Submitted:
08 February 2024
Posted:
09 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
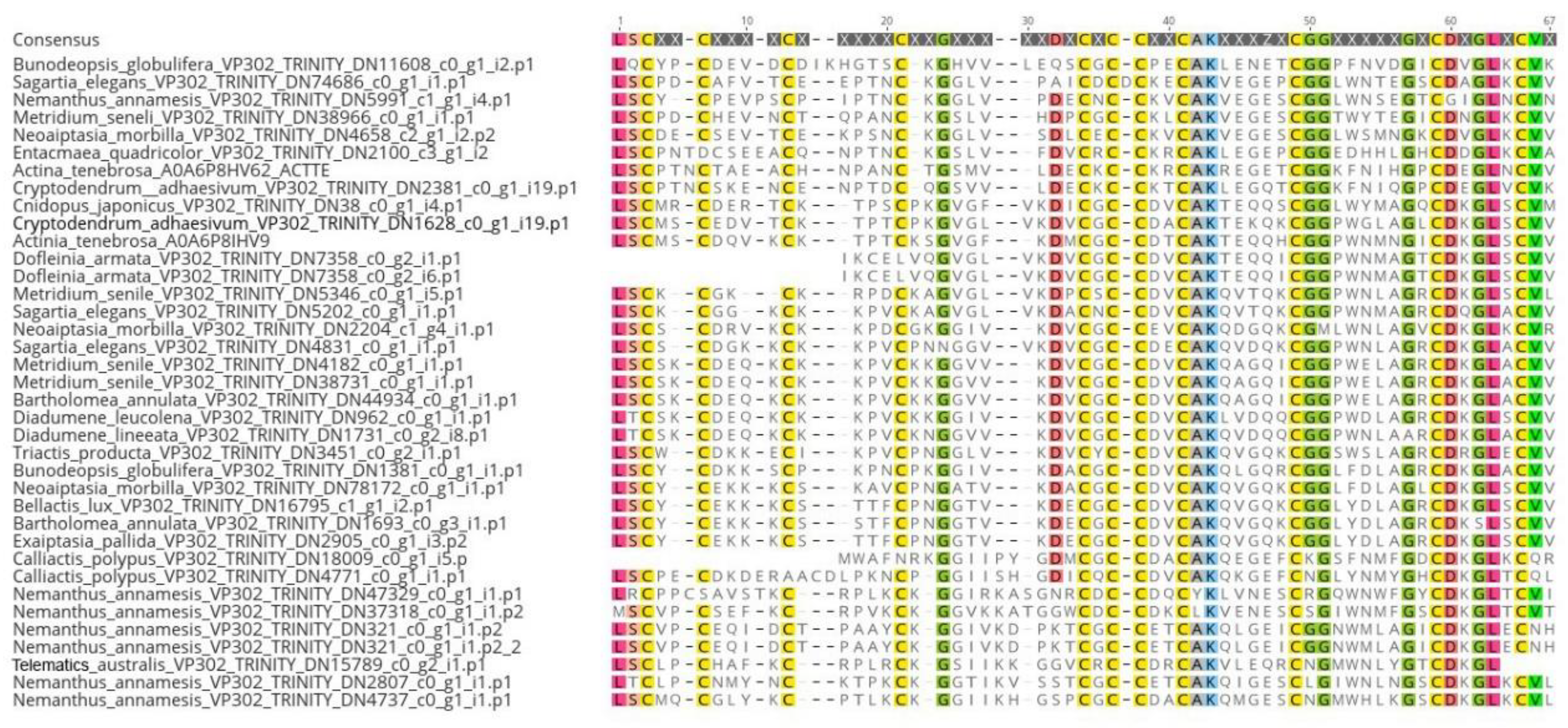
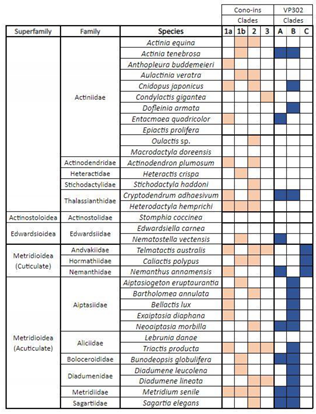
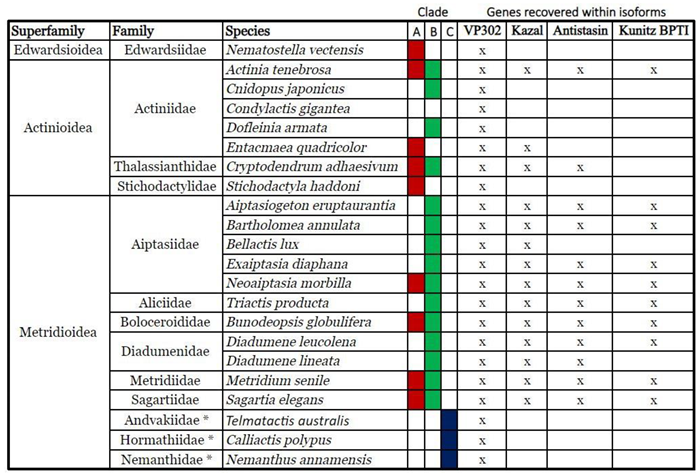
2.1. Identification
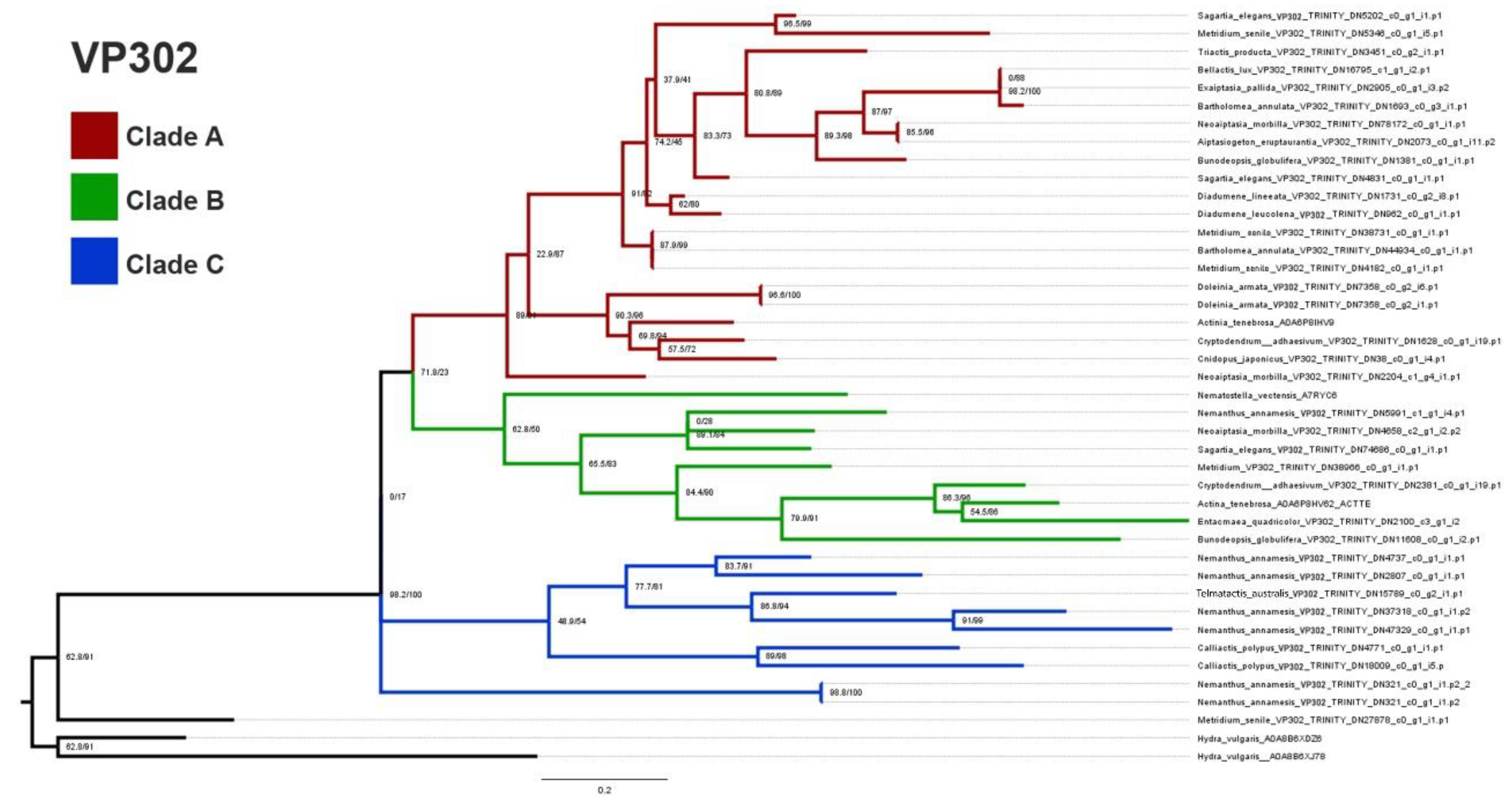
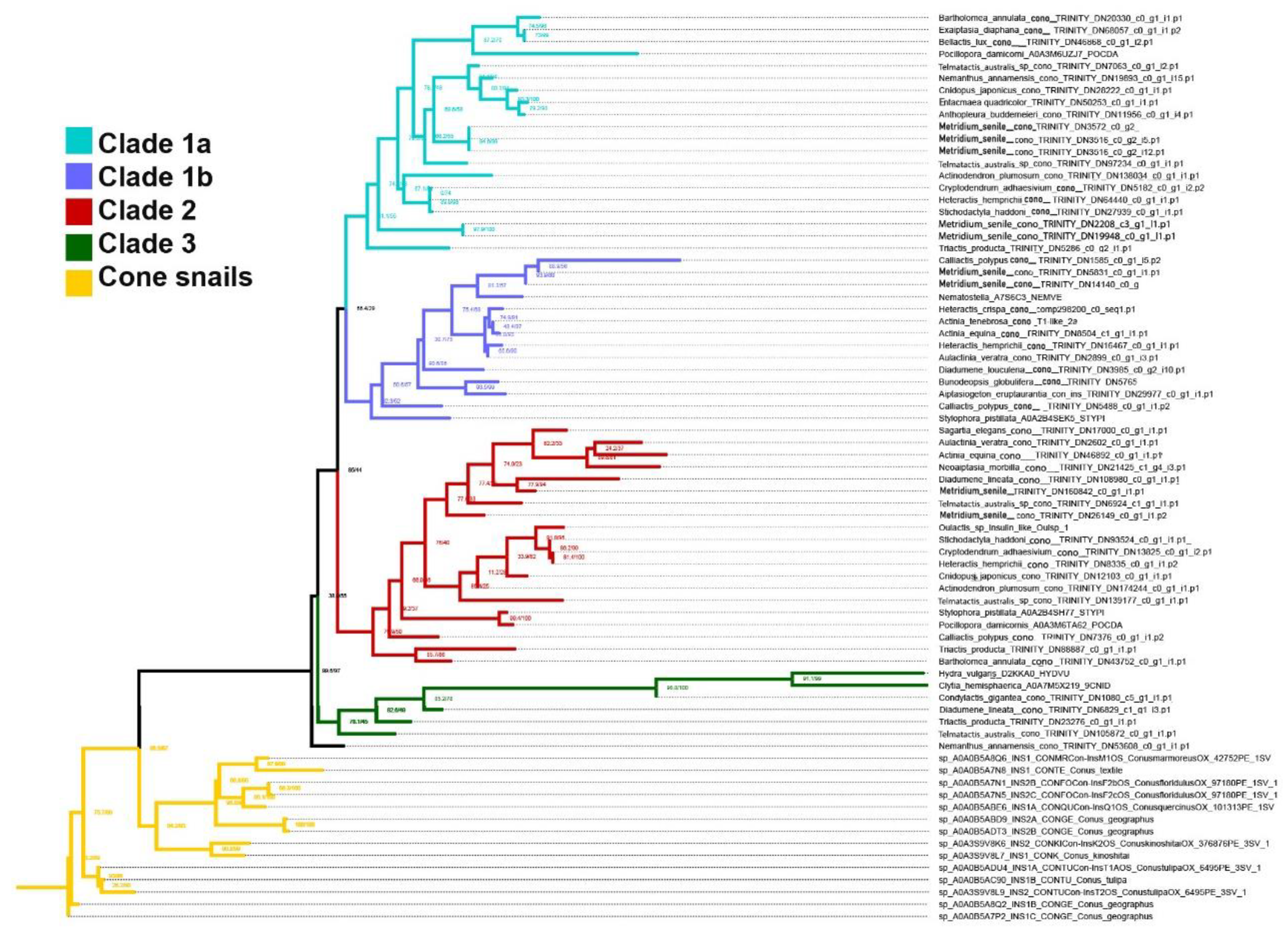
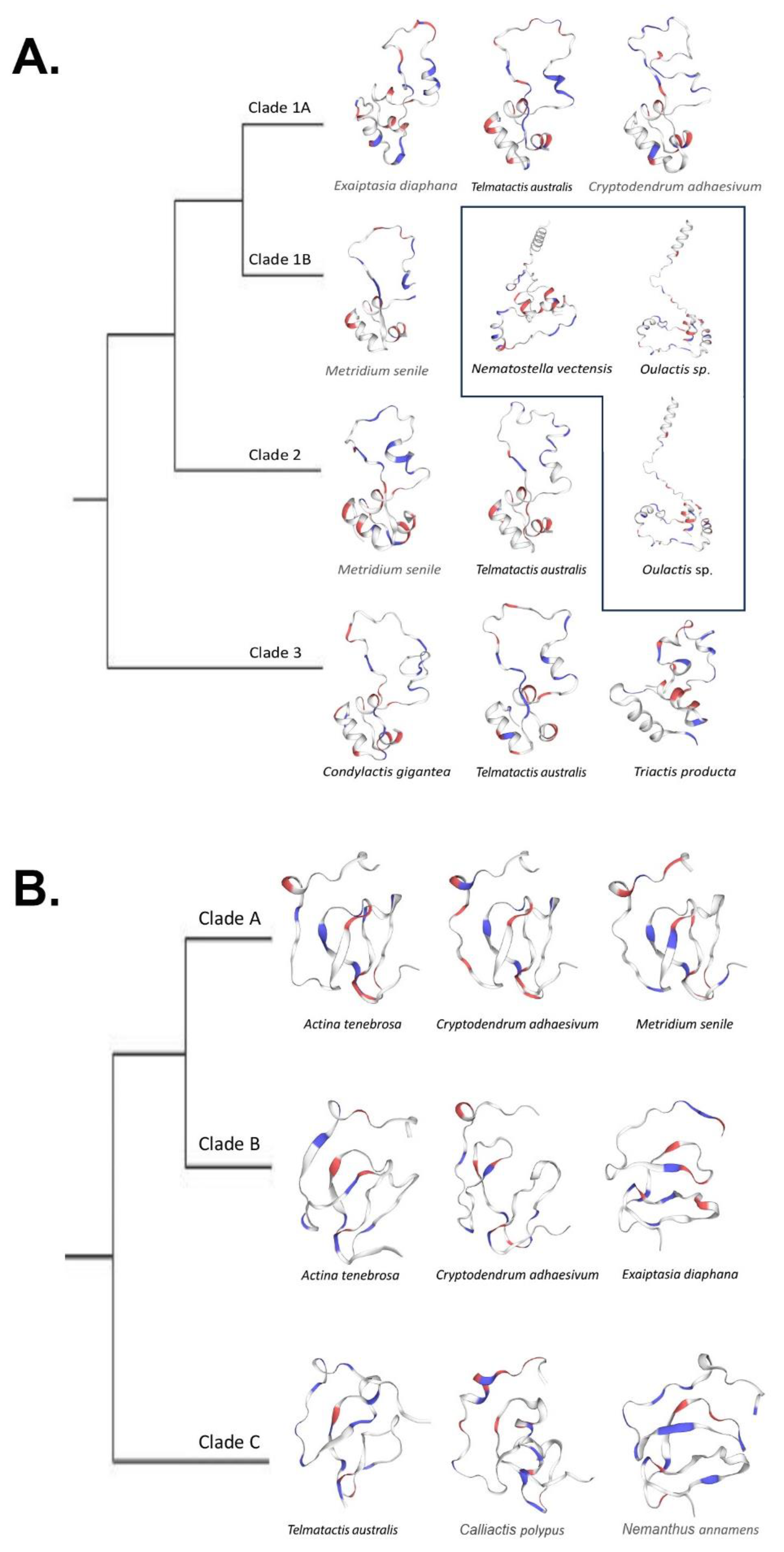
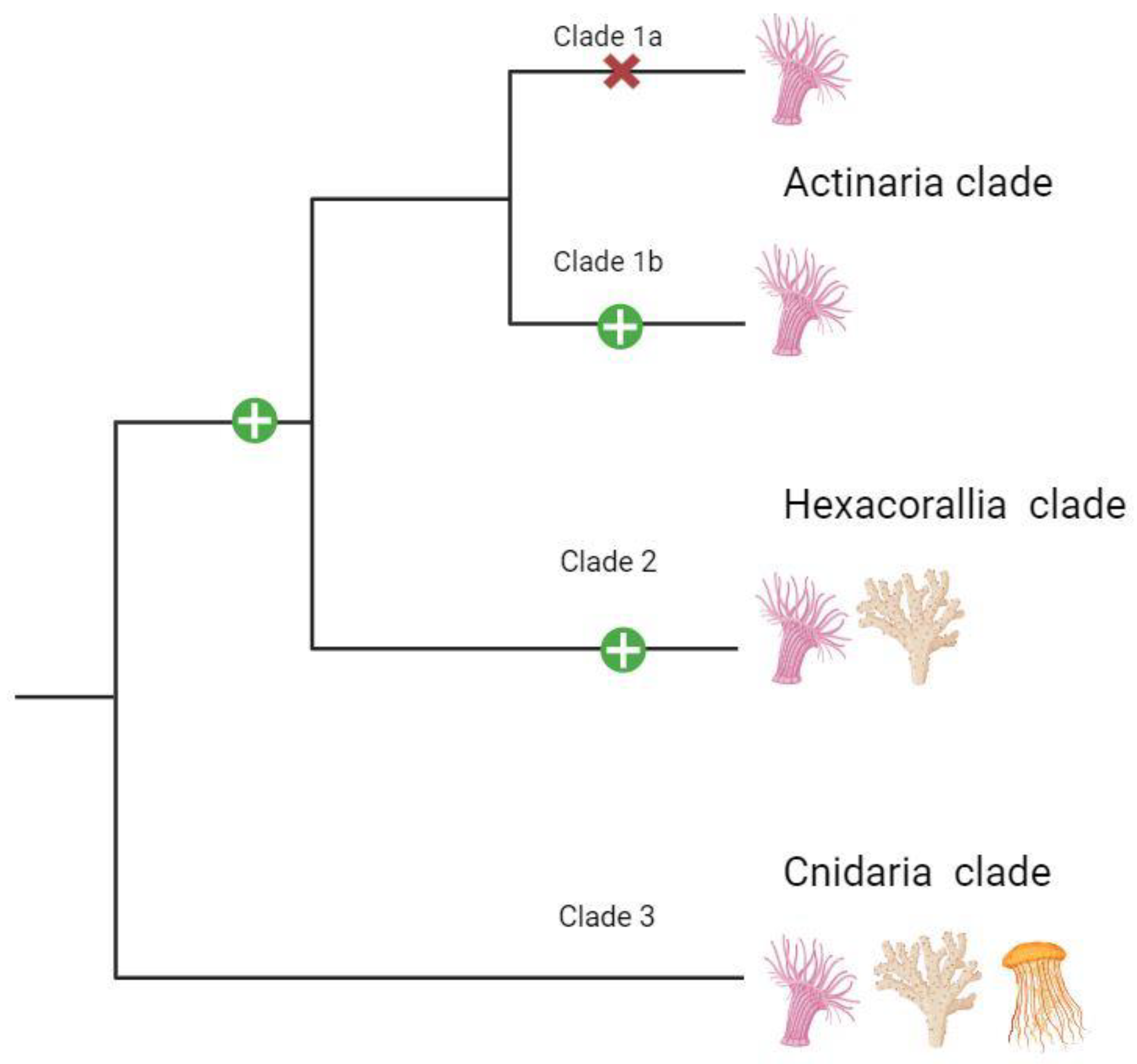
2.2. Phylogenomics
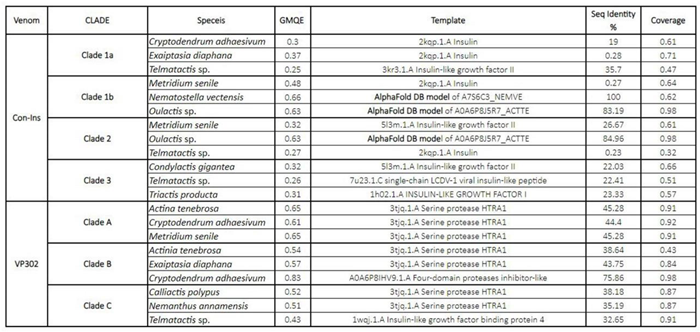
2.3. Predicted Protein Structure
3. Discussion
3.1. ILPs in Sea Anemones
3.2. Phylogenetics
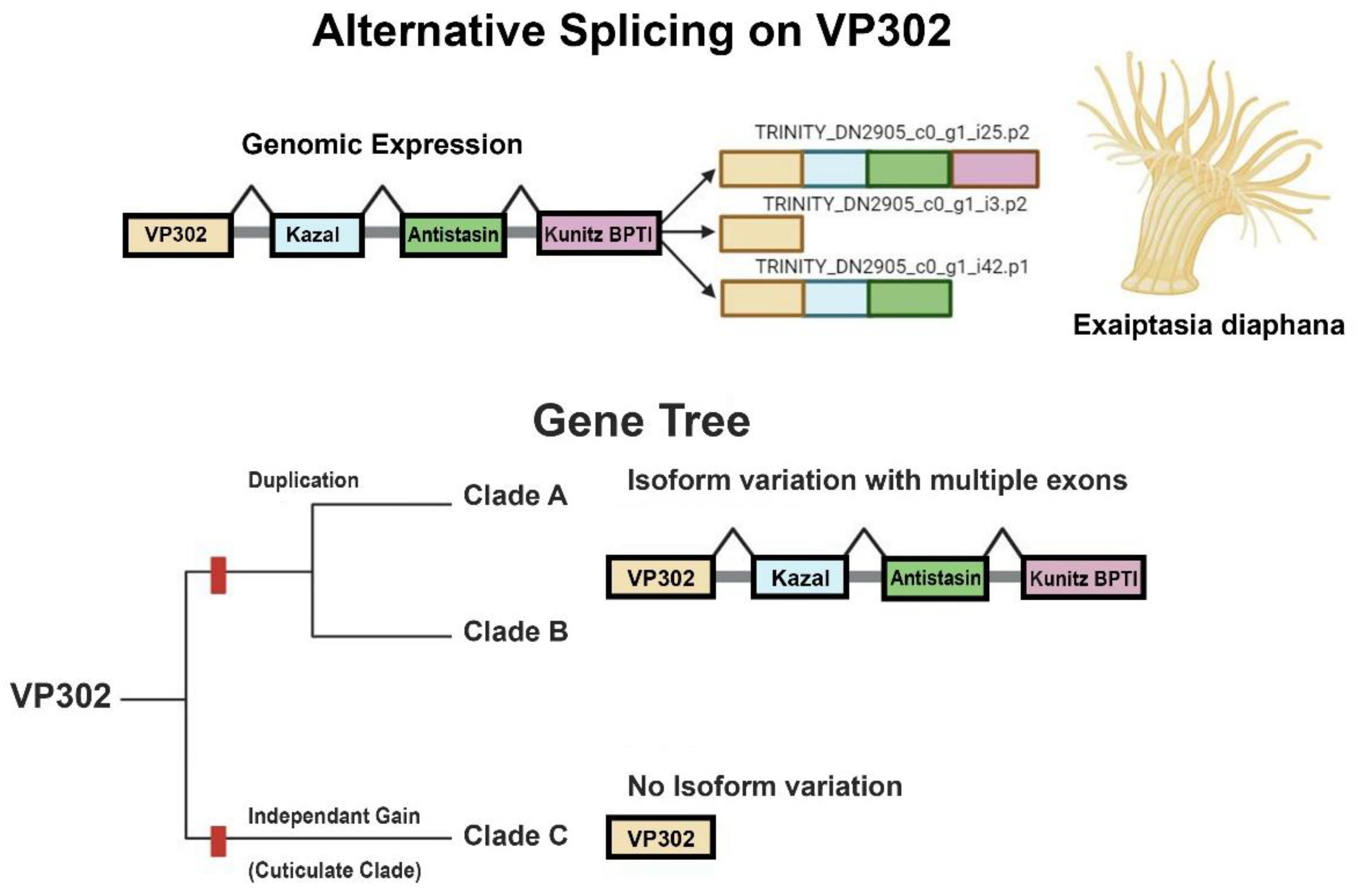
3.3. Alternative Splicing
3.4. Structural Predictions
- Selection analysis
3.5. Sites under Selection—A Scaling Issue?
4. Materials and Methods
4.1. Annotation/Identification
4.2. Phylogenomic Analyses
4.3. Structural Predictions
4.4. Mode of Selecetion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
References
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Muscatine, L. Coelenterate Biology: Reviews and New Perspectives; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Chintiroglou, C.C.; Doumenc, D.; Zamponi, M. Commented list of the Mediterranean Actiniaria and Corallimorpharia (Anthozoa). Acta Adriat. 1997, 38, 65–70. [Google Scholar]
- Daly, M. Functional and genetic diversity of toxins in sea anemones. In Evolution of Venomous Animals and Their Toxins; Malhotra, A., Gopalakrishnakone, P., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 87–104. [Google Scholar] [CrossRef]
- Madio, B.; Undheim, E.A.; King, G.F. Revisiting venom of the sea anemone Stichodactyla haddoni: Omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J. Proteom. 2017, 166, 83–92. [Google Scholar] [CrossRef]
- Macrander, J.; Broe, M.; Daly, M. Tissue-specific venom composition and differential gene expression in sea anemones. Genome Biol. Evol. 2016, 8, 2358–2375. [Google Scholar] [CrossRef] [PubMed]
- Ashwood, L.M.; Norton, R.S.; Undheim, E.A.B.; Hurwood, D.A.; Prentis, P.J. Characterising functional venom profiles of anthozoans and medusozoans within their ecological context. Mar. Drugs 2020, 18, 202. [Google Scholar] [CrossRef]
- Ashwood, L.M.; Undheim, E.A.B.; Madio, B.; Hamilton, B.R.; Daly, M.; Hurwood, D.A.; King, G.F.; Prentis, P.J. Venoms for all occasions: The functional toxin profiles of different anatomical regions in sea anemones are related to their ecological function. Mol. Ecol. 2021, 31, 866–883. [Google Scholar] [CrossRef] [PubMed]
- Delgado, A.; Benedict, C.; Macrander, J.; Daly, M. Never, ever make an enemy… out of an anemone: transcriptomic comparison of clownfish hosting sea anemone venoms. Mar. Drugs 2022, 20, 730. [Google Scholar] [CrossRef]
- Frazão, B.; Vasconcelos, V.; Antunes, A. Sea Anemone (Cnidaria, Anthozoa, Actiniaria) toxins: An overview. Mar. Drugs 2012, 10, 1812–1851. [Google Scholar] [CrossRef]
- Menezes, C.; Thakur, N.L. Sea anemone venom: Ecological interactions and bioactive potential. Toxicon 2022, 208, 31–46. [Google Scholar] [CrossRef]
- Smith, E.G.; Surm, J.M.; Macrander, J.; Simhi, A.; Amir, G.; Sachkova, M.Y.; Lewandowska, M.; Reitzel, A.M.; Moran, Y. Micro and macroevolution of sea anemone venom phenotype. Nat. Commun. 2023, 14, 249. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, M.-C.; Konaté, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or normalized counts? a comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef]
- Sunagar, K.; Moran, Y. The rise and fall of an evolutionary innovation: Contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 2015, 11, e1005596. [Google Scholar] [CrossRef]
- Jouiaei, M.; Sunagar, K.; Gross, A.F.; Scheib, H.; Alewood, P.F.; Moran, Y.; Fry, B.G. Evolution of an ancient venom: Recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemone. Mol. Biol. Evol. 2015, 32, 1598–1610. [Google Scholar] [CrossRef]
- Gibbs, H.L.; Rossiter, W. Rapid evolution by positive selection and gene gain and loss: PLA2 venom genes in closely related SISTRURUS rattlesnakes with divergent diets. J. Mol. Evol. 2008, 66, 151–166. [Google Scholar] [CrossRef]
- Modica, M.V.; Gorson, J.; E Fedosov, A.; Malcolm, G.; Terryn, Y.; Puillandre, N.; Holford, M. Macroevolutionary analyses suggest that environmental factors, not venom apparatus, play key role in terebridae marine snail diversification. Syst. Biol. 2020, 69, 413–430. [Google Scholar] [CrossRef]
- Binford, G.; Wells, M. The phylogenetic distribution of sphingomyelinase D activity in venoms of Haplogyne spiders. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2003, 135, 25–33. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Rodríguez, E.; Faircloth, B.C.; Cowman, P.F.; Brugler, M.R.; Farfan, G.A.; Hellberg, M.E.; Kitahara, M.V.; Morrison, C.L.; Paz-García, D.A.; et al. Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nat. Ecol. Evol. 2020, 4, 1531–1538. [Google Scholar] [CrossRef]
- Sachkova, M.Y.; Macrander, J.; Surm, J.M.; Aharoni, R.; Menard-Harvey, S.S.; Klock, A.; Leach, W.B.; Reitzel, A.M.; Moran, Y. Some like it hot: population-specific adaptations in venom production to abiotic stressors in a widely distributed cnidarian. BMC Biol. 2020, 18, 121. [Google Scholar] [CrossRef]
- Sachkova, M.Y.; A Singer, S.; Macrander, J.; Reitzel, A.M.; Peigneur, S.; Tytgat, J.; Moran, Y. The birth and death of toxins with distinct functions: A case study in the sea anemone Nematostella. Mol. Biol. Evol. 2019, 36, 2001–2012. [Google Scholar] [CrossRef]
- J. Prentis, P.; Pavasovic, A.; S. Norton, R. Sea Anemones: Quiet Achievers in the Field of Peptide Toxins. Toxins 2018, 10, 36. [Google Scholar] [CrossRef]
- Escoubas, P.; King, G.F. Venomics as a drug discovery platform. Expert Rev. Proteom. 2009, 6, 221–224. [Google Scholar] [CrossRef]
- Beeton, C.; W. Pennington, M.; SNorton, R. Analogs of the sea anemone potassium channel blocker ShK for the treatment of autoimmune diseases. Inflamm. Allergy Drug Targets (Former. Curr. Drug Targets Inflamm. Allergy) 2011, 10, 313–321. [Google Scholar]
- Pennington, M.W.; Lanigan, M.D.; Kalman, K.; Mahnir, V.M.; Rauer, H.; McVaugh, C.T.; Behm, D.; Donaldson, D.; Chandy, K.G.; Kem, W.R.; et al. Role of disulfide bonds in the structure and potassium channel blocking activity of ShK toxin. Biochemistry 1999, 38, 14549–14558. [Google Scholar] [CrossRef]
- Macrander, J.; Broe, M.; Daly, M. Tissue-specific venom composition and differential gene expression in sea anemones. Genome Biol. Evol. 2016, 8, 2358–2375. [Google Scholar] [CrossRef]
- Tysoe, C.; Withers, S.G. Structural dissection of Helianthamide reveals the basis of its potent inhibition of human pancreatic α-Amylase. Biochemistry 2018, 57, 5384–5387. [Google Scholar] [CrossRef]
- Sintsova, O.; Gladkikh, I.; Kalinovskii, A.; Zelepuga, E.; Monastyrnaya, M.; Kim, N.; Shevchenko, L.; Peigneur, S.; Tytgat, J.; Kozlovskaya, E.; et al. Magnificamide, a β-Defensin-Like Peptide from the Mucus of the Sea Anemone Heteractis magnifica, Is a Strong Inhibitor of Mammalian α-Amylases. Mar. Drugs 2019, 17, 542. [Google Scholar] [CrossRef]
- D’ambra, I.; Lauritano, C. A review of toxins from Cnidaria. Mar. Drugs 2020, 18, 507. [Google Scholar] [CrossRef]
- Irwin, D.M. Evolution of the mammalian insulin (Ins) gene; changes in proteolytic processing. Peptides 2021, 135, 170435. [Google Scholar] [CrossRef]
- Rotwein, P. Diversification of the insulin-like growth factor 1 gene in mammals. PLoS ONE 2017, 12, e0189642. [Google Scholar] [CrossRef]
- Chowański, S.; Walkowiak-Nowicka, K.; Winkiel, M.; Marciniak, P.; Urbański, A.; Pacholska-Bogalska, J. Insulin-like peptides and cross-talk with other factors in the regulation of insect metabolism. Front. Physiol. 2021, 12, 701203. [Google Scholar] [CrossRef]
- Semaniuk, U.; Piskovatska, V.; Strilbytska, O.; Strutynska, T.; Burdyliuk, N.; Vaiserman, A.; Bubalo, V.; Storey, K.B.; Lushchak, O. Drosophila insulin-like peptides: from expression to functions—A review. Entomol. Exp. Appl. 2021, 169, 195–208. [Google Scholar] [CrossRef]
- Wu, Q.; Brown, M.R. Signaling and function of insulin-like peptides in insects. Annu. Rev. Èntomol. 2006, 51, 1–24. [Google Scholar] [CrossRef]
- Zheng, S.; Chiu, H.; Boudreau, J.; Papanicolaou, T.; Bendena, W.; Chin-Sang, I. A functional study of all 40 Caenorhabditis elegans insulin-like peptides. J. Biol. Chem. 2018, 293, 16912–16922. [Google Scholar] [CrossRef]
- Smit, A.; VAN Kesteren, R.; Li, K.; VAN Minnen, J.; Spijker, S.; VAN Heerikhuizen, H.; Geraerts, W. Towards Understanding the Role of Insulin in the Brain: Lessons from Insulin-related Signaling Systems in the Invertebrate Brain. Prog. Neurobiol. 1998, 54, 35–54. [Google Scholar] [CrossRef]
- Hassan, A.K.; El-Kotby, D.A.; Tawfik, M.M.; Badr, R.E.; Bahgat, I.M. Antidiabetic effect of the Egyptian honey bee (Apis mellifera) venom in alloxan-induced diabetic rats. J. Basic Appl. Zoöl. 2019, 80, 58. [Google Scholar] [CrossRef]
- Wong, E.S.W.; Morgenstern, D.; Mofiz, E.; Gombert, S.; Morris, K.M.; Temple-Smith, P.; Renfree, M.B.; Whittington, C.M.; King, G.F.; Warren, W.C.; et al. Proteomics and deep sequencing comparison of seasonally active venom glands in the platypus reveals novel venom peptides and distinct expression profiles. Mol. Cell. Proteom. 2012, 11, 1354–1364. [Google Scholar] [CrossRef]
- Wong, E.S.W.; Nicol, S.; Warren, W.C.; Belov, K. Echidna Venom gland transcriptome provides insights into the evolution of monotreme venom. PLoS ONE 2013, 8, e79092. [Google Scholar] [CrossRef]
- Conlon, J.; Attoub, S.; Musale, V.; Leprince, J.; Casewell, N.R.; Sanz, L.; Calvete, J.J. Isolation and characterization of cytotoxic and insulin-releasing components from the venom of the black-necked spitting cobra Naja nigricollis (Elapidae). Toxicon X 2020, 6, 100030. [Google Scholar] [CrossRef]
- HR Fagundes, F.; Aparicio, R.; L dos Santos, M.; BSDiz Filho, E.; CBOliveira, S.; OToyama, D.; HToyama, M. A Catalytically Inactive Lys49 PLA2 Isoform from Bothrops jararacussu venom that Stimulates Insulin Secretion in Pancreatic Beta Cells. Protein Pept. Lett. 2011, 18, 1133–1139. [Google Scholar] [CrossRef]
- Furman, B.L. The development of Byetta (exenatide) from the venom of the Gila monster as an anti-diabetic agent. Toxicon 2012, 59, 464–471. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Gajewiak, J.; Karanth, S.; Robinson, S.D.; Ueberheide, B.; Douglass, A.D.; Schlegel, A.; Imperial, J.S.; Watkins, M.; Bandyopadhyay, P.K.; et al. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc. Natl. Acad. Sci. USA 2015, 112, 1743–1748. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Hu, H.; Gorasia, D.G.; Bandyopadhyay, P.K.; Veith, P.D.; Young, N.D.; Reynolds, E.C.; Yandell, M.; Olivera, B.M.; Purcell, A.W. Combined Proteomic and Transcriptomic Interrogation of the Venom Gland of Conus geographus Uncovers Novel Components and Functional Compartmentalization. Mol. Cell. Proteom. 2014, 13, 938–953. [Google Scholar] [CrossRef]
- Robinson, S.D.; Safavi-Hemami, H. Insulin as a weapon. Toxicon 2016, 123, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.L.; Hossain, M.A.; Lin, F.; Pinheiro-Junior, E.L.; Peigneur, S.; Wai, D.C.C.; Delaine, C.; Blyth, A.J.; Forbes, B.E.; Tytgat, J.; et al. Identification, Synthesis, Conformation and Activity of an Insulin-like Peptide from a Sea Anemone. Biomolecules 2021, 11, 1785. [Google Scholar] [CrossRef]
- Fu, J.; He, Y.; Peng, C.; Tang, T.; Jin, A.; Liao, Y.; Shi, Q.; Gao, B. Transcriptome Sequencing of the pale anemone (Exaiptasia diaphana) Revealed functional peptide gene resources of sea anemon. Front. Mar. Sci. 2022, 9. [Google Scholar] [CrossRef]
- Darling, J.A.; Reitzel, A.R.; Burton, P.M.; Mazza, M.E.; Ryan, J.F.; Sullivan, J.C.; Finnerty, J.R. Rising starlet: the starlet sea anemone, Nematostella vectensis. BioEssays 2005, 27, 211–221. [Google Scholar] [CrossRef]
- Ruiming, Z.; Yibao, M.; Yawen, H.; Zhiyong, D.; Yingliang, W.; Zhijian, C.; Wenxin, L. Comparative venom gland transcriptome analysis of the scorpion Lychas mucronatus reveals intraspecific toxic gene diversity and new venomous components. BMC Genom. 2010, 11, 452. [Google Scholar] [CrossRef]
- Klompen, A.M.L.; Macrander, J.; Reitzel, A.M.; Stampar, S.N. Transcriptomic Analysis of Four Cerianthid (Cnidaria, Ceriantharia) Venoms. Mar. Drugs 2020, 18, 413. [Google Scholar] [CrossRef]
- Chapman, J.A.; Kirkness, E.F.; Simakov, O.; Hampson, S.E.; Mitros, T.; Weinmaier, T.; Rattei, T.; Balasubramanian, P.G.; Borman, J.; Busam, D.; et al. The dynamic genome of Hydra. Nature 2010, 464, 592–596. [Google Scholar] [CrossRef]
- Jin, A.-H.; Muttenthaler, M.; Dutertre, S.; Himaya, S.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Conotoxins: Chemistry and Biology. Chem. Rev. 2019, 119, 11510–11549. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.S.; Olivera, B.M. Conotoxins down under. Toxicon 2006, 48, 780–798. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Oda-Ueda, N.; Hisata, K.; Nakamura, H.; Chijiwa, T.; Hattori, S.; Isomoto, A.; Yugeta, H.; Yamasaki, S.; Fukumaki, Y.; et al. Alternative mRNA Splicing in Three Venom Families Underlying a Possible Production of Divergent Venom Proteins of the Habu Snake, Protobothrops flavoviridis. Toxins 2019, 11, 581. [Google Scholar] [CrossRef]
- Ye, X.; He, C.; Yang, Y.; Sun, Y.H.; Xiong, S.; Chan, K.C.; Si, Y.; Xiao, S.; Zhao, X.; Lin, H.; et al. Comprehensive isoform-level analysis reveals the contribution of alternative isoforms to venom evolution and repertoire diversity. Genome Res. 2023, 33, 1554–1567. [Google Scholar] [CrossRef] [PubMed]
- Haney, R.A.; Matte, T.; Forsyth, F.S.; Garb, J.E. Alternative Transcription at Venom Genes and Its Role as a Complementary Mechanism for the Generation of Venom Complexity in the Common House Spider. Front. Ecol. Evol. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, I.W.; McArdle, A.J.; Coulson, R.M.; A Wagner, M.; Schmid, R.; Hirai, H.; Hoffmann, K.F. Developmentally regulated expression, alternative splicing and distinct sub-groupings in members of the Schistosoma mansoni venom allergen-like (SmVAL) gene family. BMC Genom. 2008, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Zhijian, C.; Feng, L.; Yingliang, W.; Xin, M.; Wenxin, L. Genetic mechanisms of scorpion venom peptide diversification. Toxicon 2006, 47, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Viala, V.L.; Hildebrand, D.; Trusch, M.; Fucase, T.M.; Sciani, J.M.; Pimenta, D.C.; Arni, R.K.; Schlüter, H.; Betzel, C.; Mirtschin, P.; et al. Venomics of the Australian eastern brown snake (Pseudonaja textilis): Detection of new venom proteins and splicing variants. Toxicon 2015, 107, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Ashwood, L.M.; Elnahriry, K.A.; Stewart, Z.K.; Shafee, T.; Naseem, M.U.; Szanto, T.G.; van der Burg, C.A.; Smith, H.L.; Surm, J.M.; Undheim, E.A.B.; et al. Genomic, functional and structural analyses elucidate evolutionary innovation within the sea anemone 8 toxin family. BMC Biol. 2023, 21, 121. [Google Scholar] [CrossRef] [PubMed]
- Heldenbrand, J.; Ren, Y.; Asmann, Y.; Mainzer, L.S. Step-by-step guide for downloading very large datasets to a supercomputer using the SRA Toolkit. 2017. [Google Scholar]
- Grabherr, M.G. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Mahram, A.; Herbordt, M.C. NCBI BLASTP on High-Performance Reconfigurable Computing Systems. ACM Trans. Reconfigurable Technol. Syst. 2015, 7, 33:1–33:20. [Google Scholar] [CrossRef]
- Reeks, T.A.; Fry, B.G.; Alewood, P.F. Privileged frameworks from snake venom. Cell. Mol. Life Sci. 2015, 72, 1939–1958. [Google Scholar] [CrossRef]
- Tamaoki, H.; Miura, R.; Kusunoki, M.; Kyogoku, Y.; Kobayashi, Y.; Moroder, L. Folding motifs induced and stabilized by distinct cystine frameworks. Protein Eng. Des. Sel. 1998, 11, 649–659. [Google Scholar] [CrossRef]
- Chen, Z.; Cao, Z.; Li, W.; Wu, Y. Cloning and characterization of a novel Kunitz-type inhibitor from scorpion with unique cysteine framework. Toxicon 2013, 72, 5–10. [Google Scholar] [CrossRef]
- Fainzilber, M.; Nakamura, T.; Gaathon, A.; Lodder, J.C.; Kits, K.S.; Burlingame, A.L.; Zlotkin, E. A new cysteine framework in sodium channel blocking conotoxins. Biochemistry 1995, 34, 8649–8656. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega. Current Protocols in Bioinformatics 2014, 48, 3.13.1–3.13.16. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Baumgarten, S.; Simakov, O.; Esherick, L.Y.; Liew, Y.J.; Lehnert, E.M.; Michell, C.T.; Li, Y.; Hambleton, E.A.; Guse, A.; Oates, M.E.; et al. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. 2015, 112, 11893–11898. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Kiefer, F.; Arnold, K.; Künzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009, 37, D387–D392. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository—New features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Pond, S.L.K. Detecting Individual Sites Subject to Episodic Diversifying Selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Pond, S.L.K.; Scheffler, K. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef]
- Delport, W.; Poon, A.F.Y.; Frost, S.D.W.; Pond, S.L.K. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 2010, 26, 2455–2457. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Frost, S.D.W. Not So Different After All: A Comparison of Methods for Detecting Amino Acid Sites Under Selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





