Submitted:
13 February 2024
Posted:
13 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Subjects
2.2. Human Tissue Samples
2.3. Animals
2.4. Next-Generation Sequencing
2.5. Variant Prioritization
2.6. Fluorescence Immunohistochemistry (FIHC)
2.7. ADAMTSL4 Cloning and Site-Directed Mutagenesis
2.8. Expression of Recombinant Proteins in Human Cells in Culture and Analysis of Endoplasmic Reticulum Stress
2.9. Functional Analyses in Zebrafish
2.10. Morphological Characterization of Zebrafish Anterior Segment Phenotypes
3. Results
3.1. Mutation Burden Analysis
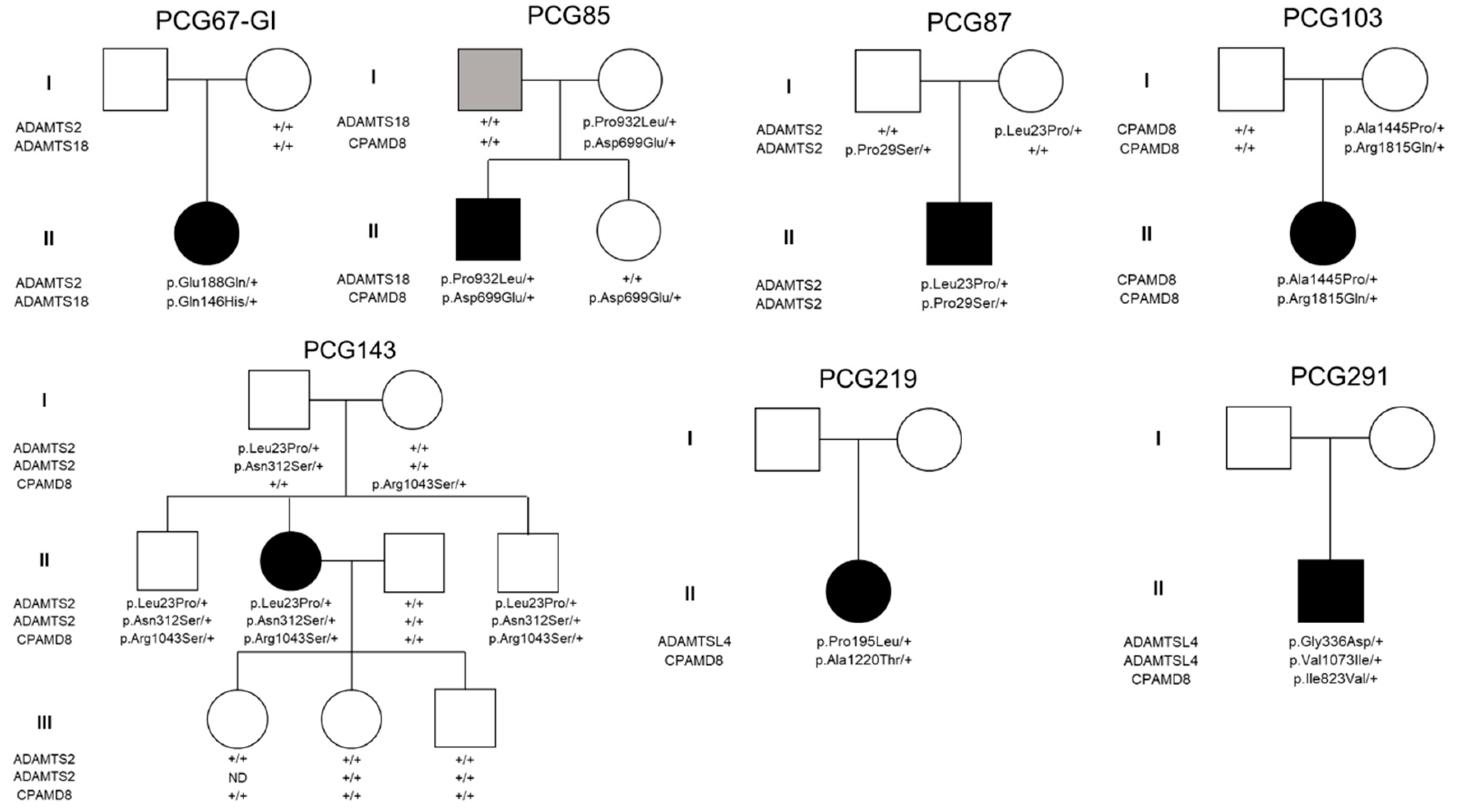
3.2. Genotype-Phenotype Correlation
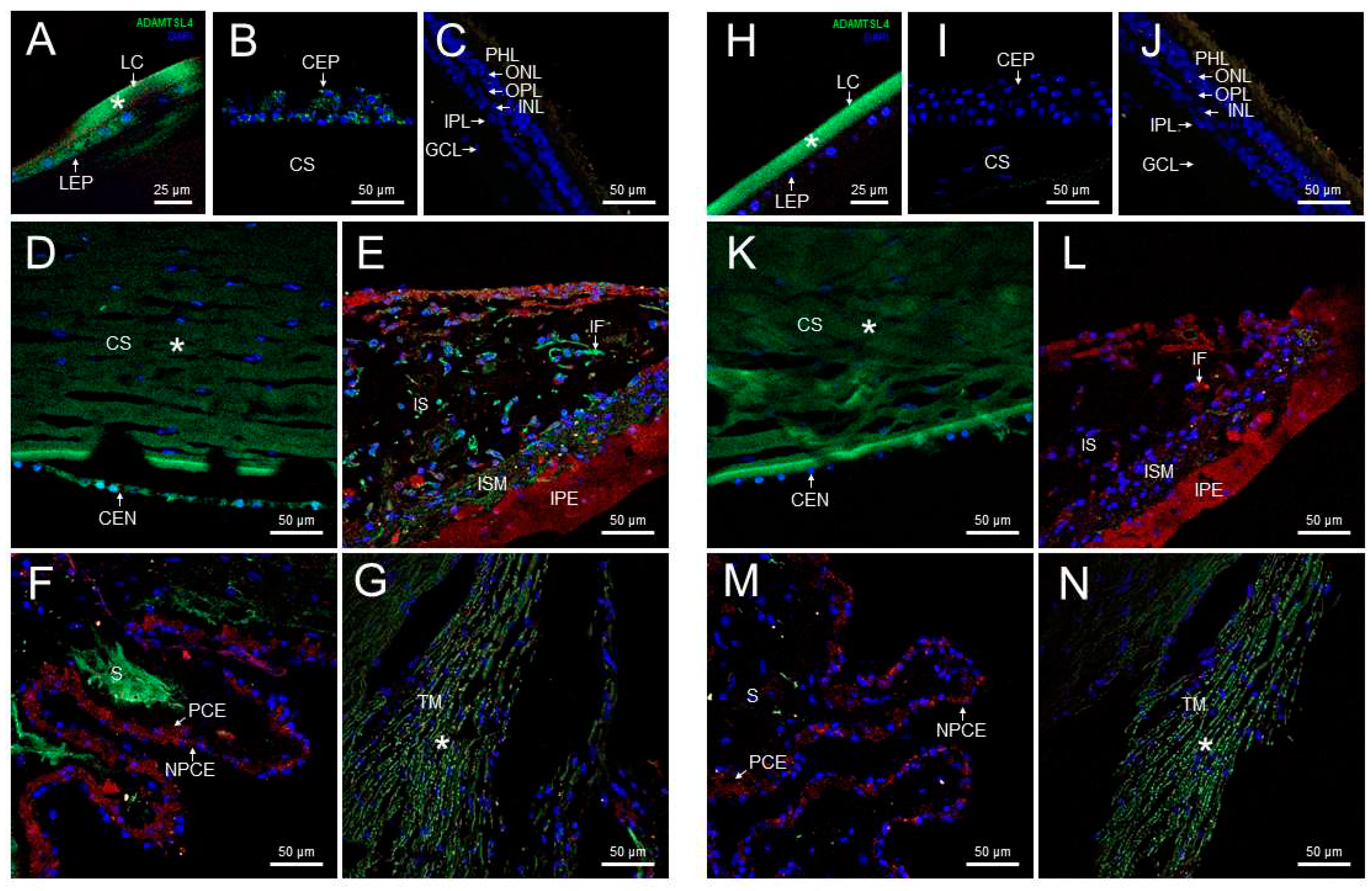
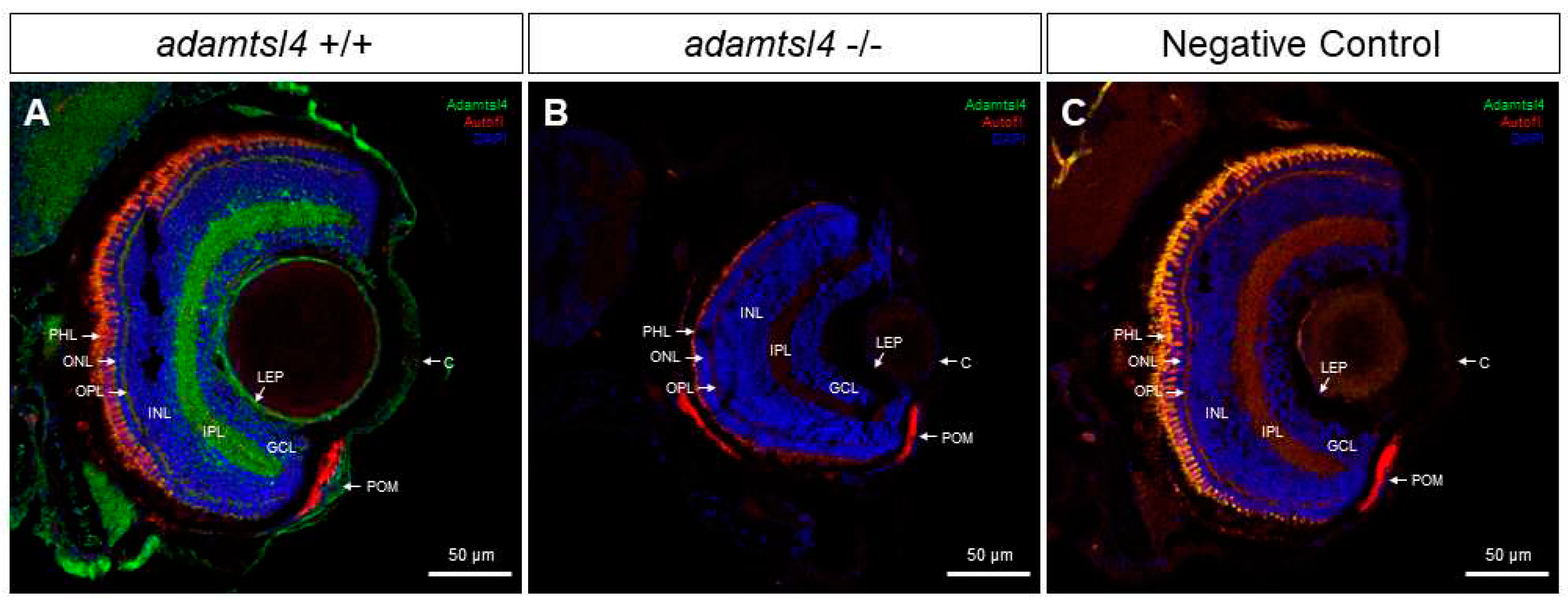
3.3. Expression of ADAMTSL4 in Human and Zebrafish Ocular Tissues
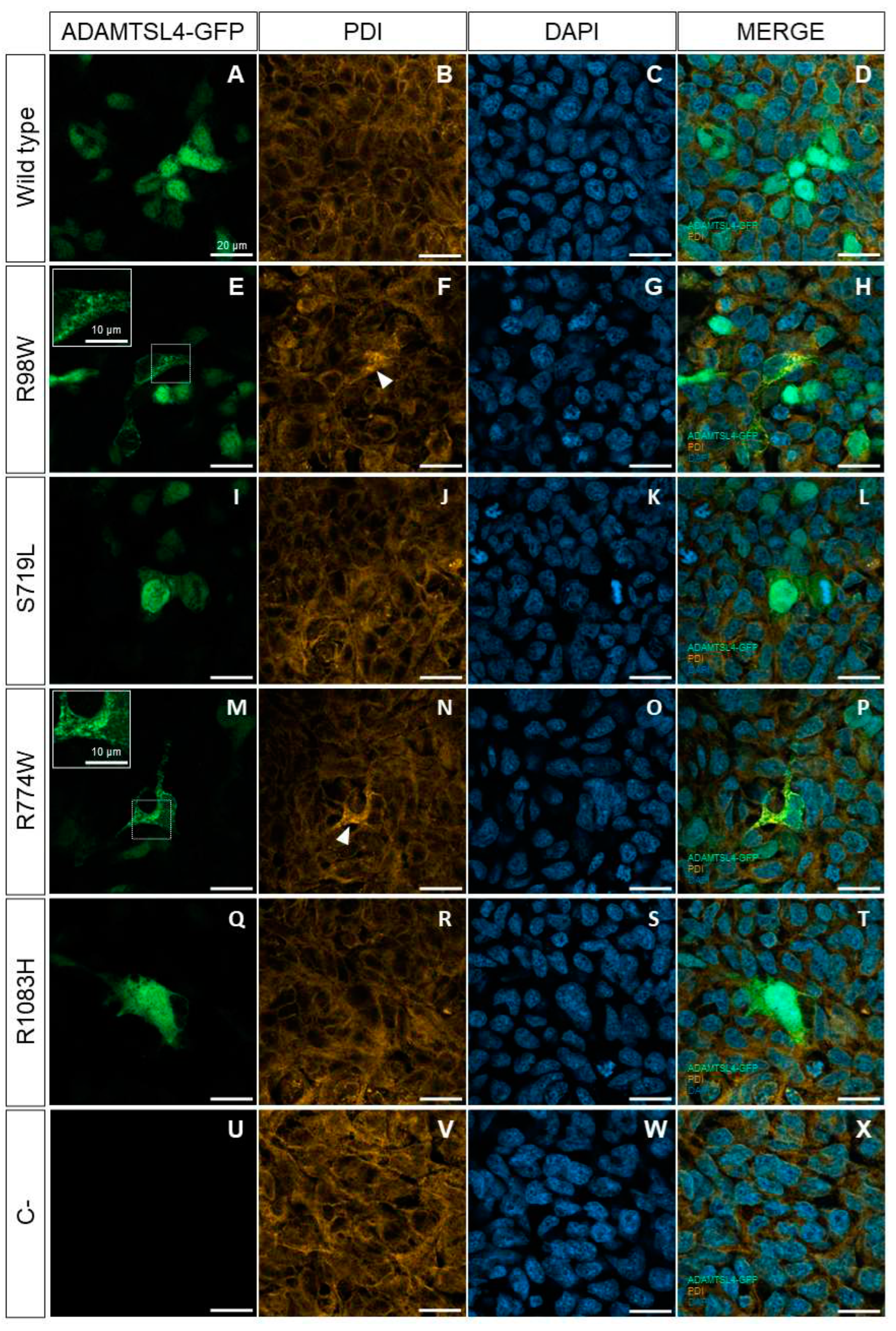
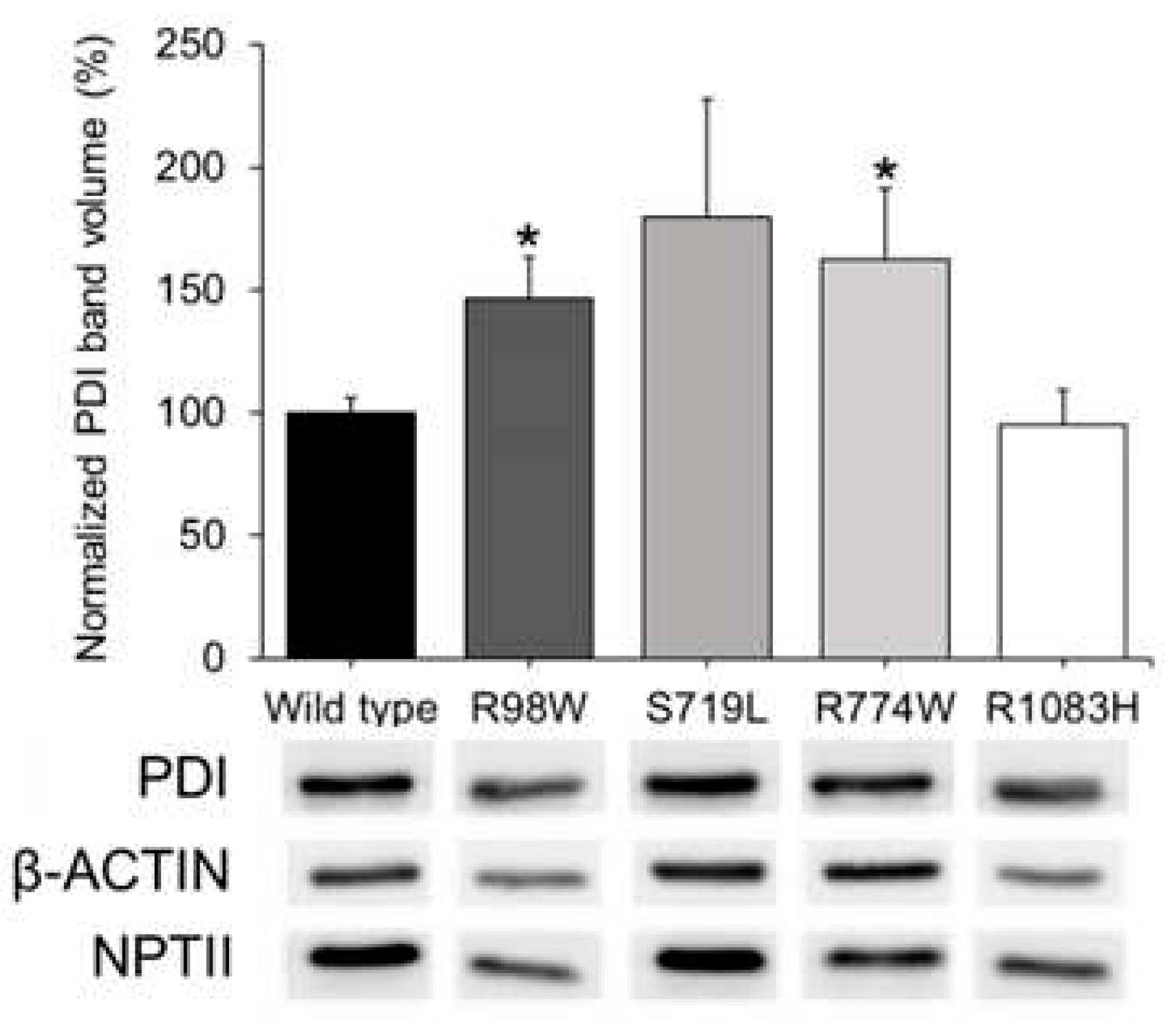
3.4. Functional Analysis of Selected ADAMTSL4 Variants in Cells in Culture and in Zebrafish Embryos
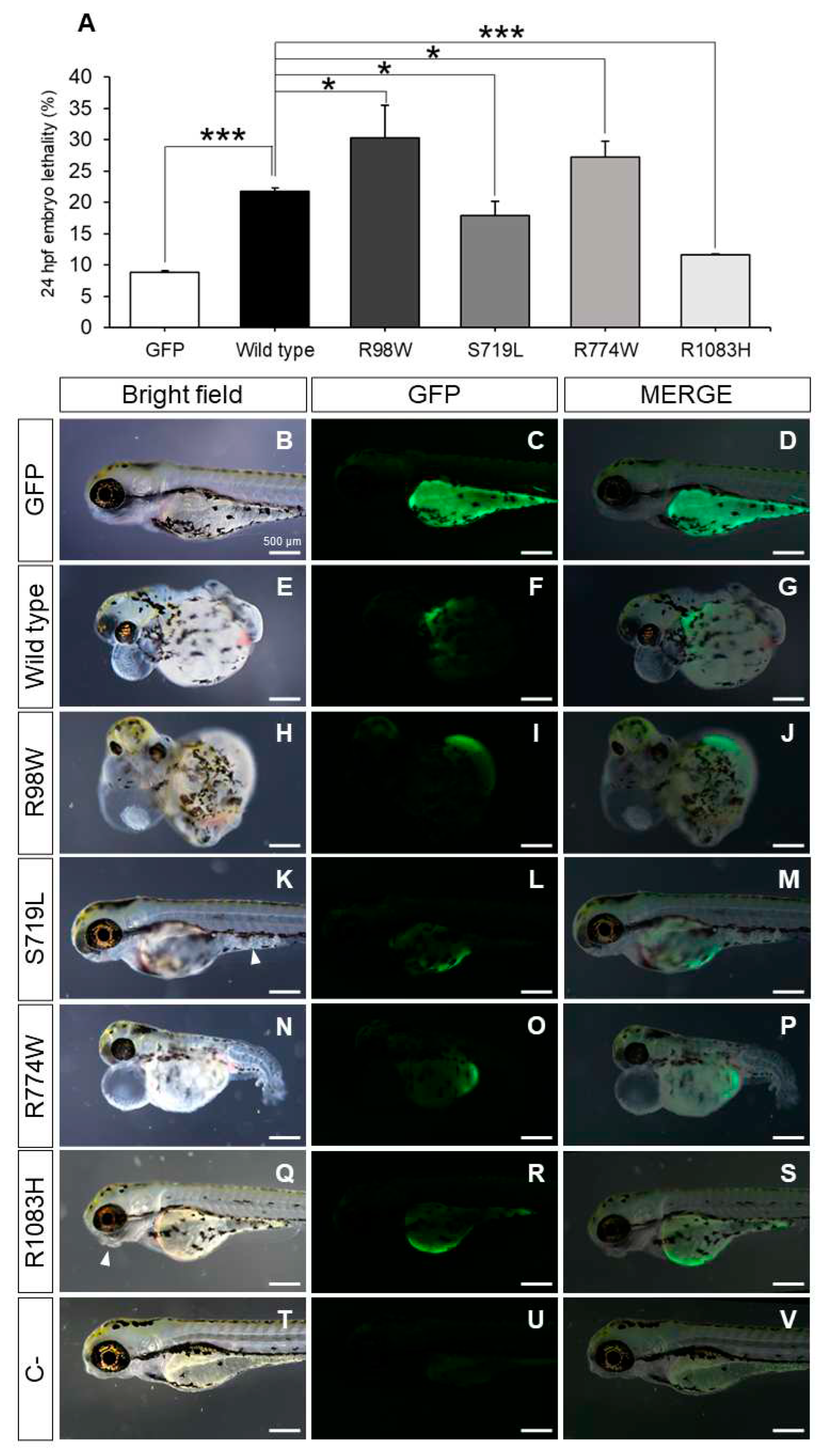
3.5. Functional Testing in Zebrafish of the Hypothesis on the Mutation Burden of MMP-Related Genes in Glaucoma
4. Discussion
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Allen Beck TCPC, Sharon Freedman. Childhood Glaucoma. The 9th Consensus Report of the World Glaucoma Association. In: Grajewski AL, Papoudopoulos M, Grigg J, Freedman S, eds.Childhood Glaucoma. The 9th Consensus Report of the World Glaucoma Association, Ámsterdam, The Netherlands: Kugler Publications, 2013; 3-10.
- Selvan H, Gupta S, Wiggs JL, Gupta V. Juvenile-onset open-angle glaucoma - A clinical and genetic update. Surv Ophthalmol 2022; 67: 1099-117. [CrossRef]
- Quigley HA. Ganglion cell death in glaucoma: pathology recapitulates ontogeny. AustNZJOphthalmol 1995; 23: 85-91. [CrossRef]
- Taylor RH, Ainsworth JR, Evans AR, Levin AV. The epidemiology of pediatric glaucoma: the Toronto experience. J AAPOS 1999; 3: 308-15. [CrossRef]
- Chan TCP, Brookes J, Cavuoto K, Bitrian E, Grajewski AL. Primary congenital glaucoma and juvenile open-angle glaucoma. In: Weinreb RN, Grajewski AL, Papadopoulos M, Grigg J, Freedman S, eds.Primary congenital glaucoma and juvenile open-angle glaucoma, Ámsterdam, The Netherlands: Kluger Publications, 2013; 137-53. K: Netherlands.
- Fung DS, Roensch MA, Kooner KS, Cavanagh HD, Whitson JT. Epidemiology and characteristics of childhood glaucoma: results from the Dallas Glaucoma Registry. Clin Ophthalmol 2013; 7: 1739-46. [CrossRef]
- Gould DB, John SW. Anterior segment dysgenesis and the developmental glaucomas are complex traits. HumMolGenet 2002; 11: 1185-93. [CrossRef]
- Anderson DR. The development of the trabecular meshwork and its abnormality in primary infantile glaucoma. TransAmOphthalmolSoc 1981; 79: 458-85.
- Gupta V, Srivastava RM, Rao A, Mittal M, Fingert J. Clinical correlates to the goniodysgensis among juvenile-onset primary open-angle glaucoma patients. Graefes Arch Clin Exp Ophthalmol 2013; 251: 1571-6. 10.1007/s00417-013-2262-2.
- García-Antón MT, Salazar JJ, de Hoz R, Rojas B, Ramírez AI, Triviño A, Aroca-Aguilar JD, García-Feijoo J, Escribano J, Ramírez JM. Goniodysgenesis variability and activity of CYP1B1 genotypes in primary congenital glaucoma. PLoS One 2017; 12: e0176386. [CrossRef]
- Hollander DA, Sarfarazi M, Stoilov I, Wood IS, Fredrick DR, Alvarado JA. Genotype and phenotype correlations in congenital glaucoma. TransAmOphthalmolSoc 2006; 104: 183-95.
- Bonet-Fernández JM, Aroca-Aguilar JD, Corton M, Ramírez AI, Alexandre-Moreno S, García-Antón MT, Salazar JJ, Ferre-Fernández JJ, Atienzar-Aroca R, Villaverde C, Iancu I, Tamayo A, Méndez-Hernández CD, Morales-Fernández L, Rojas B, Ayuso C, Coca-Prados M, Martinez-de-la-Casa JM, García-Feijoo J, Escribano J. CPAMD8 loss-of-function underlies non-dominant congenital glaucoma with variable anterior segment dysgenesis and abnormal extracellular matrix. Hum Genet 2020. [CrossRef]
- Tawara A, Inomata H. Developmental immaturity of the trabecular meshwork in juvenile glaucoma. Am J Ophthalmol 1984; 98: 82-97. [CrossRef]
- Weinreb RN, Robinson MR, Dibas M, Stamer WD. Matrix Metalloproteinases and Glaucoma Treatment. J Ocul Pharmacol Ther 2020; 36: 208-28. [CrossRef]
- Keller KE, Bradley JM, Acott TS. Differential effects of ADAMTS-1, -4, and -5 in the trabecular meshwork. Invest Ophthalmol Vis Sci 2009; 50: 5769-77.
- Apte SS. ADAMTS Proteins: Concepts, Challenges, and Prospects. Methods Mol Biol 2020; 2043: 1-12. [CrossRef]
- Hendee K, Wang LW, Reis LM, Rice GM, Apte SS, Semina EV. Identification and functional analysis of an ADAMTSL1 variant associated with a complex phenotype including congenital glaucoma, craniofacial, and other systemic features in a three-generation human pedigree. Hum Mutat 2017; 38: 1485-90. [CrossRef]
- Medina-Trillo C, Aroca-Aguilar JD, Ferre-Fernández JJ, Alexandre-Moreno S, Morales L, Méndez-Hernández CD, García-Feijoo J, Escribano J. Role of FOXC2 and PITX2 rare variants associated with mild functional alterations as modifier factors in congenital glaucoma. PLoS One 2019; 14: e0211029. [CrossRef]
- Ferre-Fernández JJ, Aroca-Aguilar JD, Medina-Trillo C, Bonet-Fernández JM, Méndez-Hernández CD, Morales-Fernández L, Corton M, Cabañero-Valera MJ, Gut M, Tonda R, Ayuso C, Coca-Prados M, García-Feijoo J, Escribano J. Whole-Exome Sequencing of Congenital Glaucoma Patients Reveals Hypermorphic Variants in GPATCH3, a New Gene Involved in Ocular and Craniofacial Development. Sci Rep 2017; 7: 46175. [CrossRef]
- Chakrabarti S, Pyatla G, Zhang W, Dixit R, Moreno L, Mandal AK, Senthil S, Bera S, Kumari S, Kabra M, Rathi S, Hameed S, Khanna SC, Kaur I, Khanna H. Complex Molecular Mechanisms Underlie Primary Congenital Glaucoma Due to the Involvement of Multiple Biochemical Pathways In.Complex Molecular Mechanisms Underlie Primary Congenital Glaucoma Due to the Involvement of Multiple Biochemical Pathways Vol. 61, Baltimore, MD, US: Investigative Ophthalmology & Visual Science 2020; 3516. I: MD, US.
- Lopez-Garrido MP, Medina-Trillo C, Morales-Fernandez L, Garcia-Feijoo J, Martinez-de-la-Casa JM, Garcia-Anton M, Escribano J. Null CYP1B1 genotypes in primary congenital and nondominant juvenile glaucoma. Ophthalmology 2013; 120: 716-23. [CrossRef]
- Sarfarazi M, Stoilov I. The Third Genetic Locus (GLC3C) for Primary Congenital Glaucoma (PCG) Maps to Chromosome 14q24.3. Am J HumGenet 2002; 71(4): (Supplement). A455.
- Stoilova D, Child A, Brice G, Desai T, Barsoum-Homsy M, Ozdemir N, Chevrette L, Adam MF, Garchon HJ, Pitts CR, Sarfarazi M. Novel TIGR/MYOC mutations in families with juvenile onset primary open angle glaucoma. JMedGenet 1998; 35: 989-92. [CrossRef]
- Campos-Mollo E, Sanchez-Sanchez F, Lopez-Garrido MP, Lopez-Sanchez E, Lopez-Martinez F, Escribano J. MYOC gene mutations in Spanish patients with autosomal dominant primary open-angle glaucoma: a founder effect in southeast Spain. Mol Vis 2007; 13: 1666-73.
- Sarfarazi M, Stoilov I. Molecular genetics of primary congenital glaucoma. Eye 2000; 14 ( Pt 3B): 422-8. [CrossRef]
- Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. HumMolGenet 1997; 6: 641-7.
- Weisschuh N, Wolf C, Wissinger B, Gramer E. A clinical and molecular genetic study of German patients with primary congenital glaucoma. AmJOphthalmol 2009; 147: 744-53.
- Colomb E, Kaplan J, Garchon HJ. Novel cytochrome P450 1B1 (CYP1B1) mutations in patients with primary congenital glaucoma in France. HumMutat 2003; 22: 496.
- Campos-Mollo E, Lopez-Garrido MP, Blanco-Marchite C, Garcia-Feijoo J, Peralta J, Belmonte-Martinez J, Ayuso C, Escribano J. CYP1B1 mutations in Spanish patients with primary congenital glaucoma: phenotypic and functional variability. Mol Vis 2009; 15: 417-31.
- Ali M, McKibbin M, Booth A, Parry DA, Jain P, Riazuddin SA, Hejtmancik JF, Khan SN, Firasat S, Shires M, Gilmour DF, Towns K, Murphy AL, Azmanov D, Tournev I, Cherninkova S, Jafri H, Raashid Y, Toomes C, Craig J, Mackey DA, Kalaydjieva L, Riazuddin S, Inglehearn CF. Null mutations in LTBP2 cause primary congenital glaucoma. AmJHumGenet 2009; 84: 664-71.
- Kaur K, Reddy AB, Mukhopadhyay A, Mandal AK, Hasnain SE, Ray K, Thomas R, Balasubramanian D, Chakrabarti S. Myocilin gene implicated in primary congenital glaucoma. ClinGenet 2005; 67: 335-40.
- Medina-Trillo C, Sánchez-Sánchez F, Aroca-Aguilar JD, Ferre-Fernández JJ, Morales L, Méndez-Hernández CD, Blanco-Kelly F, Ayuso C, García-Feijoo J, Escribano J. Hypo- and hypermorphic FOXC1 mutations in dominant glaucoma: transactivation and phenotypic variability. PLoS One 2015; 10: e0119272. [CrossRef]
- Medina-Trillo C, Aroca-Aguilar JD, Mendez-Hernandez CD, Morales L, Garcia-Anton M, Garcia-Feijoo J, Escribano J. Rare FOXC1 variants in congenital glaucoma: identification of translation regulatory sequences. Eur J Hum Genet 2016; 24: 672-80. [CrossRef]
- Souma T, Tompson SW, Thomson BR, Siggs OM, Kizhatil K, Yamaguchi S, Feng L, Limviphuvadh V, Whisenhunt KN, Maurer-Stroh S, Yanovitch TL, Kalaydjieva L, Azmanov DN, Finzi S, Mauri L, Javadiyan S, Souzeau E, Zhou T, Hewitt AW, Kloss B, Burdon KP, Mackey DA, Allen KF, Ruddle JB, Lim SH, Rozen S, Tran-Viet KN, Liu X, John S, Wiggs JL, Pasutto F, Craig JE, Jin J, Quaggin SE, Young TL. Angiopoietin receptor TEK mutations underlie primary congenital glaucoma with variable expressivity. J Clin Invest 2016; 126: 2575-87. [CrossRef]
- Morales-Camara S, Alexandre-Moreno S, Bonet-Fernandez JM, Atienzar-Aroca R, Aroca-Aguilar JD, Ferre-Fernandez JJ, Mendez CD, Morales L, Fernandez-Sanchez L, Cuenca N, Coca-Prados M, Martinez-de-la-Casa JM, Garcia-Feijoo J, Escribano J. Role of GUCA1C in Primary Congenital Glaucoma and in the Retina: Functional Evaluation in Zebrafish. Genes (Basel) 2020; 11. [CrossRef]
- Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science 1997; 275: 668-70.
- Wiggs JL, Allingham RR, Vollrath D, Jones KH, De La PM, Kern J, Patterson K, Babb VL, Del Bono EA, Broomer BW, Pericak-Vance MA, Haines JL. Prevalence of mutations in TIGR/Myocilin in patients with adult and juvenile primary open-angle glaucoma. AmJHumGenet 1998; 63: 1549-52.
- Allen KF, Gaier ED, Wiggs JL. Genetics of Primary Inherited Disorders of the Optic Nerve: Clinical Applications. Cold Spring Harb Perspect Med 2015; 5: a017277. [CrossRef]
- Collantes ERA, Delfin MS, Fan B, Torregosa JMR, Siguan-Bell C, Vincent de Guzman Florcruz N, Martinez JMD, Joy Masna-Hidalgo B, Guzman VPT, Anotado-Flores JF, Levina FD, Hernandez SRC, Collantes AA, Sibulo MC, Rong S, Wiggs JL. EFEMP1 rare variants cause familial juvenile-onset open-angle glaucoma. Hum Mutat 2022; 43: 1343. [CrossRef]
- Mackay DS, Bennett TM, Shiels A. Exome Sequencing Identifies a Missense Variant in EFEMP1 Co-Segregating in a Family with Autosomal Dominant Primary Open-Angle Glaucoma. PLoS One 2015; 10: e0132529. [CrossRef]
- Monte W. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). 5th edn.: University of Oregon Press, Eugene, 2013.
- Colige A, Nuytinck L, Hausser I, van Essen AJ, Thiry M, Herens C, Ades LC, Malfait F, Paepe AD, Franck P, Wolff G, Oosterwijk JC, Smitt JH, Lapiere CM, Nusgens BV. Novel types of mutation responsible for the dermatosparactic type of Ehlers-Danlos syndrome (Type VIIC) and common polymorphisms in the ADAMTS2 gene. J Invest Dermatol 2004; 123: 656-63.
- Okumura M, Kadokura H, Inaba K. Structures and functions of protein disulfide isomerase family members involved in proteostasis in the endoplasmic reticulum. Free Radic Biol Med 2015; 83: 314-22. [CrossRef]
- Chakravarti A. Magnitude of Mendelian versus complex inheritance of rare disorders. Am J Med Genet A 2021; 185: 3287-93. [CrossRef]
- Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet 2007; 80: 727-39. [CrossRef]
- Aldahmesh MA, Khan AO, Mohamed JY, Alkuraya H, Ahmed H, Bobis S, Al-Mesfer S, Alkuraya FS. Identification of ADAMTS18 as a gene mutated in Knobloch syndrome. J Med Genet 2011; 48: 597-601. [CrossRef]
- Colige A, Li SW, Sieron AL, Nusgens BV, Prockop DJ, Lapiere CM. cDNA cloning and expression of bovine procollagen I N-proteinase: a new member of the superfamily of zinc-metalloproteinases with binding sites for cells and other matrix components. Proc Natl Acad Sci U S A 1997; 94: 2374-9. [CrossRef]
- Gabriel LA, Wang LW, Bader H, Ho JC, Majors AK, Hollyfield JG, Traboulsi EI, Apte SS. ADAMTSL4, a secreted glycoprotein widely distributed in the eye, binds fibrillin-1 microfibrils and accelerates microfibril biogenesis. Invest Ophthalmol Vis Sci 2012; 53: 461-9. [CrossRef]
- Chandra A, Jones M, Cottrill P, Eastlake K, Limb GA, Charteris DG. Gene expression and protein distribution of ADAMTSL-4 in human iris, choroid and retina. Br J Ophthalmol 2013; 97: 1208-12. [CrossRef]
- Colige A, Sieron AL, Li SW, Schwarze U, Petty E, Wertelecki W, Wilcox W, Krakow D, Cohn DH, Reardon W, Byers PH, Lapiere CM, Prockop DJ, Nusgens BV. Human Ehlers-Danlos syndrome type VII C and bovine dermatosparaxis are caused by mutations in the procollagen I N-proteinase gene. Am J Hum Genet 1999; 65: 308-17. [CrossRef]
- Akar A, D Gl, Erdem Z, Sarathchandra P, Tysoe C, Pope M. Acrogeric Ehlers-Danlos syndrome type IV: report of a new patient with additional findings. Eur J Dermatol 2002; 12: 428-31.
- Mitra A, Ramakrishnan R, Kader MA. Open angle glaucoma in a case of Type IV Ehler Danlos syndrome: a rarely reported association. Indian J Ophthalmol 2014; 62: 880-4. [CrossRef]
- Nie J, Zhang W. Secreted protease ADAMTS18 in development and disease. Gene 2023; 858: 147169. [CrossRef]
- Aldahmesh MA, Alshammari MJ, Khan AO, Mohamed JY, Alhabib FA, Alkuraya FS. The syndrome of microcornea, myopic chorioretinal atrophy, and telecanthus (MMCAT) is caused by mutations in ADAMTS18. Hum Mutat 2013; 34: 1195-9. [CrossRef]
- Ahram D, Sato TS, Kohilan A, Tayeh M, Chen S, Leal S, Al-Salem M, El-Shanti H. A homozygous mutation in ADAMTSL4 causes autosomal-recessive isolated ectopia lentis. Am J Hum Genet 2009; 84: 274-8. [CrossRef]
- Li ZF, Wu XH, Engvall E. Identification and characterization of CPAMD8, a novel member of the complement 3/alpha2-macroglobulin family with a C-terminal Kazal domain. Genomics 2004; 83: 1083-93. [CrossRef]
- Rehman AA, Ahsan H, Khan FH. α-2-Macroglobulin: a physiological guardian. J Cell Physiol 2013; 228: 1665-75. [CrossRef]
- Peters JC, Bhattacharya S, Clark AF, Zode GS. Increased Endoplasmic Reticulum Stress in Human Glaucomatous Trabecular Meshwork Cells and Tissues. Invest Ophthalmol Vis Sci 2015; 56: 3860-8. [CrossRef]

| Childhood glaucoma Alleles (%) [n=202] | ESP6500* Alleles (%) [n=8600] | gnomAD* v2.1.1 Alleles (%) [n=104400] | p-Value (Chi-square test) (CG vs. ESP6500) | p-Value (Chi-square test) (CG vs. gnomAD v2.1.1) | Odds Ratio (95% CI) (CG vs. ESP6500) | Odds Ratio (95% CI) (CG vs. gnomAD v2.1.1 | ||
|---|---|---|---|---|---|---|---|---|
| Aggregate genes | Rare frameshift, non-sense, missense and donor/acceptor splicing sites | 59 (29.20) | 854 (10.03) | 9489 (9.13) | <1E-15 | <1E-15 | 3.49 (2.56-4.77) | 4.06 (3.00-5.50) |
| Rare Synonymous | 17 (8.42) | 773 (9.05) | 10584 (9.83) | 6.39E-01 | 5.77E-01 | 0.86 (0.52-1.42) | 0.84 (0.51-1.39) | |
| ADAMTS2 | Rare frameshift, non-sense, missense and donor/acceptor splicing sites | 14 (6.93) | 140 (1.63) | 1611 (1.56) | 6.52E-08 | 5.00E-09 | 4.49 (2.55-7.93) | 4.70 (2.72-8.10) |
| Rare Synonymous | 2 (0.99) | 184 (2.15) | 2616 (2.43) | 3.79E-01 | 2.70E-01 | 0.46 (0.11-1.85) | 0.40 (0.10-1.61) | |
| ADAMTS18 | Rare frameshift, non-sense, missense and donor/acceptor splicing sites | 14 (6.93) | 178 (2.07) | 2953 (2.70) | 9.47E-06 | 5.01E-04 | 3.52 (2.01-6.18) | 2.68 (1.56-4.62) |
| Rare Synonymous | 3 (1.49) | 158 (1.84) | 2110 (1.93) | 9.17E-01 | 8.36E-01 | 0.81 (0.25-2.55) | 0.76 (0.24-2.39) | |
| ADAMTSL4 | Rare frameshift, non-sense, missense and donor/acceptor splicing sites | 16 (7.92) | 176 (2.05) | 2213 (2.06) | 6.97E-08 | 2.08E-08 | 4.10 (2.41-6.99) | 4.10 (2.45-6.84) |
| Rare Synonymous | 3 (1.49) | 83 (0.97) | 1545 (1.41) | 7.09E-01 | 8.31E-01 | 1.54 (0.48-4.91) | 1.05 (0.34-3.29) | |
| CPAMD8 | Rare frameshift, non-sense, missense and donor/acceptor splicing sites | 15 (7.43) | 360 (4.34) | 2712 (2.84) | 5.24E-02 | 2.13E-04 | 1.77 (1.04-3.03) | 2.75 (1.62-4.65) |
| Rare Synonymous | 9 (4.46) | 348 (4.19) | 4313 (4.20) | 9.92E-01 | 9.94E-01 | 1.07 (0.54-2.10) | 1.06 (0.55-2.08) |
| Patient | ADAMTS2 | ADAMTS18 | ADAMTSL4 | CPAMD8 | Total number ofvariants per patient | Age at diagnosis (months) | IOP at diagnosis (mm Hg) (RE/LE) | Cup/discratio(RE/LE) | Surgical treatment (# surgeries) | Gender/laterality | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCG67-GI | p.Glu188Gln | p.Gln146His | 2 | 144 | 47/37 | 0.4/0.3 | T (4), C/T (6), V, C | ||||||||||
| PCG85 | p.Pro932Leu | p.Asp746Glu | 2 | 10 | NA | NA | G/G (2), T (2) | M/B | |||||||||
| PCG87 | p.Pro29Serp.Leu23Pro | 2 | 16 | 48/42 | 0.8/0.8 | G/G | M/B | ||||||||||
| PCG103 | p.Arg1862Glnp.Ala1492Pro | 2 | 8 | NA | NA | G/G | F/B | ||||||||||
| PCG143 | p.Asn312Serp.Leu23Pro | p.Arg1090Ser | 3 | 60 | NA | NA | None | F/B | |||||||||
| PCG219 | p.Pro195Leu | p.Ala1267Thr | 2 | 5 | NA | NA | T/T | F/B | |||||||||
| PCG291 | p.Val1073Ilep.Gly336Asp | p.Ile823Val | 3 | 36 | NA | NA | G/G | M/B | |||||||||
| Variable | MMP-related gene carriers (A) | MMP-related gene non-carriers (B) | PCGs with null CYP1B1 genotypes (n=37) (C) | P (A vs. B) | p (A vs. C) | p (B vs. C) |
|---|---|---|---|---|---|---|
| aAge at diagnosis (months)(mean±SD) | 17.1±37.4 (n=50) | 12.18±25.2 (n=28) | 1.9±5.2 | ns | 0.0075 | 0.0089 |
| Number of surgeries per eye(mean±SD) | 1.9±1.4 (n=47) | 0.8±1.40 (n=31) | 3.1±1.7 | 0.001 | 0.0014 | 0.00000019 |
| IOP at diagnosis (mm Hg) (mean±SD) | 30.0±1.4(n=48) | 30.5±9.3(n=30) | 28.0±5.5 | ns | ns | ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





