Classic compartmental models (CM) describing epidemic spreading assume that any individual in the infective compartment can “infect” any other in the susceptible compartment [
1]. This assumption called the mean-field approximation, ignores the network effects in favor of analytical tractability [
1]. However, social interactions do not randomly occur. They are structured along social links between specific individuals based on social interactions, e.g., love, friendship, work, etc. CMs, including explicit representations of network topologies, have been advocated as a necessary improvement of classical CMs since early 2000 [
2,
3].
In the last two decades, the research has recognized the importance of networks in epidemiology, and this has led to relevant contributions, such as the finding that the vaccination thresholds strongly depend on network topology [
4], or that network community structure, i.e., the presence of groups of nodes/individuals strongly connected among them in real social networks, has a significant impact on disease dynamics [
5].
Despite this evidence, during the COVID-19 pandemic crisis, CMs have mainly been used to predict the macroscopic dynamics of infections and deaths and to assess different policies to curb the pace of the person-to-person contagion dynamic [
3]. For example, Ferguson et colleagues [
6] ‘s famous study, which was the basis of the first containment measures to halt COVID-19 spreading, analyzed the epidemic dynamic using a simple Susceptible-Infectious-Recovered (SIR) model neglecting the social network topological structure. Kissler et al. [
7] used an ordinary differential equation mathematical analysis adapted from a Susceptible-Exposed-Infectious-Recovered (SEIR) model to simulate the transmission of SARS-CoV-2 and the hospital care capacity of the United States.
Here, we comment on research manuscripts with classic, significant, or interesting insights explaining how coupling networks and CM may improve the description of epidemic spreading. Then, we discuss the research perspective for further enhancing network use in epidemiology.
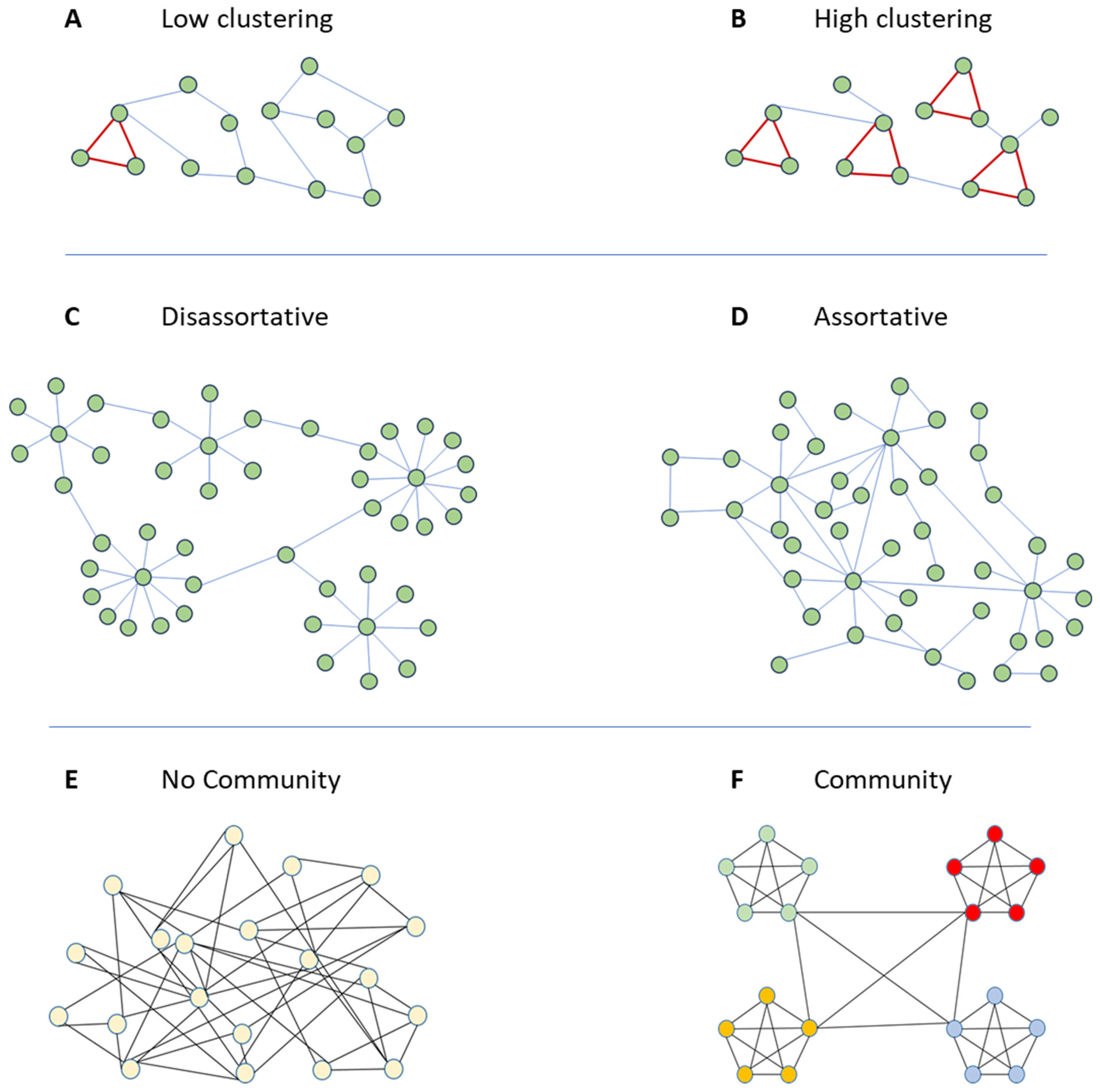
2.2. Spreading, Node Clustering Coefficient, and Node Assortativity
CM assumes that epidemic transmission passes through interactions occurring randomly in groups, with all individuals potentially interacting with all other individuals at an equal rate [
10]. However, social interactions among individuals do not occur randomly, and nodes in real social networks unveil preferential ways of connecting with other nodes, making non-random peculiar structural features [
11,
12]. Network science uses mathematical approaches to describe these structural features of real social networks.
The network clustering coefficient is an indicator that counts node triplets in the network. A triplet (or triangle) is a set of three nodes. A closed triplet is a full network of three nodes, i.e., a set of three nodes in which a link connects each node with the others. In other words, a triplet is three nodes connected by either two (open triplet) or three (closed triplet) links.
The ‘local clustering coefficient’
of node
is defined as:
where
is the number of closed triplets centered on node
, and
is the total number of triplets (both open and closed) centered on node
[
13]. The node clustering coefficient is also named node transitivity [
12]. Node transitivity is related to the binary transitive relation in mathematics, stating that a set
is transitive if, for all elements
in
, whenever
relates to
and
to
, then also
relates to
. Translating in network science terms, a network is transitive whether, for the connected node pairs
and
it is likely that nodes
and
are also connected.
Computing the triplets over the whole network, we can define the binary global clustering coefficient by generalizing Eq. 1:
where
is the number of closed triplets, and
is the total number of triplets (both open and closed) in the network [
13].
Calculating the clustering coefficient is the simplest method to investigate the presence of node communities in the network, i.e., node communities are groups of nodes that are densely connected among them. The clustering coefficient evaluates the local group cohesiveness accounting for the fraction of connected neighbors, and for this, it evaluates the tendency of network nodes to form communities. In other words, it is a measure of the magnitude to which nodes tend to form strongly connected communities characterized by a higher density of links than the average probability of links randomly drawn among nodes [
14,
15].
Figure 1A depicts a toy model network with a lower clustering coefficient, and
Figure 1B shows a network with a higher clustering coefficient.
Assortativity is a network structure indicator that evaluates to what extent nodes in a network associate with other nodes in the network, being of similar sort or being of opposing sort. Generally, the assortativity of a network is determined by the degree of the nodes in the network [
16]. The notion of assortativity was introduced by Newman [
17] and was widely used in network science for many different applications [
16].
The node degree assortativity
is defined as:
is the standard deviation of the excess degree distribution,
is the fraction of links connecting nodes of degree
and
, and
,
are the excess degree of nodes of degree
and
. On other terms, degree assortativity is a measure of its degree correlation, describing how nodes in the network associate based on their number of connections. In
Figure 1C we show a disassortative network in which higher degree nodes tend to be connected preferentially with lower degree nodes. On the opposite, in
Figure 1D, we draw an assortative network in which nodes of higher degrees are connected preferentially with nodes of higher degrees, and, therefore, the nodes of lower degrees are more likely to relate to other lower-degree nodes. Social networks are typically thought to be distinct from other networks in being assortative (possessing positive degree correlations); well-connected individuals associate with other well-connected individuals, and less connected individuals associate with each other [
18].
The node degree assortativity is an essential concept in the field of epidemiology since it can affect the spread of disease [
19].
Badham and Stocker [
19] investigate how node degree assortativity and clustering coefficient affect the spread of the SIR epidemic. They built model networks using an algorithm to tune node degree assortativity and clustering coefficient. To incorporate a real-world social network structure, the authors used a degree distribution based on the number of friends in a friendship social network of young children [
20]. To evaluate the extent of the epidemic spreading, the authors account for the final size (as a proportion of nodes infected at the end of the simulation) and whether an epidemic occurred (at least 25 nodes ever become infected, representing at least some secondary infections).
They found that there is no consistent trend in epidemic proportion related to either the degree assortativity or the clustering coefficient. However, low epidemic occurrence is associated with a high value of either feature.
Further, the authors discovered that the final size of the epidemic decreases when the assortativity or clustering coefficient increases. This means that the final number of infected nodes would be lower if connected nodes/individuals are preferentially connected with nodes of similar degrees.
Based on their results, Badham and Stocker [
19] stated that the structural properties identified by social network researchers are relevant for epidemiology, and systematic research is necessary to shed light on the potential size of the effect of the network epidemic spreading.
Volz et al. [
21] conducted mathematical and numerical analyses of the SIR model investigating the effect of node clustering over the epidemic spreading entity on networks. They found that in most cases, node clustering is correlated with a lower final extent of the spreading, i.e., networks with a higher clustering presented a lower number of infected at the end of the SIR simulations. The finding of Volz et al. [
21] corroborates the Badham and Stocker [2010] results by outlining the importance of considering the structural features of the networks when the aim is to predict the epidemic spreading. It is important for network interventions to halt epidemics, such as concerning the vaccination of individuals [
22] or performing social distancing [
23], to consider the extent of node clustering.
2.3. Spreading and Community Structure
Real social networks show marked patterns of community structure; that is, social networks present groups of nodes/individuals that are more connected among them [
24]. In
Figure 1E we depict a random network that does not present a community structure; at the opposite, in
Figure 1F we show a network with a strong community structure. The presence of communities of individuals highly connected among them may change the epidemic spreading dynamics. Salathè and Jones [
25] investigated the spread of disease in networks with community structure. They simulate SIR epidemic dynamics over both real and model networks. The authors assembled model networks with community structure by joining different subnetworks with randomly drawn edges. Then, they correlate the epidemic spreading pace with the modularity coefficient
[
26], which evaluates the magnitude of the community structure of the network.
The network modularity
measures how good the division of two node communities is or how separated the different node communities are from each other [
26].
The modularity indicator
is defined as:
where
is the total number of links in the network,
is the element
, of the adjacency matrix, equal to 1 if nodes
and
j are connected, and 0 otherwise,
, and
are the degrees of nodes
and
, respectively,
, and
are the modules (or community) of node
and
respectively, and
is 1 if
and 0 otherwise. The modularity
represents the fraction of the links that fall within the given community minus the expected fraction if links are drowned at random. Positive
indicates that the number of links within communities exceeds the randomly expected number by chance, the maximum possible value of
is 1, nonzero values indicate deviations from randomness, and values around 0.3 or more usually indicate good divisions.
Salathè and Jones [
25] found that community structure has a major impact on disease dynamics in a peculiar way; in networks with a strong community structure, an infected individual is more likely to infect members of the same community than members outside the community. Therefore, in a network with strong community structure, local outbreaks may extinguish before spreading to other communities.
Further, Salathè and Jones [
25] investigated how individuals’ immunization (vaccination) curbs the epidemic’s spread. Vaccination corresponds to removing nodes or setting nodes in a recovered (not infectious) state [
27,
28,
29]. Salathè and Jones [
25] showed that networks with strong community structure, immunization interventions targeted at individuals bridging different communities are more effective than those simply targeting highly connected individuals [
25]. These results have implications for the design of control strategies.
2.4. Effective Network Size (ENS)
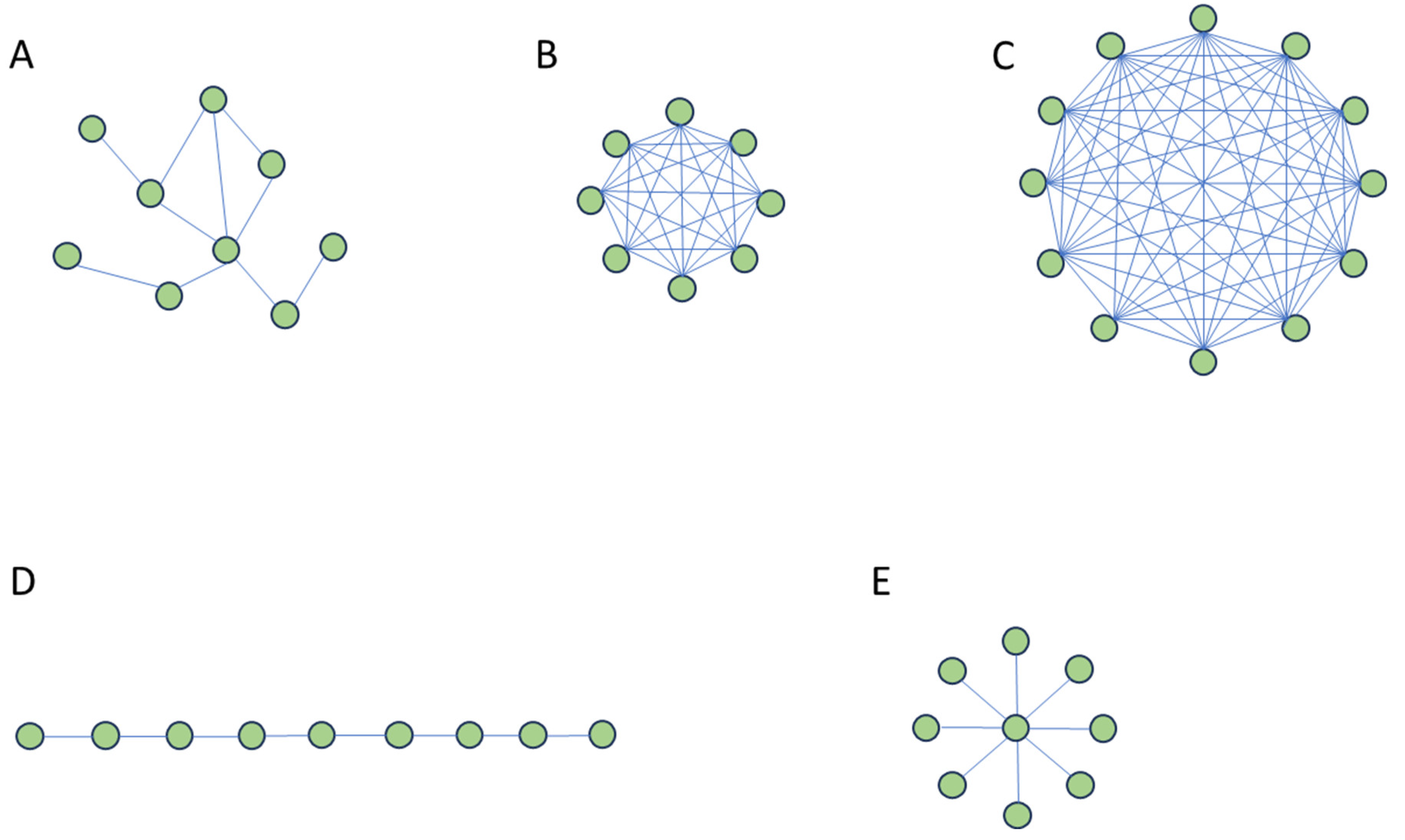
Transposing the mean-field approach of the classic CM epidemic dynamics to a network model, one should use a “complete network” in which all nodes/individuals interact with each other.
In this spirit, McCabe and Nunn [
10] compare the SI/SIR spreading pace of i) complete networks, ii) Erdos-Renyi (ER) random networks, and iii) real primate networks of the same size (number of nodes). The ER random network is a classic model for generating a random network with only two parameters, i.e., the number of nodes (
) and a fixed probability (
) for links being present or absent, independently of the other links.
The primate networks are empirically observed networks of social interactions among primates (Pan troglodytes), and they are valuable frameworks for investigating disease spreading in nature.
The authors use the ‘outbreak duration’, i.e., the number of days until the simulation ended, i.e., when all the individuals were recovered and/or infected, as a proxy of the spreading pace.
They find that outbreak durations of simulations on ER networks are more variable than those on complete networks, whereas they show similar mean durations of disease spread. This result indicates that including a simple structural feature, such as removing a fraction of the possible link/interactions, as passing from a complete network to an ER network, can increase the variability of the outbreak duration. On the other hand, real primate networks show a longer outbreak duration concerning a complete network, suggesting how the mean-field approach overestimates the spreading pace.
Bearing on these results, the authors propose a measure to account for such heterogeneity: ‘
effective network size’ (ENS), which refers to the size of a complete network (i.e., unstructured, where all individuals interact with all others equally) that corresponds to the outbreak characteristics of a given heterogeneous, structured network. The ENS of real primate networks are always higher than their real network size, meaning that the CM models with infection probability parameter values of the real network will overestimate the pace of the epidemic spreading. In
Figure 2 A-C we explain the rationale of the McCabe and Nunn [
10] analyses.
The article has excellent merit in showing in a simple way how to assume the mean field interaction may produce an erroneous description of the disease spreading in real social networks.
2.5. The Case of the COVID-19 Spreading
A recent paper by Thurner et al. [
1] adopts a network approach to investigate the spread of the COVID-19 epidemic. The authors point out that traditional CM epidemiological models cannot explain how the COVID-19 infection curves for many countries reveal a remarkable linear growth over extended periods. Using the salient real network features and a SIR model, the authors explain that linear growth can emerge naturally in real social networks. Traditional CM typically ignore the structure of real contact networks that are essential in the characteristic spreading dynamics of COVID-19.
The authors consider structural features of empirical social contact networks, including node degree heterogeneity (heterogeneity in the number of social links), the fact that people tend to live in small groups (families or communities), and bridge links connecting distant groups (such as that work and leisure links/relations). They show that in these realistic social network structures, a critical number of social contacts () exists for any given transmission rate, below which linear growth and low infection prevalence must occur.
Calibrating the SIR model to empirical estimates of the Covid-19 transmission rate and the number of days being individually contagious, the authors found ∼7.2, i.e., the node degree indicating the number of social contact links should be above 7.2 to produce a super linear epidemic growth. Realistic contact networks show a node degree of about five, and lockdown measures would reduce the social interactions to household size (∼2.5). Therefore, the real social contact networks may reproduce the empirical infection curves with significant precision without additional model assumptions or fine-tuning of parameters.
The probability of observing linear growth with standard CM is practically zero. For this, Thurner et al. [
1] question the applicability of standard CM to describe the COVID-19 containment phase. Further, the effect of non-pharmaceutical interventions (NPIs), like national lockdowns, can be modeled with a remarkable degree of precision by coupling a proper network approach to standard epidemiological CM models.
2.6. Predict Epidemic Spreading in Real-World Social Networks
Bellingeri et al. [
30] investigated the effect of the network structure on the spread of the epidemic. They simulate SIR spreading over a dataset of 50 real-world complex systems from different fields of science.
To model the effect of the network structure on the epidemic spreading, the authors considered 40 different network structure indexes (NSIs) to test which were the best predictors of the SIR model epidemic spreading. The NSIs covered the relevant network structural features, such as community structure, link density, node distance, node assortativity, etc.
They found that the ‘
average node distance’, or a derived notion such as the “average normalized node closeness”, are the best predictors of the initial spreading pace. The ‘distance’
between nodes
and
is the minimum length of a path joining them, i.e., the minimum number of links to travel between the nodes. The average node distance for undirected networks is defined as:
The average node distance
measures the mean number of links to travel along the shortest path among node pairs in the network [
31]. The authors find that the higher
, the lower the spreading pace.
Further, indexes of ‘topological complexity’ of the network that consider both the node degree and the node distance are the best predictors of both the epidemic peak’s value and the spreading’s final extent. The
index, as the ratio of the average node degree
(i.e., the average number of links per node) and the average node distance
, was introduced in mathematical graph theory to evaluate the topological complexity of the network [
32]. The
index, which is a derivation of the
index using the farness at the place of the node distance, produced the best fitting of the SIR epidemic peak and total number of infected individuals at the end of the epidemic.
Very important, Bellingeri et colleagues [
30] research outlines that the most usual NSI evaluating the connectivity level of the network, such as the average node degree
, returns a scarce prediction of the network spreading for all the three spreading indicators adopted in the study. Network structures with the same average node degree and, for this, the same connectivity level may show very different epidemic spreading paces (See
Figure 2D and 2E).
The authors point out that most of the non-pharmaceutical interventions (NPIs) implemented to curb the SARS-Cov2 epidemic follow the rationale of reducing social interactions, which is equivalent to decreasing the number of social network links. Nonetheless, the Bellingeri et al. [
30] study unveils that considering the distance among nodes is more important than focusing on their connectivity level to predict network spreading. These findings suggest that performing a reliable social network disease spreading model is necessary to account for the node distance. Therefore, implementing NPIs to space out the nodes/individuals, i.e., increasing the node distance in the network, would be a more effective strategy to halt the epidemic.