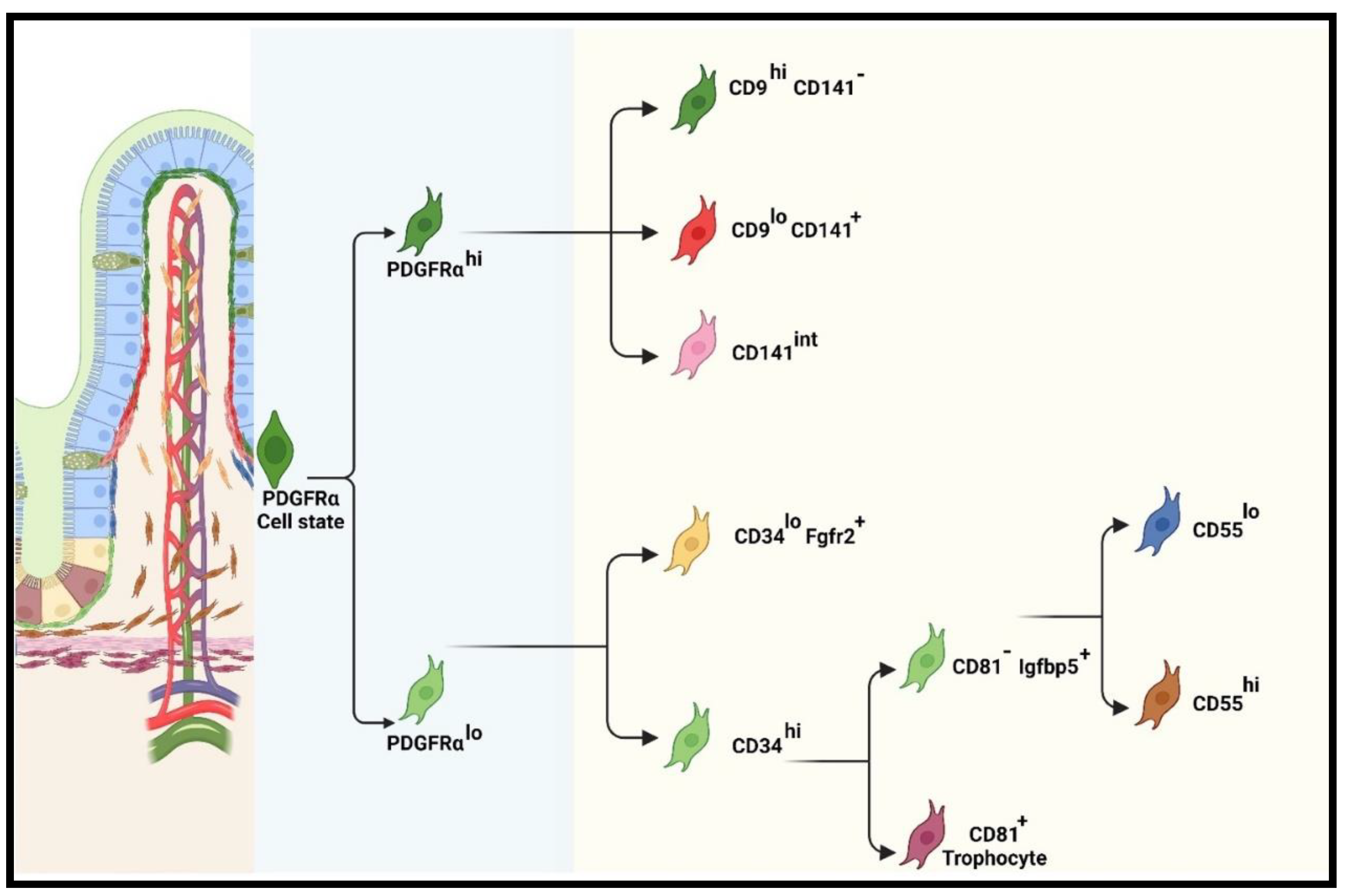
4.2.1. Recent classification of iMSC lineage
Functions of pericryptal (PDGFRαlo or CD34+Gp38+) subpopulations
Functions of PDGFRαhi subpopulations
4.3. Roles of intestinal mesenchymal stromal cells during intestinal injury and repair
5. Conclusion and Future Perspectives
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Corominas-Murtra, B.; Hannezo, E. Modelling the dynamics of mammalian gut homeostasis. In Proceedings of the Seminars in Cell & Developmental Biology; 2022. [Google Scholar]
- Gehart, H.; Clevers, H. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol 2019, 16, 19–34. [Google Scholar] [CrossRef]
- Guiu, J.; Hannezo, E.; Yui, S.; Demharter, S.; Ulyanchenko, S.; Maimets, M.; Jorgensen, A.; Perlman, S.; Lundvall, L.; Mamsen, L.S.; et al. Tracing the origin of adult intestinal stem cells. Nature 2019, 570, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Mannino, G.; Russo, C.; Maugeri, G.; Musumeci, G.; Vicario, N.; Tibullo, D.; Giuffrida, R.; Parenti, R.; Lo Furno, D. Adult stem cell niches for tissue homeostasis. J Cell Physiol 2022, 237, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Buczacki, S.J.; Zecchini, H.I.; Nicholson, A.M.; Russell, R.; Vermeulen, L.; Kemp, R.; Winton, D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 2013, 495, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef]
- van der Flier, L.G.; van Gijn, M.E.; Hatzis, P.; Kujala, P.; Haegebarth, A.; Stange, D.E.; Begthel, H.; van den Born, M.; Guryev, V.; Oving, I.; et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 2009, 136, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Beumer, J.; Clevers, H. Cell fate specification and differentiation in the adult mammalian intestine. Nat Rev Mol Cell Biol 2021, 22, 39–53. [Google Scholar] [CrossRef]
- Hu, D.; Yan, H.; He, X.C.; Li, L. Recent advances in understanding intestinal stem cell regulation. F1000Res 2019, 8. [Google Scholar] [CrossRef]
- Santos, A.J.M.; Lo, Y.H.; Mah, A.T.; Kuo, C.J. The Intestinal Stem Cell Niche: Homeostasis and Adaptations. Trends Cell Biol 2018, 28, 1062–1078. [Google Scholar] [CrossRef]
- Spit, M.; Koo, B.K.; Maurice, M.M. Tales from the crypt: intestinal niche signals in tissue renewal, plasticity and cancer. Open Biol 2018, 8, 180120. [Google Scholar] [CrossRef]
- Felsenthal, N.; Vignjevic, D.M. Stand by me: Fibroblasts regulation of the intestinal epithelium during development and homeostasis. Curr Opin Cell Biol 2022, 78, 102116. [Google Scholar] [CrossRef]
- Kabiri, Z.; Greicius, G.; Madan, B.; Biechele, S.; Zhong, Z.; Zaribafzadeh, H.; Edison; Aliyev, J.; Wu, Y.; Bunte, R.; et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development 2014, 141, 2206–2215. [Google Scholar] [CrossRef]
- Miyoshi, H. Wnt-expressing cells in the intestines: guides for tissue remodeling. J Biochem 2017, 161, 19–25. [Google Scholar] [CrossRef]
- Sasaki, N.; Sachs, N.; Wiebrands, K.; Ellenbroek, S.I.; Fumagalli, A.; Lyubimova, A.; Begthel, H.; van den Born, M.; van Es, J.H.; Karthaus, W.R.; et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc Natl Acad Sci U S A 2016, 113, E5399–5407. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.Y.; Blutt, S.E.; Zeng, X.L.; Chen, M.S.; Lo, Y.H.; Castillo-Azofeifa, D.; Klein, O.D.; Shroyer, N.F.; Donowitz, M.; Estes, M.K. Epithelial WNT Ligands Are Essential Drivers of Intestinal Stem Cell Activation. Cell Rep 2018, 22, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Bahar Halpern, K.; Massalha, H.; Zwick, R.K.; Moor, A.E.; Castillo-Azofeifa, D.; Rozenberg, M.; Farack, L.; Egozi, A.; Miller, D.R.; Averbukh, I.; et al. Lgr5+ telocytes are a signaling source at the intestinal villus tip. Nat Commun 2020, 11, 1936. [Google Scholar] [CrossRef] [PubMed]
- Bernier-Latmani, J.; Mauri, C.; Marcone, R.; Renevey, F.; Durot, S.; He, L.; Vanlandewijck, M.; Maclachlan, C.; Davanture, S.; Zamboni, N.; et al. ADAMTS18(+) villus tip telocytes maintain a polarized VEGFA signaling domain and fenestrations in nutrient-absorbing intestinal blood vessels. Nat Commun 2022, 13, 3983. [Google Scholar] [CrossRef]
- Maimets, M.; Pedersen, M.T.; Guiu, J.; Dreier, J.; Thodberg, M.; Antoku, Y.; Schweiger, P.J.; Rib, L.; Bressan, R.B.; Miao, Y.; et al. Mesenchymal-epithelial crosstalk shapes intestinal regionalisation via Wnt and Shh signalling. Nat Commun 2022, 13, 715. [Google Scholar] [CrossRef]
- Kurokawa, K.; Hayakawa, Y.; Koike, K. Plasticity of Intestinal Epithelium: Stem Cell Niches and Regulatory Signals. Int J Mol Sci 2020, 22. [Google Scholar] [CrossRef] [PubMed]
- Onfroy-Roy, L.; Hamel, D.; Malaquin, L.; Ferrand, A. Colon Fibroblasts and Inflammation: Sparring Partners in Colorectal Cancer Initiation? Cancers (Basel) 2021, 13, 1749. [Google Scholar] [CrossRef]
- Chen, L.; Dupre, A.; Qiu, X.; Pellon-Cardenas, O.; Walton, K.D.; Wang, J.; Perekatt, A.O.; Hu, W.; Spence, J.R.; Verzi, M.P. TGFB1 Induces Fetal Reprogramming and Enhances Intestinal Regeneration. bioRxiv 2021. [Google Scholar] [CrossRef]
- Iqbal, S.; Andersson, S.; Nestaite, E.; Pentinmikko, N.; Kumar, A.; Borshagovski, D.; Webb, A.; Saarinen, T.; Juuti, A.; Ori, A.; et al. Fetal-like reversion in the regenerating intestine is regulated by mesenchymal Asporin. bioRxiv 2021. [Google Scholar] [CrossRef]
- Brugger, M.D.; Basler, K. The diverse nature of intestinal fibroblasts in development, homeostasis, and disease. Trends Cell Biol 2023, 33, 834–849. [Google Scholar] [CrossRef] [PubMed]
- Fink, M.; Wrana, J.L. Regulation of homeostasis and regeneration in the adult intestinal epithelium by the TGF-beta superfamily. Dev Dyn 2023, 252, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Baulies, A.; Angelis, N.; Li, V.S.W. Hallmarks of intestinal stem cells. Development 2020, 147. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Y.G. Intestinal epithelial plasticity and regeneration via cell dedifferentiation. Cell Regen 2020, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, O.H.; Katajisto, P.; Lamming, D.W.; Gultekin, Y.; Bauer-Rowe, K.E.; Sengupta, S.; Birsoy, K.; Dursun, A.; Yilmaz, V.O.; Selig, M.; et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 2012, 486, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Escudero, S.; Shivdasani, R.A. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc Natl Acad Sci U S A 2012, 109, 3932–3937. [Google Scholar] [CrossRef]
- Durand, A.; Donahue, B.; Peignon, G.; Letourneur, F.; Cagnard, N.; Slomianny, C.; Perret, C.; Shroyer, N.F.; Romagnolo, B. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc Natl Acad Sci U S A 2012, 109, 8965–8970. [Google Scholar] [CrossRef] [PubMed]
- Ayyaz, A.; Kumar, S.; Sangiorgi, B.; Ghoshal, B.; Gosio, J.; Ouladan, S.; Fink, M.; Barutcu, S.; Trcka, D.; Shen, J.; et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 2019, 569, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Abud, H.E.; Chan, W.H.; Jarde, T. Source and Impact of the EGF Family of Ligands on Intestinal Stem Cells. Front Cell Dev Biol 2021, 9, 685665. [Google Scholar] [CrossRef]
- Jones, J.C.; Brindley, C.D.; Elder, N.H.; Myers, M.G., Jr.; Rajala, M.W.; Dekaney, C.M.; McNamee, E.N.; Frey, M.R.; Shroyer, N.F.; Dempsey, P.J. Cellular Plasticity of Defa4(Cre)-Expressing Paneth Cells in Response to Notch Activation and Intestinal Injury. Cell Mol Gastroenterol Hepatol 2019, 7, 533–554. [Google Scholar] [CrossRef]
- Koch, U.; Lehal, R.; Radtke, F. Stem cells living with a Notch. Development 2013, 140, 689–704. [Google Scholar] [CrossRef]
- Meyer, A.R.; Brown, M.E.; McGrath, P.S.; Dempsey, P.J. Injury-Induced Cellular Plasticity Drives Intestinal Regeneration. Cell Mol Gastroenterol Hepatol 2022, 13, 843–856. [Google Scholar] [CrossRef]
- Kraiczy, J.; McCarthy, N.; Malagola, E.; Tie, G.; Madha, S.; Boffelli, D.; Wagner, D.E.; Wang, T.C.; Shivdasani, R.A. Graded BMP signaling within intestinal crypt architecture directs self-organization of the Wnt-secreting stem cell niche. Cell Stem Cell 2023, 30, 433–449 e438. [Google Scholar] [CrossRef]
- Paerregaard, S.I.; Wulff, L.; Schussek, S.; Niss, K.; Morbe, U.; Jendholm, J.; Wendland, K.; Andrusaite, A.T.; Brulois, K.F.; Nibbs, R.J.B.; et al. The small and large intestine contain related mesenchymal subsets that derive from embryonic Gli1(+) precursors. Nat Commun 2023, 14, 2307. [Google Scholar] [CrossRef] [PubMed]
- Sylvestre, M.; Di Carlo, S.E.; Peduto, L. Stromal regulation of the intestinal barrier. Mucosal Immunol 2023, 16, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Pentinmikko, N.; Iqbal, S.; Mana, M.; Andersson, S.; Cognetta, A.B., 3rd; Suciu, R.M.; Roper, J.; Luopajarvi, K.; Markelin, E.; Gopalakrishnan, S.; et al. Notum produced by Paneth cells attenuates regeneration of aged intestinal epithelium. Nature 2019, 571, 398–402. [Google Scholar] [CrossRef]
- Van Es, J.H.; Sato, T.; Van De Wetering, M.; Lyubimova, A.; Yee Nee, A.N.; Gregorieff, A.; Sasaki, N.; Zeinstra, L.; Van Den Born, M.; Korving, J. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature cell biology 2012, 14, 1099–1104. [Google Scholar] [CrossRef]
- Das, S.; Feng, Q.; Balasubramanian, I.; Lin, X.; Liu, H.; Pellon-Cardenas, O.; Yu, S.; Zhang, X.; Liu, Y.; Wei, Z.; et al. Colonic healing requires Wnt produced by epithelium as well as Tagln+ and Acta2+ stromal cells. Development 2022, 149. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.M.; Di Carlo, S.E.; Stzepourginski, I.; Lepelletier, A.; Ndiaye, P.D.; Varet, H.; Legendre, R.; Kornobis, E.; Benabid, A.; Nigro, G.; et al. PDGFRalpha-induced stromal maturation is required to restrain postnatal intestinal epithelial stemness and promote defense mechanisms. Cell Stem Cell 2022, 29, 856–868 e855. [Google Scholar] [CrossRef] [PubMed]
- Stzepourginski, I.; Nigro, G.; Jacob, J.M.; Dulauroy, S.; Sansonetti, P.J.; Eberl, G.; Peduto, L. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc Natl Acad Sci U S A 2017, 114, E506–E513. [Google Scholar] [CrossRef] [PubMed]
- Greicius, G.; Kabiri, Z.; Sigmundsson, K.; Liang, C.; Bunte, R.; Singh, M.K.; Virshup, D.M. PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc Natl Acad Sci U S A 2018, 115, E3173–E3181. [Google Scholar] [CrossRef] [PubMed]
- Kuhnert, F.; Davis, C.R.; Wang, H.T.; Chu, P.; Lee, M.; Yuan, J.; Nusse, R.; Kuo, C.J. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A 2004, 101, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, G.; Banziger, C.; Basler, K. Helping Wingless take flight: how WNT proteins are secreted. Nat Rev Mol Cell Biol 2007, 8, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.S.; Janda, C.Y.; Chang, J.; Zheng, G.X.Y.; Larkin, K.A.; Luca, V.C.; Chia, L.A.; Mah, A.T.; Han, A.; Terry, J.M.; et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature 2017, 545, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, D.J.; Austin, C.R.; Vincan, E.; Phesse, T.J. Wnt Signalling in Gastrointestinal Epithelial Stem Cells. Genes (Basel) 2018, 9, 178. [Google Scholar] [CrossRef]
- Bottcher, A.; Buttner, M.; Tritschler, S.; Sterr, M.; Aliluev, A.; Oppenlander, L.; Burtscher, I.; Sass, S.; Irmler, M.; Beckers, J.; et al. Author Correction: Non-canonical Wnt/PCP signalling regulates intestinal stem cell lineage priming towards enteroendocrine and Paneth cell fates. Nat Cell Biol 2021, 23, 566–576. [Google Scholar] [CrossRef]
- Flores-Hernandez, E.; Velazquez, D.M.; Castaneda-Patlan, M.C.; Fuentes-Garcia, G.; Fonseca-Camarillo, G.; Yamamoto-Furusho, J.K.; Romero-Avila, M.T.; Garcia-Sainz, J.A.; Robles-Flores, M. Canonical and non-canonical Wnt signaling are simultaneously activated by Wnts in colon cancer cells. Cell Signal 2020, 72, 109636. [Google Scholar] [CrossRef]
- Werner, J.; Boonekamp, K.E.; Zhan, T.; Boutros, M. The Roles of Secreted Wnt Ligands in Cancer. Int J Mol Sci 2023, 24, 5349. [Google Scholar] [CrossRef]
- Halpern, K.B.; Massalha, H.; Zwick, R.K.; Moor, A.E.; Castillo-Azofeifa, D.; Rozenberg, M.; Farack, L.; Egozi, A.; Miller, D.R.; Averbukh, I.; et al. Lgr5+ telocytes are a signaling hub at the intestinal villus tip. bioRxiv 2020, 850909. [Google Scholar] [CrossRef]
- McCarthy, N.; Kraiczy, J.; Shivdasani, R.A. Cellular and molecular architecture of the intestinal stem cell niche. Nat Cell Biol 2020, 22, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Bonis, V.; Rossell, C.; Gehart, H. The Intestinal Epithelium - Fluid Fate and Rigid Structure From Crypt Bottom to Villus Tip. Front Cell Dev Biol 2021, 9, 661931. [Google Scholar] [CrossRef]
- Corda, G.; Sala, A. Non-canonical WNT/PCP signalling in cancer: Fzd6 takes centre stage. Oncogenesis 2017, 6, e364. [Google Scholar] [CrossRef] [PubMed]
- De, A. Wnt/Ca2+ signaling pathway: a brief overview. Acta Biochim Biophys Sin (Shanghai) 2011, 43, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Hingole, S.; Chaudhary, V. The Emerging Mechanisms of Wnt Secretion and Signaling in Development. Front Cell Dev Biol 2021, 9, 714746. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, U.K.; Kini, S.G.; Garg, V.; Agrawal, S.; Tomar, P.K.; Pathak, P.; Chaudhary, A.; Gupta, P.; Malik, A. JNK pathway signaling: a novel and smarter therapeutic targets for various biological diseases. Future Med Chem 2015, 7, 2065–2086. [Google Scholar] [CrossRef] [PubMed]
- Walton, K.D.; Freddo, A.M.; Wang, S.; Gumucio, D.L. Generation of intestinal surface: an absorbing tale. Development 2016, 143, 2261–2272. [Google Scholar] [CrossRef]
- Walton, K.D.; Kolterud, A.; Czerwinski, M.J.; Bell, M.J.; Prakash, A.; Kushwaha, J.; Grosse, A.S.; Schnell, S.; Gumucio, D.L. Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc Natl Acad Sci U S A 2012, 109, 15817–15822. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Y.G. BMP signaling in homeostasis, transformation and inflammatory response of intestinal epithelium. Sci China Life Sci 2018, 61, 800–807. [Google Scholar] [CrossRef]
- Madison, B.B.; Braunstein, K.; Kuizon, E.; Portman, K.; Qiao, X.T.; Gumucio, D.L. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development 2005, 132, 279–289. [Google Scholar] [CrossRef]
- Walton, K.D.; Gumucio, D.L. Hedgehog Signaling in Intestinal Development and Homeostasis. Annu Rev Physiol 2021, 83, 359–380. [Google Scholar] [CrossRef]
- McCarthy, N.; Manieri, E.; Storm, E.E.; Saadatpour, A.; Luoma, A.M.; Kapoor, V.N.; Madha, S.; Gaynor, L.T.; Cox, C.; Keerthivasan, S.; et al. Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell Stem Cell 2020, 26, 391–402 e395. [Google Scholar] [CrossRef]
- Orzechowska-Licari, E.J.; Bialkowska, A.B.; Yang, V.W. Sonic Hedgehog and WNT Signaling Regulate a Positive Feedback Loop Between Intestinal Epithelial and Stromal Cells to Promote Epithelial Regeneration. Cell Mol Gastroenterol Hepatol 2023, 16, 607–642. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.M.; Hill, D.R.; Aurora, M.; Spence, J.R. Morphogenesis and maturation of the embryonic and postnatal intestine. Semin Cell Dev Biol 2017, 66, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Greicius, G.; Virshup, D.M. Stromal control of intestinal development and the stem cell niche. Differentiation 2019, 108, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Pellegrinet, L.; Rodilla, V.; Liu, Z.; Chen, S.; Koch, U.; Espinosa, L.; Kaestner, K.H.; Kopan, R.; Lewis, J.; Radtke, F. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 2011, 140, 1230–1240 e1231-1237. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.J.; Li, X.G.; Wang, X.Q. Notch Signaling in Mammalian Intestinal Stem Cells: Determining Cell Fate and Maintaining Homeostasis. Curr Stem Cell Res Ther 2019, 14, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Geyer, N.; Gerling, M. Hedgehog signaling in colorectal cancer: All in the stroma? International Journal of Molecular Sciences 2021, 22, 1025. [Google Scholar] [CrossRef] [PubMed]
- Degirmenci, B.; Valenta, T.; Dimitrieva, S.; Hausmann, G.; Basler, K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 2018, 558, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Fei, L.; Yin, W.C.; Coquenlorge, S.; Rao-Bhatia, A.; Zhang, X.; Shi, S.S.W.; Lee, J.H.; Hahn, N.A.; Rizvi, W.; et al. Single cell and genetic analyses reveal conserved populations and signaling mechanisms of gastrointestinal stromal niches. Nat Commun 2020, 11, 334. [Google Scholar] [CrossRef]
- Rees, W.D.; Tandun, R.; Yau, E.; Zachos, N.C.; Steiner, T.S. Regenerative Intestinal Stem Cells Induced by Acute and Chronic Injury: The Saving Grace of the Epithelium? Front Cell Dev Biol 2020, 8, 583919. [Google Scholar] [CrossRef] [PubMed]
- Baghdadi, M.B.; Ayyaz, A.; Coquenlorge, S.; Chu, B.; Kumar, S.; Streutker, C.; Wrana, J.L.; Kim, T.H. Enteric glial cell heterogeneity regulates intestinal stem cell niches. Cell Stem Cell 2022, 29, 86–100 e106. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Schewe, M.; Sacchetti, A.; Feijtel, D.; van de Geer, W.S.; Teeuwssen, M.; Sleddens, H.F.; Joosten, R.; van Royen, M.E.; van de Werken, H.J.G.; et al. Paneth Cells Respond to Inflammation and Contribute to Tissue Regeneration by Acquiring Stem-like Features through SCF/c-Kit Signaling. Cell Rep 2018, 24, 2312–2328 e2317. [Google Scholar] [CrossRef]
- Metcalfe, C.; Kljavin, N.M.; Ybarra, R.; de Sauvage, F.J. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 2014, 14, 149–159. [Google Scholar] [CrossRef]
- Wang, Y.; Chiang, I.L.; Ohara, T.E.; Fujii, S.; Cheng, J.; Muegge, B.D.; Ver Heul, A.; Han, N.D.; Lu, Q.; Xiong, S.; et al. Long-Term Culture Captures Injury-Repair Cycles of Colonic Stem Cells. Cell 2019, 179, 1144–1159 e1115. [Google Scholar] [CrossRef]
- Rees, W.D.; Telkar, N.; Lin, D.T.S.; Wong, M.Q.; Poloni, C.; Fathi, A.; Kobor, M.; Zachos, N.C.; Steiner, T.S. An in vitro chronic damage model impairs inflammatory and regenerative responses in human colonoid monolayers. Cell Rep 2022, 38, 110283. [Google Scholar] [CrossRef]
- Barry, E.R.; Morikawa, T.; Butler, B.L.; Shrestha, K.; de la Rosa, R.; Yan, K.S.; Fuchs, C.S.; Magness, S.T.; Smits, R.; Ogino, S.; et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature 2013, 493, 106–110. [Google Scholar] [CrossRef]
- Yan, K.S.; Chia, L.A.; Li, X.; Ootani, A.; Su, J.; Lee, J.Y.; Su, N.; Luo, Y.; Heilshorn, S.C.; Amieva, M.R.; et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A 2012, 109, 466–471. [Google Scholar] [CrossRef]
- Tian, H.; Biehs, B.; Warming, S.; Leong, K.G.; Rangell, L.; Klein, O.D.; de Sauvage, F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 2011, 478, 255–259. [Google Scholar] [CrossRef]
- Murata, K.; Jadhav, U.; Madha, S.; van Es, J.; Dean, J.; Cavazza, A.; Wucherpfennig, K.; Michor, F.; Clevers, H.; Shivdasani, R.A. Ascl2-Dependent Cell Dedifferentiation Drives Regeneration of Ablated Intestinal Stem Cells. Cell Stem Cell 2020, 26, 377–390 e376. [Google Scholar] [CrossRef] [PubMed]
- Bankaitis, E.D.; Ha, A.; Kuo, C.J.; Magness, S.T. Reserve Stem Cells in Intestinal Homeostasis and Injury. Gastroenterology 2018, 155, 1348–1361. [Google Scholar] [CrossRef]
- Montgomery, R.K.; Carlone, D.L.; Richmond, C.A.; Farilla, L.; Kranendonk, M.E.; Henderson, D.E.; Baffour-Awuah, N.Y.; Ambruzs, D.M.; Fogli, L.K.; Algra, S.; et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A 2011, 108, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, M.; Li, N.; Nakauka-Ddamba, A.; Wang, S.; Davidow, K.; Schoenberger, J.; Yu, Z.; Jensen, S.T.; Kharas, M.G.; Lengner, C.J. Msi RNA-binding proteins control reserve intestinal stem cell quiescence. J Cell Biol 2016, 215, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.N.; Kim, M.J.; Jung, Y.S.; Lien, E.M.; Jun, S.; Park, J.I. Quiescence Exit of Tert(+) Stem Cells by Wnt/beta-Catenin Is Indispensable for Intestinal Regeneration. Cell Rep 2017, 21, 2571–2584. [Google Scholar] [CrossRef]
- Hageman, J.H.; Heinz, M.C.; Kretzschmar, K.; van der Vaart, J.; Clevers, H.; Snippert, H.J.G. Intestinal Regeneration: Regulation by the Microenvironment. Dev Cell 2020, 54, 435–446. [Google Scholar] [CrossRef]
- Sprangers, J.; Zaalberg, I.C.; Maurice, M.M. Organoid-based modeling of intestinal development, regeneration, and repair. Cell Death Differ 2021, 28, 95–107. [Google Scholar] [CrossRef]
- Yan, K.S.; Gevaert, O.; Zheng, G.X.Y.; Anchang, B.; Probert, C.S.; Larkin, K.A.; Davies, P.S.; Cheng, Z.F.; Kaddis, J.S.; Han, A.; et al. Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell 2017, 21, 78–90 e76. [Google Scholar] [CrossRef]
- Shivdasani, R.A.; Clevers, H.; de Sauvage, F.J. Tissue regeneration: Reserve or reverse? Science 2021, 371, 784–786. [Google Scholar] [CrossRef]
- Tomic, G.; Morrissey, E.; Kozar, S.; Ben-Moshe, S.; Hoyle, A.; Azzarelli, R.; Kemp, R.; Chilamakuri, C.S.R.; Itzkovitz, S.; Philpott, A.; et al. Phospho-regulation of ATOH1 Is Required for Plasticity of Secretory Progenitors and Tissue Regeneration. Cell Stem Cell 2018, 23, 436–443 e437. [Google Scholar] [CrossRef]
- Castillo-Azofeifa, D.; Fazio, E.N.; Nattiv, R.; Good, H.J.; Wald, T.; Pest, M.A.; de Sauvage, F.J.; Klein, O.D.; Asfaha, S. Atoh1(+) secretory progenitors possess renewal capacity independent of Lgr5(+) cells during colonic regeneration. EMBO J 2019, 38, e99984. [Google Scholar] [CrossRef]
- Yu, S.; Tong, K.; Zhao, Y.; Balasubramanian, I.; Yap, G.S.; Ferraris, R.P.; Bonder, E.M.; Verzi, M.P.; Gao, N. Paneth Cell Multipotency Induced by Notch Activation following Injury. Cell Stem Cell 2018, 23, 46–59 e45. [Google Scholar] [CrossRef]
- Deng, F.; Peng, L.; Li, Z.; Tan, G.; Liang, E.; Chen, S.; Zhao, X.; Zhi, F. YAP triggers the Wnt/beta-catenin signalling pathway and promotes enterocyte self-renewal, regeneration and tumorigenesis after DSS-induced injury. Cell Death Dis 2018, 9, 153. [Google Scholar] [CrossRef]
- Hong, A.W.; Meng, Z.; Guan, K.L. The Hippo pathway in intestinal regeneration and disease. Nat Rev Gastroenterol Hepatol 2016, 13, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Palikuqi, B.; Rispal, J.; Klein, O. Good Neighbors: The Niche that Fine Tunes Mammalian Intestinal Regeneration. Cold Spring Harb Perspect Biol 2022, 14, a040865. [Google Scholar] [CrossRef]
- Chalkidi, N.; Paraskeva, C.; Koliaraki, V. Fibroblasts in intestinal homeostasis, damage, and repair. Front Immunol 2022, 13, 924866. [Google Scholar] [CrossRef] [PubMed]
- Nusse, Y.M.; Savage, A.K.; Marangoni, P.; Rosendahl-Huber, A.K.; Landman, T.A.; De Sauvage, F.J.; Locksley, R.M.; Klein, O.D. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 2018, 559, 109–113. [Google Scholar] [CrossRef] [PubMed]
- van Dop, W.A.; Heijmans, J.; Buller, N.V.; Snoek, S.A.; Rosekrans, S.L.; Wassenberg, E.A.; van den Bergh Weerman, M.A.; Lanske, B.; Clarke, A.R.; Winton, D.J.; et al. Loss of Indian Hedgehog activates multiple aspects of a wound healing response in the mouse intestine. Gastroenterology 2010, 139, 1665–1676.e10. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Peng, Y.; Li, H. The Injury-Related Activation of Hedgehog Signaling Pathway Modulates the Repair-Associated Inflammation in Liver Fibrosis. Front Immunol 2017, 8, 1450. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, M.; Zhou, G.; Lin, L.; Han, J.; Wang, Y.; Li, L.; He, Y.; Zeng, Z.; Chen, M.; et al. Emerging roles of the Hedgehog signalling pathway in inflammatory bowel disease. Cell Death Discov 2021, 7, 314. [Google Scholar] [CrossRef]
- Pasztoi, M.; Ohnmacht, C. Tissue Niches Formed by Intestinal Mesenchymal Stromal Cells in Mucosal Homeostasis and Immunity. Int J Mol Sci 2022, 23, 5181. [Google Scholar] [CrossRef]
- Owens, B.M. Inflammation, Innate Immunity, and the Intestinal Stromal Cell Niche: Opportunities and Challenges. Front Immunol 2015, 6, 319. [Google Scholar] [CrossRef]
- Roulis, M.; Flavell, R.A. Fibroblasts and myofibroblasts of the intestinal lamina propria in physiology and disease. Differentiation 2016, 92, 116–131. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.W.; Pinchuk, I.V.; Saada, J.I.; Chen, X.; Mifflin, R.C. Mesenchymal cells of the intestinal lamina propria. Annu Rev Physiol 2011, 73, 213–237. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Fawkner-Corbett, D.; Antanaviciute, A.; Parikh, K.; Jagielowicz, M.; Geros, A.S.; Gupta, T.; Ashley, N.; Khamis, D.; Fowler, D.; Morrissey, E.; et al. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell 2021, 184, 810–826 e823. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, R.I.; Maehr, R.; Mazzoni, E.O.; Melton, D.A. Wnt signaling specifies and patterns intestinal endoderm. Mech Dev 2011, 128, 387–400. [Google Scholar] [CrossRef]
- Chen, L.; Acciani, T.; Le Cras, T.; Lutzko, C.; Perl, A.K. Dynamic regulation of platelet-derived growth factor receptor alpha expression in alveolar fibroblasts during realveolarization. Am J Respir Cell Mol Biol 2012, 47, 517–527. [Google Scholar] [CrossRef]
- Korinek, V.; Barker, N.; Moerer, P.; van Donselaar, E.; Huls, G.; Peters, P.J.; Clevers, H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet 1998, 19, 379–383. [Google Scholar] [CrossRef]
- Chin, A.M.; Tsai, Y.H.; Finkbeiner, S.R.; Nagy, M.S.; Walker, E.M.; Ethen, N.J.; Williams, B.O.; Battle, M.A.; Spence, J.R. A Dynamic WNT/beta-CATENIN Signaling Environment Leads to WNT-Independent and WNT-Dependent Proliferation of Embryonic Intestinal Progenitor Cells. Stem Cell Reports 2016, 7, 826–839. [Google Scholar] [CrossRef]
- Sun, H.; Tan, J.; Chen, H.; Wu, N.; Su, B. Immune niches orchestrated by intestinal mesenchymal stromal cells lining the crypt-villus. Front Immunol 2022, 13, 1057932. [Google Scholar] [CrossRef] [PubMed]
- Haffen, K.; Kedinger, M.; Simon-Assmann, P. Mesenchyme-dependent differentiation of epithelial progenitor cells in the gut. J Pediatr Gastroenterol Nutr 1987, 6, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Fazilaty, H.; Brugger, M.D.; Valenta, T.; Szczerba, B.M.; Berkova, L.; Doumpas, N.; Hausmann, G.; Scharl, M.; Basler, K. Tracing colonic embryonic transcriptional profiles and their reactivation upon intestinal damage. Cell Rep 2021, 36, 109484. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, M.; Suzuki, N.; Wang, T.; Wright, J.A.; Lannagan, T.R.M.; Vrbanac, L.; Kobayashi, H.; Gieniec, K.A.; Ng, J.Q.; Hayakawa, Y.; et al. Stromal DLK1 promotes proliferation and inhibits differentiation of the intestinal epithelium during development. Am J Physiol Gastrointest Liver Physiol 2021, 320, G506–G520. [Google Scholar] [CrossRef] [PubMed]
- Bry, L.; Falk, P.; Huttner, K.; Ouellette, A.; Midtvedt, T.; Gordon, J.I. Paneth cell differentiation in the developing intestine of normal and transgenic mice. Proc Natl Acad Sci U S A 1994, 91, 10335–10339. [Google Scholar] [CrossRef] [PubMed]
- Dehmer, J.J.; Garrison, A.P.; Speck, K.E.; Dekaney, C.M.; Van Landeghem, L.; Sun, X.; Henning, S.J.; Helmrath, M.A. Expansion of intestinal epithelial stem cells during murine development. PLoS One 2011, 6, e27070. [Google Scholar] [CrossRef] [PubMed]
- Vannucchi, M.G.; Traini, C.; Manetti, M.; Ibba-Manneschi, L.; Faussone-Pellegrini, M.S. Telocytes express PDGFRalpha in the human gastrointestinal tract. J Cell Mol Med 2013, 17, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Cretoiu, D.; Cretoiu, S.M.; Simionescu, A.A.; Popescu, L.M. Telocytes, a distinct type of cell among the stromal cells present in the lamina propria of jejunum. Histol Histopathol 2012, 27, 1067–1078. [Google Scholar] [CrossRef]
- Hong, S.P.; Yang, M.J.; Cho, H.; Park, I.; Bae, H.; Choe, K.; Suh, S.H.; Adams, R.H.; Alitalo, K.; Lim, D.; et al. Distinct fibroblast subsets regulate lacteal integrity through YAP/TAZ-induced VEGF-C in intestinal villi. Nat Commun 2020, 11, 4102. [Google Scholar] [CrossRef]
- VanDussen, K.L.; Carulli, A.J.; Keeley, T.M.; Patel, S.R.; Puthoff, B.J.; Magness, S.T.; Tran, I.T.; Maillard, I.; Siebel, C.; Kolterud, A.; et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 2012, 139, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Valenta, T.; Degirmenci, B.; Moor, A.E.; Herr, P.; Zimmerli, D.; Moor, M.B.; Hausmann, G.; Cantu, C.; Aguet, M.; Basler, K. Wnt Ligands Secreted by Subepithelial Mesenchymal Cells Are Essential for the Survival of Intestinal Stem Cells and Gut Homeostasis. Cell Rep 2016, 15, 911–918. [Google Scholar] [CrossRef]
- Lahar, N.; Lei, N.Y.; Wang, J.; Jabaji, Z.; Tung, S.C.; Joshi, V.; Lewis, M.; Stelzner, M.; Martin, M.G.; Dunn, J.C. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One 2011, 6, e26898. [Google Scholar] [CrossRef]
- Guenin-Mace, L.; Konieczny, P.; Naik, S. Immune-Epithelial Cross Talk in Regeneration and Repair. Annu Rev Immunol 2023, 41, 207–228. [Google Scholar] [CrossRef]
- Poggi, L.; Casarosa, S.; Carl, M. An Eye on the Wnt Inhibitory Factor Wif1. Front Cell Dev Biol 2018, 6, 167. [Google Scholar] [CrossRef] [PubMed]
- Melissari, M.T.; Henriques, A.; Tzaferis, C.; Prados, A.; Sarris, M.E.; Chalkidi, N.; Mavroeidi, D.; Chouvardas, P.; Grammenoudi, S.; Kollias, G.; et al. Col6a1(+)/CD201(+) mesenchymal cells regulate intestinal morphogenesis and homeostasis. Cell Mol Life Sci 2021, 79, 1. [Google Scholar] [CrossRef]
- Chivukula, R.R.; Shi, G.; Acharya, A.; Mills, E.W.; Zeitels, L.R.; Anandam, J.L.; Abdelnaby, A.A.; Balch, G.C.; Mansour, J.C.; Yopp, A.C.; et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell 2014, 157, 1104–1116. [Google Scholar] [CrossRef] [PubMed]
| Epithelial cells | Cell Markers | Ligands | Functions | Signaling pathways | References |
|---|---|---|---|---|---|
| Paneth cells | Defa4 expressing cell | Wnt3a, Wnt9b, Wnt11 | Support regeneration | Notch | [35] |
| pS6+ | Notum | Wnt inhibitor | Non-canonical Wnt/mTORC1 | [41] | |
| Progenitor cells | Dll+ | Dll1, dll4 | Support regeneration | Notch | [42] |
| Secretory lineage | Unknown | Egf, Tgfa, | Promote IEC homeostasis | EGFR/RAS | [34] |
| Colonic Paneth cells | REG4+ | Dll1, Egf | Promote stemness | Notch | [43] |
| +4 quiescent cells | BMI1, HOPX, MTERT | Unknown | Support regeneration | Hippo | [21,33] |
| Tuft cells | DCLK1 | Dll1 | Promote regeneration | Notch | [37] |
| Non-epithelial cells | Cell Markers | Ligands | Functions | Signaling pathways | References |
|---|---|---|---|---|---|
| PDGFRαlo Cells | CD81+, CD55hi | Wnt2b, Gremlin1/2, Rspo3 | Wnt promoters | Wnt/β-catenin | [38,66] |
| Fgfr2+, CD55lo | Wnt4, Frzb, Sfrp1 | Wnt repressors | Non-canonical Wnt/Bmp | [38,39] | |
| PDGFRαhi Cells | FOXL1+, CD9hiCD141-, CD9loCD141+, CD141int | Bmp3/4, Wnt5a/b, Dkk | BMP agonists,Wnt inhibitor | Non-canonical Wnt/Bmp | [39] |
| PDGFRα+ | PDGFRα+DLK1+ | Dlk1 | Embryonic morphogenesis | Notch | [117] |
| LTβR+ | LTβR+PDGFRαhi | Pdgf | Stromal maturation | Bmp activation | [44] |
| Smooth muscle cells | Tagln+, Acta2+,Myh11+ | Wnts | ISC integrity and wound healing | Wnt/β-catenin | [43] |
| Immune cells | ILC2, ILC3 | Il13, Il22 | Promote regeneration | Wnt/β-catenin | [126] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





