1. Introduction
The ovulation rate and litter size are genetically affected by many minor genes and also some major genes, called Fecundity (
Fec) genes [
1]. The bone morphogenetic protein receptor 1b (
BMPR1B) and growth differentiation factor 9 (
GDF9) located on ovine chromosomes 6 and 5, respectively, are two of the three major
Fec genes in the superfamily of transforming growth factor beta (TGF-β) superfamily.
The
FecB allele of the
BMPR1B gene is known to be the first major gene associated with prolificity in sheep, found in Booroola Merino breed [
2,
3,
4]. The A to G transition at nucleotide position 746 of the cDNA induces a nonconservative substitution of a glutamine with arginine at position 249 of the protein (Q249R). This mutation has been hypothesized to be a partial loss of function mutation that is less sensitive to the action of bone morphogenetic protein 4 (BMP-4) on the inhibition of progesterone production and proliferation of granulosa cells [
2,
5]. Enabling a higher sensitivity to follicular stimulating hormone (FSH) sensitivity [
6] and earlier acquisition of luteinizing hormone receptors (LH) and LH-induced responses in granulosa cells of antral follicles. The consequences are precocious follicular maturation, ovarian follicles maturing at much smaller diameters with each follicle containing fewer granulosa cells than a similar-sized follicle in the wild-type [
7].
The FecB mutation has been reported in many prolific sheep breeds distributed in several countries, including Australia, New Zealand, India, China, Indonesia and Iran [
8]. The weighted mean effects of
FecB were summarized as ewes with one copy (+/
B) increased the ovulation rate by +.26 ova (+0.8 to +2.0) and the litter size by +0.67 (+0.4 to +1.3) of the born lambs. However, the effect of a second copy (
B/
B) compared to wild-type (+/+) ewes was +3.61 for the ovulation rate and +0.77 for litter size [
9].
The GDF9 gene is associated with ovarian folliculogenesis through the organization and proliferation of the ovarian follicle component by an encoding factor that promotes the development of primordial follicles and stimulates the proliferation of granulosa cells. More than 14 positions of the
GDF9 gene mutation in sheep have been reported, but five positions are associated with ovulation rate and/or prolificacy which separate in some prolific sheep breeds. There are
FecGH (high fertility) in Belclare and Cambridge [
10],
FecGT (Thoka) in Thoka Cheviot sheep [
11],
FecGE (Embrapa) in Santa Inês [
12],
FecGV (Vecaria) in Ile de France breed [
13], and
FecGNWS or
FecGF (Norwegian White Sheep, NWS; Finnish Landrace sheep, F) [
14,
15].
The FecGE mutation was first found in the prolific Santa Inés sheep breed in Brazil. The transition from T to G at nucleotide position 1034 of cDNA represented a nonconservative amino acid change at position 345 from phenylalanine to cysteine (F345C). The ovulation rate of homozygous ewes increased by 65% and 82% compared to heterozygous ewes and wild-type ewes, respectively. The prolificacy of homozygous ewes increased by 23.6% and 57.5% compared to heterozygous ewes and wild-type ewes, respectively [
12]. Recently, the
FecGE mutation was also found in Mexican Pelibuey sheep [
16].
The number of sheep and sheep farms in Thailand has continuously increased, from 42,040 heads and 5,170 farms in 2013 to 126,836 heads and 8,472 farms in 2022 [
17]. Sheep farming in Thailand can be classified into breeder sheep farms, which mainly focus on breeding purebred meat-typed sheep such as Dorper, Santa Inês, and Katahdin, and commercial sheep farms for meat market, which raise mostly crossbred sheep. To increase the profit and production of sheep farms, increasing reproductive efficiency in sheep farms such as litter size (prolificacy) is important. To alleviate the inefficiency and long cycle length of traditional breeding, marker-assisted selection (MAS) for
Fec alleles through molecular genetic for genetic improvement of reproduction efficiency was used [
18].
The distribution of FecB in several breeds and countries of sheep, the raising of the Santa Inês sheep breed in Thailand and the determination to increase the size of the litter for the sheep farm led the study to examine the exhibit of FecB and FecGE in the sheep population of Thailand.
2. Materials and Methods
2.1. Animals and Sample Collection
454 blood samples of crossbred sheep were collected from 430 female and 24 male sheep, raised by 21 sheep farms located in four major sheep-raising provinces in Thailand. Kanchanaburi, Suphanburi, Nakhon Pathom, and Kamphaeng Phet. Approximately 3 to 6 milliliters of blood samples were collected from jugular vein from each animal using an aseptic technique, then transferred to an EDTA blood collection tube and stored in an ice box during transport to the laboratory. The samples were stored in a -20°C refrigerator until DNA extraction and PCR-RFLP were performed.
2.2. DNA Extraction
DNA extraction from white blood cells from blood sample was performed with the proteinase-K, silica-based membrane column method using the FavorPrepTM tissue genomic DNA extraction mini kit according to the manufacturer's instruction protocol.
2.3. Primers Designing
The PCR-RFLP primer set for the identification of
FecB has been cited by Davis et al. [
19]. For
FecGE, the set of primers to differentiate the FecG
E genotype was designed using the website
http://primer1.soton.ac.uk/primer1.html and then the outer set selected which can be used for PCR-RFLP (
Table 1). The
GDF9 ovine gene was recovered from the NCBI database (accession number NM_001142888.2).
2.4. Polymerase Chain Reaction (PCR) Condition
Each PCR reaction was performed in total volume 20 µl containing 2 µl of 10X PCR buffer, 200 µM of each dNTP, 0.5 µM of each primer, 50 – 100 ng of ovine genomic DNA template and 0.5 U of DreamTaq DNA Polymerase (Thermo Scientific™). Optimized annealing conditions for both FecB and FecGE primers were determined by gradient PCR. PCR was performed with the following condition: Initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing for 30 s at 60°C for FecB or 52°C for FecGE, extension at 72°C for 30 s for FecB or 60 s for FecGE, and final extension at 72°C for 5 min.
2.5. Restriction Fragment Length Polymorphism (RFLP) and Gel Electrophoresis
FastDigest AvaII Enzyme (Themo Scientific™) and FastDigest TscAI Enzyme (Themo Scientific™) were used to differentiate genotypes of FecB and FecGE genotypes, respectively. Follow the protocol according to the manufacturer's instructions. Samples containing FecB mutation will be digested into 30 and 160 bp fragments, whereas non-carrier products remained uncut at 190 bp. For FecGE, the TspRI enzyme will digest the FecGE PCR product samples of non-carrier sheep into two fragments of 42 bp and 415 bp, while the samples containing the FecGE mutation will be digested into three fragments of 42, 173 and 242 bp. All digested RFLP products were electrophoresed by running 10 µl of the product through 2% agarose gel stained with RedSafeTM nucleic acid staining solution at 100 volt for 20 minutes. Fragment specific sizes were distinguished using the ExcelBandTM 100 bp+3K DNA ladder. The gel was visualized under ultraviolet (UV) light and imaged to differentiate the genotype of the samples by using the GelDoc Go imaging system (Bio-Rad Laboratories, Inc.).
2.6. Direct DNA Sequencing of PCR Products
To verify the identified polymorphisms from PCR-RFLP, some
FecB and
FecGE carrier PCR product samples were submitted for DNA purification and DNA sequencing using the Sanger Method (Celemics, Inc., Seoul, South Korea) by U2Bio (Thailand) Co., Ltd. The nucleotide sequence data and the chromatogram were analyzed by Bioedit v7.2.5 [
20].
2.7. Data Collection and Statistical Analysis
The history of ewe birth types was collected from the owners to analyze the association between
Fec genotypes and the history of ewe birth types using Fisher’s exact test. The genotype distribution was analyzed using the Chi-square test to test the deviation from the Hardy-Weinberg equilibrium. All statistical analyzes were analyzed using the R-studio software version 4.1.3 [
21].
3. Results
3.1. PCR-RFLP Results of FecB and FecGE
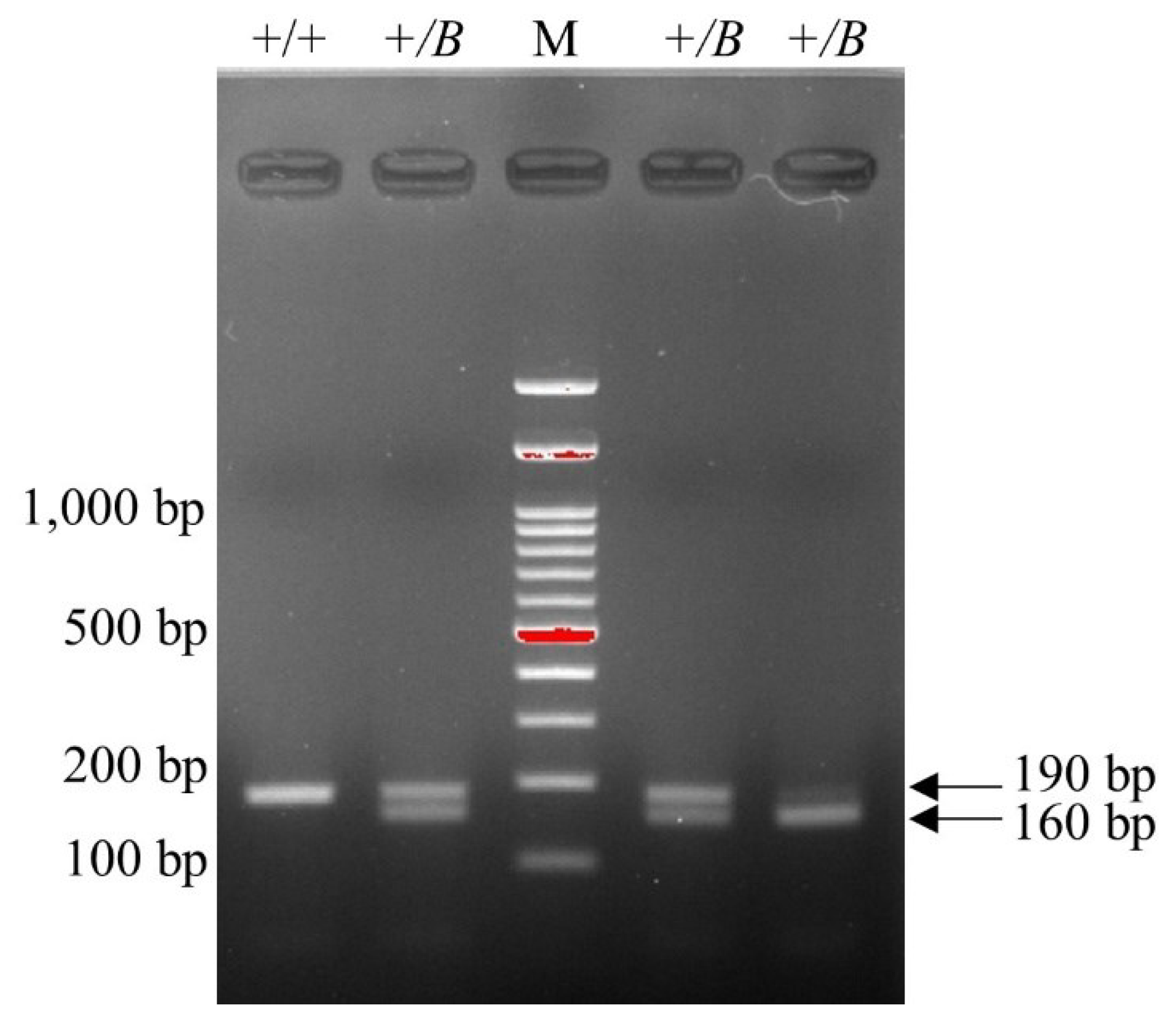
FecB-carrying samples displayed a 160 bp band, while noncarrier samples displayed a 190 bp band on gel electrophoresis after RFLP (
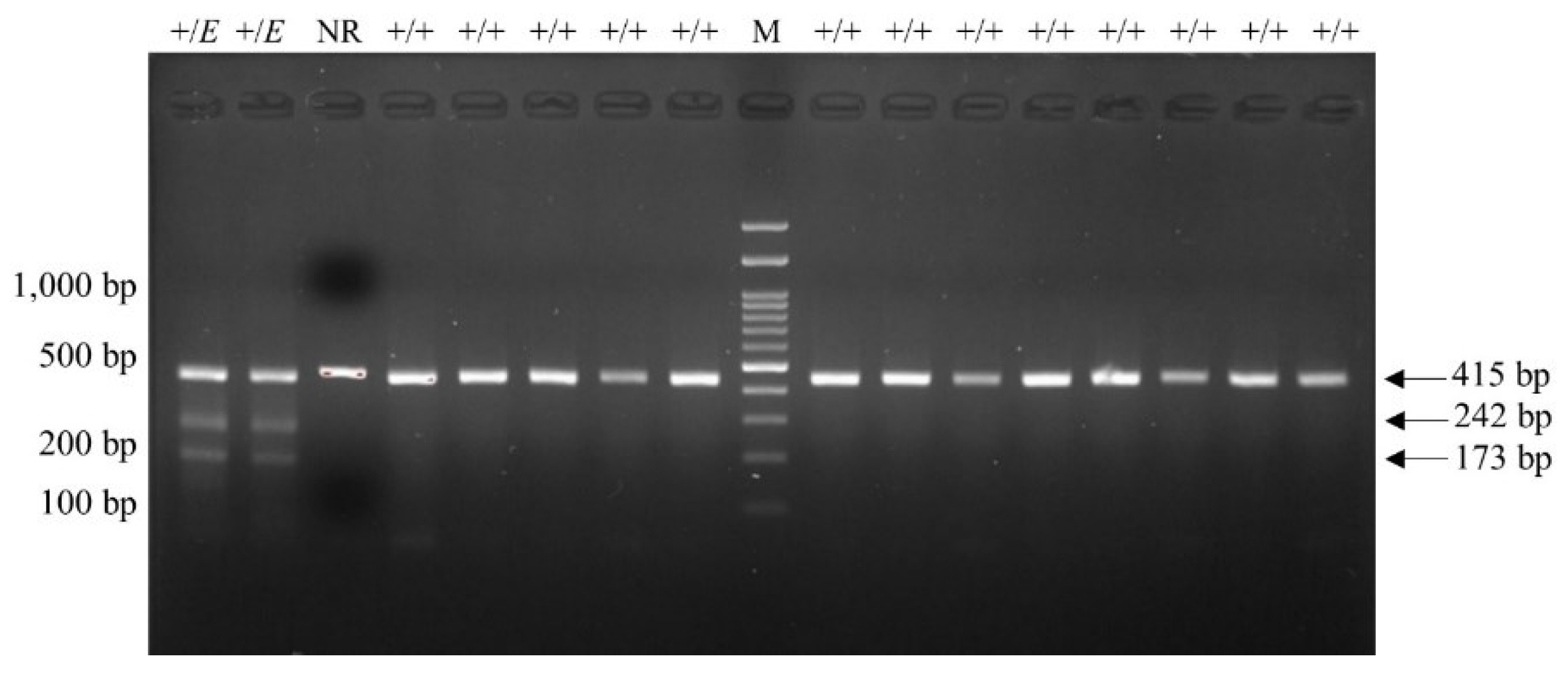
Figure 1). For FecG
E, carrying samples displayed 42, 173 and 242 bp bands on gel electrophoresis following RFLP, whereas non-carrier samples displayed 42 and 415 bp bands. (
Figure 2)
The genotypic and allele frequencies of FecB and FecG
E from the PCR-RFLP method are shown in
Table 2. The genotypic distribution of FecB deviated from the Hardy-Weinberg equilibrium (p<0.01). On the other hand, the genotypic distribution of FecG
E was in Hardy-Weinberg equilibrium (p=0.97). Furthermore, when combined the FecB and FecG
E genotypes, two of these ewes possess both heterozygous FecB (+/B) and heterozygous FecG
E (+/E) genotypes, and one of the ewes possesses both homozygous FecB (B/B) and heterozygous FecG
E (+/E) genotypes.
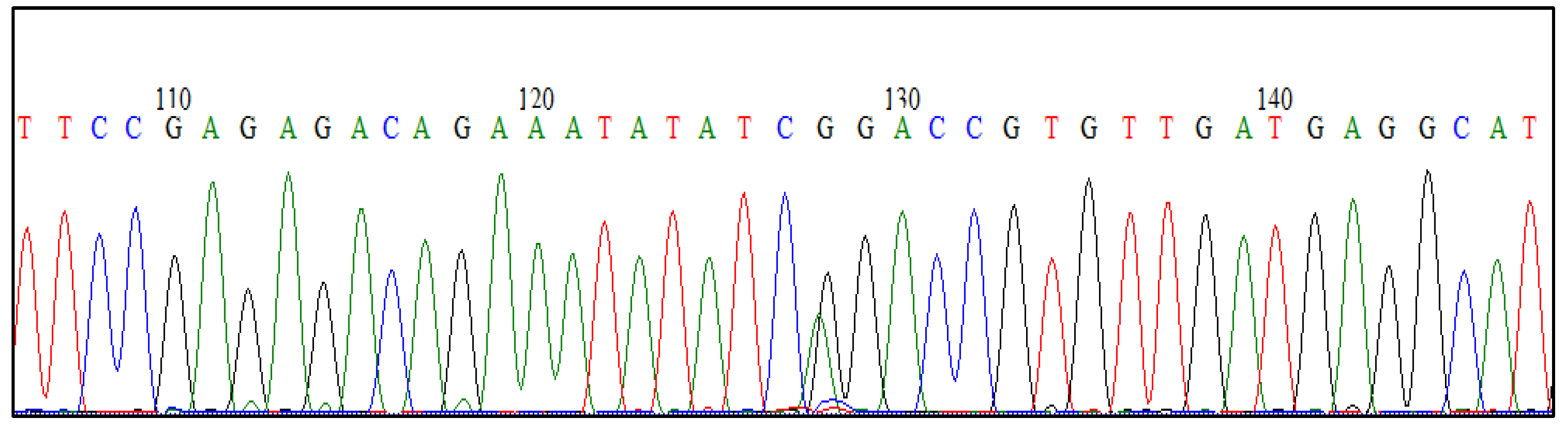
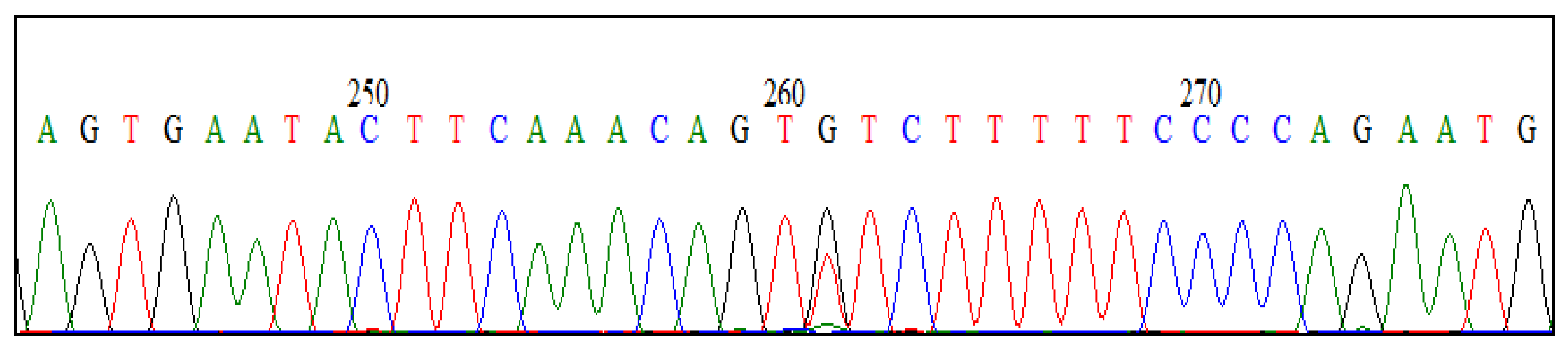
3.2. Direct Sequencing of PCR Products
Eight identified FecB carrier and three identified FecG
E carrier PCR products were submitted for direct DNA sequencing and the PCR-RFLP results were verified (
Figure 3 and
Figure 4).
3.3. History of Birth Types of Ewe and Association between Fec Genotypes
The history of ewe birth types was derived from owners, but litter size records of all parity were obtained from only one farm in Kamphaeng Phet Province. The history of Ewe birth types classified by combined FecB and FecG
E genotypes is shown in
Table 3.
To examine the association between individual Fec allele and birth type history, data regarding another Fec allele were excluded to separate the influence of Fec alleles. Statistical analysis using Fisher’s exact test (
Table 4) revealed a statistically significant association between heterozygous FecB genotypes (+/B) and birth type history (p<0.01). On the contrary, there was no association between heterozygous FecG
E genotypes (+/E) and birth-type history (p>0.05).
4. Discussion
The PCR-RFLP technique is one of the conventional methods for genotyping single nucleotide polymorphisms (SNPs). The PCR-RFLP technique to genotype
FecB was first developed by Wilson et al. [
3]. The alternative primer set for amplifying the 190 bp PCR product was first used by Devis et al. [
19] and subsequently used in several studies. The DNA sequencing results verified the competence of this protocol similar to previous studies. For
FecGE, a new alternative set of PCR-RFLP primers was designed and can be used to differentiate the
FecGE genotype.
The possible origin of the
FecB allele in crossbred sheep in Thailand could be attributed to two primary causes: the introduction of discharged imported Merino sheep from tourist attractions, and the transportation and trafficking of sheep among Myanmar, China, and Thailand. This later activity could have led to the intermingling of
FecB-containing ewes from India or China with the local sheep population in Thailand. However, the potential origin of the
FecGE allele in crossbred sheep in Thailand could possibly be related to the introduction of Brazilian Santa Inês sheep since 1997 [
22], which were imported into breeder farms. It should be noted that some of these imported sheep possessed
FecGE. Subsequently, these imported sheep were marketed as breeders and bred with the local sheep population in Thailand.
The genotypic distribution of
FecB in our study resembled several breeds such as Banyabulak [
23], Wadi [
24], Lori [
25], Zandi [
8], and Poll Dorset [
26]. Our study found that almost
FecB-carrier ewes exhibited a previous record of multiple births of lambs. This finding corresponds to the summary from Potki et al. [
8] that all breeds carrying the
FecB allele showed a significant relationship between that allele and higher fertility traits, except the Bonapala breed from India. The average effect of
FecB on litter size could not be determined in our study due to the limited number of
FecB carrier ewes identified and limitation of data recording. However, the mean litter size of heterozygous
FecB (+/
B) ewe recorded on the farm in Kamphaeng Phet province was 1.33 which was lower than any previous reports. The lower twinning rate of crossbred ewes in Thailand could be due to ewes' undernutrition, resulting from lower feed quality and poor feed management practices, particularly in free-range commercial sheep farms.
The result that there was no association between
FecGE genotypes and birth type history (p=0.098) in this study corresponds to the result from Silva et al. [
12] which found that only homozygous
FecGE group (
E/
E, n=9) showed a genotype effect on the frequency of twinning per ewe (44%) (p=0.014). The heterozygous
FecGE group (+/E, n = 15) did not show differences in the frequency of twinning (14%) compared to wild-type ewes (-/-, n=15, 0%). Furthermore, they also found that the heterozygous
FecGE group (+ / E) did not present differences (p = 0.612) in the average number of CL (1.34±0.08) or in the frequency of ewes with multiple ovulations (31.8%), compared to wild-type ewes (1.22±0.11 and 14.6%, respectively). However, they found the difference (p<0.001) of the mean prolificacy of the Santa Inês flock not selected for F1 between the heterozygous
FecGE group (
+/
E, n=102) and the wild type group (+/+, n=219) which is 1.44 and 1.13, respectively. Therefore, it was possible that the absence of differences in the frequency of twinning between heterozygous and wild-type ewes in this study might be due to the small sample sizes.
From our findings, three ewes possessed FecB and FecGE carriers were discovered in one farm located in Kanchanaburi province. This finding suggests that both FecB and FecGE alleles are intermingled with cross-bred sheep in Thailand. The coexistence of FecB and FecGE alleles might potentially increase the ovulation rate together. However, there was no previous report on the influence of coexisting FecB and FecGE alleles on ovulation rate.
5. Conclusions
In this study, both FecB and FecGE mutations were identified in crossbred sheep in Thailand. The presence of the FecB allele was associated with multiple birth of lambs, which was consistent with several previous reports. Therefore, sheep that contain the FecB allele would be a better choice to improve the prolificacy of crossbred sheep in Thailand more than FecGE. The discovery of FecB and FecGE carrier sheep suggested that some of the sheep population in Thailand may have crossed over with the Booroola Merino breed or other previously reported FecB containing sheep, as well as the Santa Inês breed for FecGE. The coexistence of the FecB and FecGE allele mutations was also discovered and should be studied for the effect on ovulation rate and prolificacy in future research.
Author Contributions
Conceptualization, P.S., S.T. and T.R.; methodology, P.S., S.T.; formal analysis, P.S..; writing—original draft preparation, P.S., S.T.; writing—review and editing, T.R..; visualization, P.S..; supervision, T.R.; project administration, P.S.; funding acquisition, T.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research and the APC was funded by Faculty of Veterinary Medicine, Kasetsart University.
Institutional Review Board Statement
The present study was conducted under animal care approved by the Institutional Animal Care and Use Committee in accordance to the guidelines of animal care and use under the Ethics Board of the Office of National Research Council of Thailand (NRCT) for the conduction of the scientific research. The approved number was ACKU66-VET-058.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.
Acknowledgments
This work was partially funded by the Faculty of Veterinary Medicine, Kasetsart University. The authors also thank Kasetsart University Veterinary Teaching Hospital, Kamphaeng Saen Campus, and the Molecular Laboratory Unit, Kamphaeng Saen Veterinary Diagnostic Center (KVDC) for providing laboratory facilities for this research.
Conflicts of Interest
The authors declare that there is no conflict of interest with any financial organization with respect to the material discussed in the manuscript.
References
- Drouilhet, L.; Lecerf, F.; Bodin, L.; Fabre, S. Fine mapping of the FecL locus influencing prolificacy in Lacaune sheep. Anim. Genet., 2009, 40, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Mulsant, P.; Lecerf, F.; Fabre, S.; Schibler, L.; Monget, P.; Lanneluc, I.; Pisselet, C.; Riquet, J.; Monniaus, D.; Callebaut, I.; Cribiu, E.; Thimonier, J.; Teyssier, J.; Bodin, L.; Cognie, Y.; Chitour, N.; Elsen, J.M. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Mérino ewes. Proc. Natl. Acad. Sci. USA, 2001, 98, 5104–5109. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.; Wu, X.Y.; Juengel, J.L.; Ross, I.K.; Lumsden, J.M.; Lord, E.A.; Dodds, K.G.; Walling, G.A.; McEwan, J.C.; O’Connell, A.R.; McNatty, K.P.; Montgomery, G.W. Highly Prolific Booroola Sheep Have a Mutation in the Intracellular Kinase Domain of Bone Morphogenetic Protein IB Receptor (ALK-6) That Is Expressed in Both Oocytes and Granulosa Cells1. Biol. Reprod., 2001, 64, 1225–1235. [Google Scholar] [CrossRef]
- Davis, G.H.; Montgomery, G.W.; Allison, A.J.; Kelly, R.W.; Bray, A.R. Segregation of a major gene influencing fecundity in progeny of Booroola sheep. New Zealand J. Agric. Res., 1982, 25, 525–529. [Google Scholar] [CrossRef]
- Fabre, S.; Pierre, A.; Pisselet, C.; Mulsant, P.; Lecerf, F.; Pohl, J.; Monget, P.; Monniaux, D. The Booroola mutation in sheep is associated with an alteration of the bone morphogenetic protein receptor-IB functionality. J. Endocrinol., 2003, 177, 435–444. [Google Scholar] [CrossRef]
- Montgomery, G.W.; McNatty, K.P.; Davis, G.H. Physiology and molecular genetics of mutations that increase ovulation rate in sheep. Endocr. Rev., 1992, 13, 309–28. [Google Scholar] [CrossRef]
- McNatty, K.P.; Henderson, K.M. , Gonadotrophins, fecundity genes and ovarian follicular function. J. Steroid Biochem., 1987, 27, 365–73. [Google Scholar] [CrossRef] [PubMed]
- Potki, P.; Mirhoseini, S.Z.; Afraz, F.; Vahidi, S.M.F. A Profile of Single Nucleotide Polymorphisms in Fecundity Genes Among Iranian Sheep Breeds by Using Polymerase Chain Reaction–Restriction Fragment Length Polymorphism (PCR-RFLP) Method. Iranian J. Applied Anim. Sci., 2020, 10, 265–285. [Google Scholar]
- Fogarty, N. A review of the effects of the Booroola gene (FecB) on sheep production. Small Rumin. Res., 2009, 85, 75–84. [Google Scholar] [CrossRef]
- Hanrahan, J.P.; Gregan, S.M.; Mulsant, P.; Mullen, M.; Davis, G.H.; Powell, R.; Galloway, S.M. Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries). Biol. Reprod., 2004, 70, 900–909. [Google Scholar] [CrossRef]
- Nicol, L.; Bishop, S.C.; Pong-Wong, R.; Bendixen, C.; Holm, L.E.; Rhind, S.M.; McNeilly, A.S. Homozygosity for a single base-pair mutation in the oocyte-specific GDF9 gene results in sterility in Thoka sheep. Reproduction, 2009, 138, 921–933. [Google Scholar] [CrossRef]
- Silva, B.D.M.; Castro, E.A.; Souza, C.J.H.; Paiva, S.R.; Sartori, R.; Franco, M.M.; Azevedo, H.C.; Silva, T.A.S.N.; Vieira, A.M.C.; Neves, J.P.; Melo, E.O. A new polymorphism in the Growth and Differentiation Factor 9 (GDF9) gene is associated with increased ovulation rate and prolificacy in homozygous sheep. Anim. Genet., 2011, 42, 89–92. [Google Scholar] [CrossRef]
- Souza, C.J.H.; McNeilly, A.S.; Benavides, M.V.; Melo, E.O.; Moraes, J.C.F. Mutation in the protease cleavage site of GDF9 increases ovulation rate and litter size in heterozygous ewes and causes infertility in homozygous ewes. Anim. Genet., 2014, 45, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Mullen, M.P.; Hanrahan, J.P. Direct Evidence on the Contribution of a Missense Mutation in GDF9 to Variation in Ovulation Rate of Finnsheep. PLOS ONE, 2014, 9, e95251. [Google Scholar] [CrossRef] [PubMed]
- Våge, D.I.; Husdal, M.; Kent, M.P.; Klemetsdal, G.; Boman, I.A. A missense mutation in growth differentiation factor 9 (GDF9) is strongly associated with litter size in sheep. BMC Genetics, 2013, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ruiz, E.; Gallegos-Sánchez, J.; Cortez-Romero, C.; Segura-León; Salinas-Ruíz, J.; Salazar-Ortiz, J. FecGE mutation in pelibuey sheep. Anim. Genet., 2020, 51, 346–347. [Google Scholar] [CrossRef] [PubMed]
- DLD. National Animal Statistics. 2022. Available online: http://ict.dld.go.th/webnew/index.php/th/service-ict/report/247-report-thailand-livestock.
- Abdoli, R.; Zamani, P.; Deljou, A.; Rezvan, H. Association of BMPR-1B and GDF9 genes polymorphisms and secondary protein structure changes with reproduction traits in Mehraban ewes. Gene, 2013, 524, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.H.; Galloway, S.M.; Ross, I.K.; Gregan, S.M.; Ward, J.; Nimbkar, B.V.; Ghalsasi, P.M.; Nimbkar, C.; Grey, G.D.; Subandriyo; Inoumu, I.; Tiesnamurti, B.; Martyniuk, E.; Eythorsdottir, E.; Mulsant, P.; Lecerf, F.; Hanrahan, J.P.; Bradford, G.E.; Wilson, T. DNA tests in prolific sheep from eight countries provide new evidence on origin of the Booroola (FecB) mutation. Biol. Reprod., 2002, 66, 1869–1874. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser., 1999, 41, 95–98. [Google Scholar]
- Team, R., RStudio: Integrated Development Environment for R. 2022, RStudio, PBC: Boston, MA.
- DLD, Sheep Farming. 1 ed. Guide Book, DLD, Ministry of Agriculture and Cooperatives, ed. S. Banlengthong. 2548 (2005 A.D.), The Agricultural Cooperative Federation of Thailand., Limited, Bangkok: Publishing and Public Relations Group, Division of Livestock Development and Technology Transfer, DLD, Bangkok 36.
- Zuo, B.; Qian, H.; Wang, Z.; Wang, X.; Nisa, N.; Bayier, A.; Ying, S.; Hu, X.; Gong, C.; Guo, Z.; Wang, F. A Study on BMPR-IB Genes of Bayanbulak Sheep. Asian-Australas. J. Anim. Sci. 2013, 26, 36–42. [Google Scholar] [CrossRef]
- Zhang, C.S.; Geng, L.Y.; Du, L.X.; Liu, X.Z.; Fu, Z.X.; Feng, M.S.; Gong, Y.F. Polymorphic Study of FecXG, FecGH and FecB Mutations in Four Domestic Sheep Breeds in the Lower Yellow River Valley of China. J. Anim. Vet. Adv, 2011, 10, 2198–2201. [Google Scholar] [CrossRef]
- Nanekarani, S.; Goodarzi, M.; Khederzadeh, S. Detection of polymorphism in booroola gene and growth differentiation factor 9 in Lori sheep breed. Trop. J. Pharm. Res., 2016, 15, 1605–16011. [Google Scholar] [CrossRef]
- Jia, C.; Li, N.; Zhao, X.; Zhu, X.; Jia, Z. Association of Single Nucleotide Polymorphisms in Exon 6 Region of BMPRIB Gene with Litter Size Traits in Sheep. Asian-Australas. J. Anim. Sci., 2005, 18, 1375–1378. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).