1. Introduction
Association rule mining is a prominent data mining method for object or item set relationships [
1,
2,
3]. Today, association rule mining requires massive data. The popular association rule mining method is Apriori [
4].
Most of the medical data is generated continuously and moves in streams. Doctors carry diagnoses and prognoses. Continuously prescribes the medicines. The doctors need to be altered instantly when negative associations are found among the drugs prescribed by them.
Many Applications in modern days require capturing continuous data generated by man-held wearable devices. The data flowing through those streams must be processed, and any negative associations must be reported to the people concerned. Since data is detected and transported quickly, instream data is huge. Unlike databases, since instream data is never kept raw, it can only be scanned once.
Sensors and other streaming devices collect data now. The processed findings are briefly kept for analysis and decision-making, not the instream data. The instream data must be processed sequentially to uncover regular and frequent patterns with negative relationships.
Positive association rule mining is used for online log data, census data, biological data, fraud detection, and more. The number of negative associations decreases as the frequency of the Item set increases. Negative association rule mining can help improve healthcare crime data analysis decision support systems [
5]. A basic technique mines too many patterns, creating too many association rules in which consumers may not be interested [
6].
Di-Sets helps a vertical data presentation, which helps in comparing candidate patterns and reduces vertical table entry memory Mohammed J. Zaki [
7].
Although FP-Tree [
8] and RP-Tree are more stable and effective, many algorithms have been described. Several methods were developed to find sequential, irregular, unusual, etc. patterns. There’s little focus on uncovering rare negative patterns. Real-time settings are important for finding negative patterns, such as penguins being birds but not flying.
Patterns are item sets that occur in multiple transactions. Processing enormous numbers of patterns will take time. As the database grows, so do patterns. Frequent patterns matter more. Selecting patterns that fulfil the user-specified minimum threshold value finds frequent patterns. The user-selected support value is intriguing. The pattern frequency is how often it appears in the database. No timeframe is set here. Sporadic frequent patterns may not have a regular occurrence behavior.
Regular patterns can be inconsistent with frequent ones. Sometimes, both must be considered. Association between mined patterns is crucial. Positive connections between regular or regular-frequent, regular-frequent-maximal-closed patterns are commonly considered.
Some patterns are negatively correlated, meaning they conflict. When dealing with a medicine with a different chemical composition or weather forecast, one pattern contradicts another, leading to incorrect conclusions and actions. Negative associations are sometimes more essential than good ones and require clear examinations.
Based on the data to be processed, pattern discovery varies substantially. Streamed data must be handled as it flows and is not stored, although non-streamed data can be processed multiple times. Data flows are recognized, and actions are taken when recognized. Data streams are dynamic, with many variables and complicated objects. Such an environment benefits from the sliding window technique.
Mining common items and favorable relationships dominated the research. In addition to item set frequency, item regularity is significant. Negative associations show patterns of enemies, whereas positive relationships are desirable. Stream data flow is important, especially for medical data.
Negative associations among chemicals of various drugs are dangerous as they lead to adverse effects on the patients. Medical data moves in streams continuously from the hospitals, which needs to be processed soon after the data is captured, and the negative associations are to be found and reported immediately. The item sets which are frequent and regular, which meet threshold criteria, must be found, and see if the item sets form any negative associations.
Create a database and example set to investigate negative associations between regular, frequent, closed, and maximal items.
To create a mechanism for processing medical institution data streams.
Develop an algorithm to identify frequent, regular item sets and negative correlations among them.
Determine optimal frequency and regularity thresholds for accurate negative connections.
2. Related work
Many techniques for mining frequent data sets from static databases are published. Data flows in streams with the internet, especially from remote sensors to cloud storage. Dynamic mining follows data streams. To satisfy the user, respond quickly to constant requests. Time-sensitive sliding windowing was utilized by Chih-Hsiang Lin et al. [
9] for frequent data stream mining. Their algorithm stores all frequent items in memory and a database containing expired data items. Based on storage space, the table can hold fewer expired data items.
One confidence threshold is typically used to identify positive and negative association rules (A ⇒ B, A ⇒ ¬B, ¬A ⇒ B, and ¬A ⇒ ¬B). In this method, the user must choose between positive and negative connections. X. Dong et al. [(CH4 ⇒ CH8).[
10] compared confidence criteria for four positive and three negative associations. The linkages between the four confidences showed that four confidence intervals should focus on the four types of associations. Considering the four confidences helps create deceptive rules. They explained how the chi-squared test mines association rules. They suggested a chi-squared test-based PNARMC algorithm with four confidence thresholds.
Many data-gathering technologies have allowed every organization to store data on every activity. Large amounts of data make it hard to mine relevant information. Large data sets allow for more realistic and informative patterns. Sequential patterns give intriguing data insights. Existing sequential mining approaches focus on positive pattern behavior to anticipate the next event after a series. Fahad Anwar et al. [
11] mine negative sequential patterns that contradict each other. Their technique finds patterned events/event sets.
Due to internet application migration, most data flows in streams. Streamed data is difficult to find fascinating. Support metric-based streamed data pattern mining is successful. However, pattern occurrence frequency is not a good measure for finding significant patterns. However, temporal regularity is best for online data stream mining for stock market applications. A pattern is regular if it appears within the user-defined period. None of the methods mine frequent patterns from online application stream data. Regular Pattern Stream tree (RPS tree) and an effective mining method for detecting interesting patterns in streamed data were developed by Syed Khairuzzaman Tanbeer et al. [
12]. They recorded stream data in the RPS tree using a sliding window approach. The tree has been updated with the current data using an efficient approach.
Shirin Mirabedini et al. [
13] reviewed all data stream pattern mining algorithms. Frequent item mining helps cluster and classify data. As network data volume has expanded, so needs to mine data streams for interesting patterns. Regular itemset mining is needed for static and streaming data. Frequent patterns might reveal business trends, scientific phenomena, etc. Pattern finding is the foundation for machine learning activities like association rule induction. Data has been scanned several times to uncover intriguing patterns. Only one scan of streamed data is possible; therefore, pattern-finding is performed in one scan. Since the stream length is unknown, such data has no closure. Before processing streaming data, an initial data set is collected and kept temporarily. VE: A review of online frequent pattern mining methods by Lee et al. [
14]. They categorised approaches by pattern, data, and time window.
DSM-Miner by Juni Yang et al. [
15] mines maximal common patterns effectively. Shirin Mirabedini et al. [
13] examined all data stream pattern mining techniques. Frequent item mining clusters and classifies data. Mining network data streams for intriguing patterns has increased as data volume has grown. Static and streaming data require consistent itemset mining. Frequent patterns may reflect business, scientific, etc., trends. Machine learning operations like association rule induction start with pattern detection. Data was scanned multiple times to find unusual patterns. Only one streamed data scan is possible; therefore, pattern-finding is done there. No closure exists for such data because the stream length is uncertain. A temporary data set is collected before processing streaming data. VE Lee et al. present a structured review of online frequent pattern mining [
14]. By pattern, data, and period, they classed approaches. Included the transaction sliding window mechanism that leverages each processing phase’s transaction count. Decaying distinguishes old and new transactions. SWM trees (Sliding window maximum frequent pattern trees) are suggested for retaining frequent patterns. SWM tree root is used as an enumerated tree root for searching.
Low interest or great confidence can make association rules stand out. The Daly et al. [
16] method assessed exceptional mining rules. They explored negative and exceptional association rules. Negative association rules generate exceptions. Also, they created a metric for anomalous rule interest. Candidate rules analyze patterns and decisions using exceptional rules and metrics.
Most literature-based tactics cut desirable decision-making patterns utilizing fascinating criteria. However, choosing an interest measure is tricky and may need trial and error. No accurate method exists for determining interesting metrics. Thiruvady 2004[
17] suggested using user inputs to determine rules and limitations. The GRD algorithm finds the most intriguing rules.
Statistical correlations determine data set connectivity. Maria-Luiza and Antonie [
18] used a correlation between two item sets to identify negative association rules. Negative rules are retrieved if item sets correlate negatively, and confidence is strong.
Negative rules are retrieved if item sets are negatively correlated, and confidence is strong. Chris Cornelis [
19] examined many algorithms that mine negative and positive association rules and identified various failures. These characteristics were used to classify and catalogue mining algorithms and identify gaps. A confidence framework-based modified Apriori mining method can find negative correlations with intriguing ones. They used upward closure to match validity definitions’ support-based negative connection interest. Dataset entries frequently have the intriguing “Support” parameter. Each level of data records has support values. Each level has multiple support values. The authors devised an Apriori-based upward closure method (PNAR) to find negative association rules. If
ℸ X passes the minimal support condition, so do Y ⊆ I, X ∩ Y =
, and
ℸ(XY).
Interesting negative and positive association rules (PNAR) and mining multiple-level support approaches have been developed. Their method mines positive and negative association rules from fascinating frequent and infrequent item sets using varying support levels. Based on item set regularity, Pavan et al. [
20] found positive and negative associations using vertical table mining.
Bagui and Dhar et al. [
21] demonstrated how to mine positive and negative association rules from MAP REDUCE data. They utilised the Apriori approach to mine several item sets, which was efficient but required much computation time.
None of the negative association rule mining studies [
22,
23] used big data. Positive and negative association rules were found in uncommon item sets. The positive association rule mining identifies commonalities. This could reject many important or low-support items. Rare commodities or item sets can trigger negative association rules despite low support. Significant negative association rule mining requires more search space than positive rule mining for low-support objects. It would make sequential Apriori algorithm implementations easier and harder on big data. Few times has negative association rule mining been used.
Simarjeet Kauri et al. [
24] reviewed AI-based illness diagnosis. However, no review has examined the medications’ effects on heart disease prediction. Jianxiong et al. [
25] developed a machine learning-based drug response risk prediction algorithm. They anticipated disease-treatment drug risks. They have not considered patient risk when administering adversely related medications.
Lu Yuwen, Shuyu et al. [
26] suggested “Prefix-Span” sequential data mining and “Proportional Report Ratio” disproportionality-based technique to detect major adverse drug responses based on casual relationships, drugs, and drug reactions. They tested single drug-to-drug reactions. Restricting adverse pharmaceutical responses has not been studied.
Yifeng Lu et al. [
27] found predicted patterns in frequent item set mining. Negative correlations between suggested medicines are discovered. However, an uncommon item collection with negative drug connections is also important. The authors used bi-directional traversal to mine uncommon, closed item sets. However, negative relationships between infrequent or frequent item sets have not been studied.
Jingzhuo Zhang et al. [
28] developed a system to obtain drug interactions for patients with various conditions. They compiled a drug–drug interaction database from medical sources. Distance monitoring was used to extract drug–drug interactions. A transformer-based bidirectional encoder representation was employed to extract drug interactions. There is no modelling to classify positive or negative interactions.
Generalised tensor decomposition was used to create drug–gene–disease connections by Yoonbee Kim et al. [
29]. Chemical structure and ATC code drug feature networks predicted drug–gene–disease relationships. They learned drug, gene, and sickness using a multi-layer perceptron neural network. They emphasised positive connections and disregarded unfavourable ones, especially among drugs.
Aan Ahmed Toor etc al., [
30] have presented a data stream mining method that includes enforcing Privacy before the same is disseminated. The data is mined from the data streams, and privacy is preserved. They primarily focussed on the drift in data Flowing in the streams due to changes in the data flow context.
U. H. W. A. Hewage et al. [
31] have conducted a literature survey of the methods used for privacy-preserving data streaming mining methods. They focus primarily on the accuracy of privacy-preserving accuracy and data stream mining accuracy. They have not considered the issue of response time.
In his thesis, Aref [
32] focused on developing a data stream association rule mining algorithm among co-occurring events, which means the kind of data stream varies occasionally. The proposed algorithm incrementally mines association rules over data streams that carry varied data.
Nannan Zhang et al. [
33] have recommended a method to mine sequential patterns from data streams. They have used the sliding window method to arrive at the latest data by updating the old data with new data. A Prefix tree generates sequential patterns. They contended that Negative associations can be mined using the Prefix tree.
Most of the presentation in the thesis concentrated on mining drift privacy preservation of the data structure variations flowing in the data streams. The sliding window technique is used to store the current state of the data and ignore the continuous data flowing in the data stream. The main drawback is the inability to process data streams as they appear, find negative associations considering even the historical data and report the same to the destination to take control of false treatments.
3. Establishing data streams
A medical database is created out of registrations and prescriptions and fetching the details from pharmaceutical companies regarding the chemical compositions of the drugs. 100,000 records have been used to create the database.
Ninety thousand records were stored into a flat file, then 10,000 were placed into another flat file using streamed data. The same server holds both flat files. This implies that a 90,000-record static database and a 10,000-record streamed database are examined. Updates to the streamed database consider one window..
The data collected over the WEB is placed. Into a window (Buffer slot at a time), while the nearest is filled with the data, the rarest window is copied into a database. The size of Each window is designed to hold 1000 records. 10 such widow slots are used to hold the instream data.
Figure 1 shows the data streaming method used to process ongoing and continuous data flow from medical establishments.
4. Streamed database algorithms for negative association finding based on item set regularity and frequency
Algorithm-A This technique generates negative patterns using sliding windowing. Data is scanned as received, and the initial data is copied into 10 windows, each window of size 1000 records. 10 such windows are buffered, after which, while the latest window is received, the oldest window is written to a database.
A concurrent process accesses window database data. The Horizontal table, which lists item details and sales transactions, is updated. The pattern generation algorithm finds transaction data in instream data and writes it to a memory buffer. A buffer large enough to hold 1000 transactions is allocated. After round-robin buffer scanning, the horizontal table was updated based on transaction table data.
From a historical database, algorithm A creates an inverted table. Instream processing and Inverted Table updates are done in algorithm B. Process C creates negative linkages between medications and their development chemicals.
Table 3.
Records from the Data stream into the window buffer
Table 3.
Records from the Data stream into the window buffer
| P.SL. No. |
Transaction ID |
Patient Number |
Disease |
Drug |
Chemicals |
Drug |
Chemicals |
| 1 |
T1 |
P100 |
DE1 |
DR1 |
CH1 |
CH2 |
CH3 |
NA |
NA |
DR2 |
CH4 |
CH5 |
CH9 |
CH10 |
| T3 |
P100 |
DE3 |
DR5 |
CH2 |
CH3 |
CH7 |
NA |
NA |
DR6 |
CH13 |
CH14 |
CH15 |
NA |
| T5 |
P223 |
DE5 |
DR9 |
CH1 |
CH3 |
CH5 |
CH16 |
CH19 |
NA |
NA |
NA |
NA |
NA |
| 4 |
T7 |
P937 |
DE7 |
DR11 |
CH2 |
CH3 |
CH7 |
CH11 |
NA |
DR12 |
CH12 |
CH13 |
NA |
NA |
| 5 |
T9 |
P119 |
DE9 |
DR15 |
CH1 |
CH3 |
CH5 |
NA |
NA |
DR16 |
CH8 |
CH9 |
NA |
NA |
| 6 |
T11 |
P1235 |
DE11 |
DR19 |
CH5 |
CH8 |
CH11 |
CH15 |
NA |
DR20 |
NA |
NA |
NA |
NA |
| 8 |
T15 |
P4573 |
DE15 |
DR27 |
CH1 |
CH3 |
CH5 |
NA |
NA |
DR28 |
CH9 |
CH11 |
NA |
NA |
| 10 |
T19 |
P10987 |
DE19 |
DR35 |
CH1 |
CH3 |
CH5 |
NA |
NA |
DR36 |
CH6 |
CH9 |
CH10 |
NA |
Table 4.
Inverted Table for Instream Data
Table 4.
Inverted Table for Instream Data
| Chemical Code |
Transaction Ids |
| CH1 |
TT1 |
TT5 |
TT9 |
TT15 |
TT19 |
|
|
| CH2 |
TT1 |
TT3 |
TT7 |
|
|
|
|
| CH3 |
TT1 |
TT3 |
TT5 |
TT7 |
TT9 |
TT15 |
TT19 |
| CH4 |
TT1 |
|
|
|
|
|
|
| CH5 |
TT1 |
TT5 |
TT9 |
TT11 |
TT15 |
TT19 |
|
| CH6 |
TT19 |
|
|
|
|
|
|
| CH7 |
TT3 |
TT7 |
|
|
|
|
|
| CH8 |
TT6 |
|
|
|
|
|
|
| CH9 |
TT1 |
TT9 |
T15 |
T19 |
|
|
|
| CH10 |
TT1 |
TT19 |
|
|
|
|
|
| CH11 |
TT11 |
TT15 |
|
|
|
|
|
| CH12 |
TT7 |
|
|
|
|
|
|
| CH13 |
TT3 |
TT7 |
|
|
|
|
|
| CH14 |
TT3 |
|
|
|
|
|
|
| CH15 |
TT3 |
TT11 |
|
|
|
|
|
| CH16 |
TT5 |
|
|
|
|
|
|
| CH19 |
TT5 |
|
|
|
|
|
|
Table 5.
Updated Medical Data (Historical Data + Instream data) (P - Patient, DE - Disease, DR = Drug, CH - Chemical in the Drug)
Table 5.
Updated Medical Data (Historical Data + Instream data) (P - Patient, DE - Disease, DR = Drug, CH - Chemical in the Drug)
| P.SL. No. |
Transaction ID |
Patient Number |
Disease |
Drug |
Chemicals |
Drug |
Chemicals |
| 1 |
T1 |
P100 |
DE1 |
DR1 |
CH1 |
CH2 |
CH3 |
NA |
NA |
DR2 |
CH4 |
CH5 |
CH9 |
CH10 |
| T2 |
P100 |
DE2 |
DR3 |
CH4 |
CH5 |
CH6 |
NA |
NA |
DR4 |
CH10 |
CH15 |
NA |
NA |
| T3 |
P100 |
DE3 |
DR5 |
CH2 |
CH3 |
CH7 |
NA |
NA |
DR6 |
CH13 |
CH14 |
CH15 |
NA |
| 2 |
T4 |
P223 |
DE4 |
DR7 |
CH5 |
CH8 |
CH10 |
NA |
NA |
DR8 |
CH11 |
CH15 |
NA |
NA |
| T5 |
P223 |
DE5 |
DR9 |
CH1 |
CH3 |
CH5 |
CH16 |
CH19 |
NA |
NA |
NA |
NA |
NA |
| 3 |
T6 |
P749 |
DE6 |
DR10 |
CH4 |
CH5 |
CH16 |
CH19 |
NA |
NA |
NA |
NA |
NA |
NA |
| 4 |
T7 |
P937 |
DE7 |
DR11 |
CH2 |
CH3 |
CH7 |
CH11 |
NA |
DR12 |
CH12 |
CH13 |
NA |
NA |
| 5 |
T8 |
P119 |
DE8 |
DR13 |
CH5 |
CH8 |
CH11 |
NA |
NA |
DR14 |
CH12 |
CH14 |
CH15 |
NA |
| T9 |
P119 |
DE9 |
DR15 |
CH1 |
CH3 |
CH5 |
NA |
NA |
DR16 |
CH8 |
CH9 |
NA |
NA |
| T10 |
P119 |
DE10 |
DR17 |
CH2 |
CH3 |
CH7 |
CH8 |
NA |
DR18 |
CH13 |
CH14 |
CH15 |
NA |
| 6 |
T11 |
P1235 |
DE11 |
DR19 |
CH5 |
CH8 |
CH11 |
CH15 |
NA |
DR20 |
NA |
NA |
NA |
NA |
| 7 |
T12 |
P11 |
DE12 |
DR21 |
CH4 |
CH5 |
CH6 |
NA |
NA |
DR22 |
CH10 |
CH15 |
NA |
NA |
| T13 |
P11 |
DE13 |
DR23 |
CH2 |
CH3 |
CH7 |
CH8 |
NA |
DR24 |
CH13 |
CH14 |
CH15 |
NA |
| T14 |
P11 |
DE14 |
DR25 |
CH5 |
CH8 |
CH11 |
CH15 |
NA |
DR26 |
NA |
NA |
NA |
NA |
| 8 |
T15 |
P4573 |
DE15 |
DR27 |
CH1 |
CH3 |
CH5 |
NA |
NA |
DR28 |
CH9 |
CH11 |
NA |
NA |
| T16 |
P4573 |
DE16 |
DR29 |
CH4 |
CH5 |
CH6 |
NA |
NA |
DR30 |
CH14 |
CH15 |
NA |
NA |
| 9 |
T17 |
P8765 |
DE17 |
DR31 |
CH2 |
CH3 |
CH6 |
CH7 |
NA |
DR32 |
CH12 |
CH13 |
NA |
NA |
| T18 |
P8765 |
DE18 |
DR33 |
CH5 |
CH8 |
CH11 |
CH12 |
NA |
DR34 |
CH14 |
CH15 |
NA |
NA |
| 10 |
T19 |
P10987 |
DE19 |
DR35 |
CH1 |
CH3 |
CH5 |
NA |
NA |
DR36 |
CH6 |
CH9 |
CH10 |
NA |
| T20 |
P10987 |
DE20 |
DR37 |
CH4 |
CH5 |
CH6 |
NA |
NA |
DR38 |
CH12 |
CH14 |
CH15 |
NA |
| T21 |
P10987 |
DE21 |
DR39 |
CH2 |
CH3 |
CH4 |
NA |
NA |
DR40 |
CH7 |
CH13 |
NA |
NA |
| T22 |
P10987 |
DE22 |
DR41 |
CH5 |
CH8 |
CH11 |
NA |
NA |
DR42 |
CH12 |
CH15 |
NA |
NA |
| T23 |
P10987 |
DE23 |
DR43 |
CH1 |
CH3 |
CH5 |
NA |
NA |
DR44 |
CH9 |
CH14 |
NA |
NA |
Table 6.
Updated Inverted Table along with filled-up frequency and Regularity.
Table 6.
Updated Inverted Table along with filled-up frequency and Regularity.
| Chemical Code |
Transaction Ids |
Maximum
Regularity (4) |
Minimum
Frequency (3) |
| CH1 |
TT1 |
TT5 |
TT9 |
TT13 |
TT17 |
TT21 |
|
|
|
|
|
|
|
|
|
|
4 |
6 |
| CH2 |
TT1 |
TT3 |
TT7 |
TT11 |
TT5 |
TT9 |
|
|
|
|
|
|
|
|
|
|
6 |
6 |
| CH3 |
TT1 |
TT3 |
TT5 |
TT7 |
TT9 |
TT11 |
TT13 |
TT15 |
TT17 |
TT19 |
TT21 |
|
|
|
|
|
2 |
11 |
| CH4 |
TT1 |
TT2 |
TT6 |
TT10 |
TT14 |
TT18 |
TT19 |
|
|
|
|
|
|
|
|
|
4 |
7 |
| CH5 |
TT1 |
TT2 |
TT4 |
TT5 |
TT6 |
TT8 |
TT9 |
TT10 |
TT12 |
TT13 |
TT14 |
TT16 |
TT17 |
TT18 |
TT20 |
TT21 |
2 |
16 |
| CH6 |
TT2 |
TT5 |
TT6 |
TT10 |
TT14 |
TT15 |
TT17 |
TT18 |
|
|
|
|
|
|
|
|
4 |
8 |
| CH7 |
TT3 |
TT7 |
TT11 |
TT15 |
TT19 |
|
|
|
|
|
|
|
|
|
|
|
4 |
5 |
| CH8 |
TT4 |
TT8 |
TT9 |
TT11 |
TT12 |
TT16 |
TT20 |
|
|
|
|
|
|
|
|
|
4 |
7 |
| CH9 |
TT1 |
TT5 |
TT9 |
TT13 |
TT17 |
TT21 |
|
|
|
|
|
|
|
|
|
|
4 |
6 |
| CH10 |
TT1 |
TT2 |
TT4 |
TT10 |
TT17 |
|
|
|
|
|
|
|
|
|
|
|
7 |
5 |
| CH11 |
TT4 |
TT7 |
TT8 |
TT12 |
TT13 |
TT16 |
TT20 |
|
|
|
|
|
|
|
|
|
4 |
7 |
| CH12 |
TT7 |
TT8 |
TT15 |
TT16 |
TT18 |
TT20 |
|
|
|
|
|
|
|
|
|
|
7 |
6 |
| CH13 |
TT3 |
TT7 |
TT11 |
TT15 |
TT19 |
|
|
|
|
|
|
|
|
|
|
|
4 |
5 |
| CH14 |
TT1 |
TT3 |
TT8 |
TT11 |
TT14 |
TT16 |
TT18 |
TT21 |
|
|
|
|
|
|
|
|
5 |
8 |
| CH15 |
TT2 |
TT3 |
TT4 |
TT6 |
TT8 |
TT10 |
TT12 |
TT14 |
TT16 |
TT18 |
TT20 |
|
|
|
|
|
7 |
11 |
Table 7.
Pruned Inverted Table for Maximum Regularity and Minimum Frequency
Table 7.
Pruned Inverted Table for Maximum Regularity and Minimum Frequency
| Chemical Code |
Transaction Ids |
Maximum
Regularity (4) |
Minimum
Frequency (3) |
| CH1 |
TT1 |
TT5 |
TT9 |
TT13 |
TT17 |
TT21 |
|
|
|
|
|
|
|
|
|
|
4 |
6 |
| CH3 |
TT1 |
TT3 |
TT5 |
TT7 |
TT9 |
TT11 |
TT13 |
TT15 |
TT17 |
TT19 |
TT21 |
|
|
|
|
|
2 |
11 |
| CH4 |
TT1 |
TT2 |
TT6 |
TT10 |
TT14 |
TT18 |
TT19 |
|
|
|
|
|
|
|
|
|
4 |
7 |
| CH5 |
TT1 |
TT2 |
TT4 |
TT5 |
TT6 |
TT8 |
TT9 |
TT10 |
TT12 |
TT13 |
TT14 |
TT16 |
TT17 |
TT18 |
TT20 |
TT21 |
2 |
16 |
| CH6 |
TT2 |
TT5 |
TT6 |
TT10 |
TT14 |
TT15 |
TT17 |
TT18 |
|
|
|
|
|
|
|
|
4 |
8 |
| CH7 |
TT3 |
TT7 |
TT11 |
TT15 |
TT19 |
|
|
|
|
|
|
|
|
|
|
|
4 |
5 |
| CH8 |
TT4 |
TT8 |
TT9 |
TT11 |
TT12 |
TT16 |
TT20 |
|
|
|
|
|
|
|
|
|
4 |
7 |
| CH9 |
TT1 |
TT5 |
TT9 |
TT13 |
TT17 |
TT21 |
|
|
|
|
|
|
|
|
|
|
4 |
6 |
| CH11 |
TT4 |
TT7 |
TT8 |
TT12 |
TT13 |
TT16 |
TT20 |
|
|
|
|
|
|
|
|
|
4 |
7 |
| CH13 |
TT3 |
TT7 |
TT11 |
TT15 |
TT19 |
|
|
|
|
|
|
|
|
|
|
|
4 |
5 |
Table 8.
Drugs mapped to Chemicals with Negative Association
Table 8.
Drugs mapped to Chemicals with Negative Association
| “Chemical” |
“Associated
Drugs” |
“Chemical” |
“Associated
Drugs” |
“Chemical” |
“Associated
Drugs” |
“Chemical” |
“Associated
Drugs” |
| CC4 |
DH3 |
CC8 |
DG7 |
CC11 |
DG11 |
|
|
| CC6 |
DG3 |
CC8 |
DG7 |
CC11 |
DG11 |
|
|
| CC4 |
DG3 |
CC6 |
DG3 |
CC8 |
DG7 |
CC11 |
DG11 |
5. Experimenting and Results
5.1. Creation of data set
A database contains patent registration, diagnostic, patient-diagnosis, chemical, drug-chemical, quantity, and prescription codes. Hospitals have contributed 100,000 patient registrations and medications to the database. Each diagnosis, drug administration, and drug chemical makeup are included in an Example set. Data items in repeating groups are encoded and replaced with codes. We sort the sample set, compute the frequency and regularity of each item set, update the database, and import 100,000 Flat file entries. These records were processed using this paper’s algorithm. No standard data set exists with the data items needed to discover negative connections.
5.2. Results
The procedure was applied on the data above, yielding the following results.
-
Step-1
Read the Historical Data Contained in a Database and add transaction IDs for each transaction. Details of the data fetched from the Database is shown in
Table 1.
-
Step-2
Construct an Inverted Table in vertical format for the Transactions shown in
Table 8 and
Table 2, along with the frequency of occurrence and the Regularity. Regularity is based on database record placements, and frequency is based on occurrence count.
-
Step-3
Read the Data from the Input data stream and load the same into a Temporary buffer. After filling the Temporary buffer, transfer the Records to window-1 of the window buffer after the last window is written to the database. The Records are read from the window Buffer.
Table 3 shows the records from the data stream
-
Step 4
Develop an Inverted Table from the buffered data.
Table 4 shows the Inverted Table for records read from the window buffer. Here, the issue of frequency and regularity has not been used. The data is retrieved from online transactions.
-
Step 5
Update the Historical Inverted Table with the data in the inverted table relating to the data stream. The frequencies and the regularity are updated simultaneously. The updated Transaction and Inverted are shown in
Table 5 and
Table 6.
-
Step -6
Prune the records which do not meet the Maximum Frequency and Minimum regulatory.
Records that do not satisfy the user’s criterion are pruned using the maximum regularity and minimum frequency of 4 and 3, respectively.
Table 3 reveals that Chemical codes CH2, CH10, CH12, CH14, and CH15 were trimmed because they do not fulfil the Regularity and Frequency criterion.
Table 7 lists leftover records. By this criteria, 5 chemicals were pruned. Yellow records are trimmed because they don’t meet thresholds.
-
Step-7
Make adverse chemicals. Find Negative Associations with Records with No Common Transactions (Nill Common Items). Application of record intersection Produces unfavourable associations like
(CC4 ⇒ CC8, CC11), (CC4 ⇒ CC8), (CC6 ⇒ CC8), (CC4 ⇒ CC11), (ACC6 ⇒ CC11), (CC8 ⇒ CC4, CC6), (CC6 ⇒ CC8, CC11), (CC11 ⇒ CC4, CC6), (CC4, CC6), ⇒ CC8, CC11)
-
Step 8 Find Negatively Associated Drugs
As stated in
Table 8, map back the chemicals connected with the medications to determine the negatively associated pharmaceuticals.
The unfavourable associations show that DG3 should not be used with DG7 or DG11 because they conflict.
6. Discussion
Data Analysis has been conducted considering different sample sizes of 20,000, 50,000 and 80,000 Examples. The analysis is carried out considering thresholds in respect of frequencies and Regularities. While the sample sizes are used for the historical database, the Instream data size is fixed at 10,000 examples.
Table 9 shows the number of negative associations considering the sample size of 20000 and data stream size of 10,000 Examples.
Data gathering yielded 100,000 cases evaluated for 30,000 sample sizes. 50,000, and 70,000). We set regularity and support thresholds and the number of unfavorable Associations observed.
Table 9 shows frequent, regular, negative connections from 30,000 samples varying in regularity between 2.0 and 3.0 and frequency between 1.5 and 2.0.
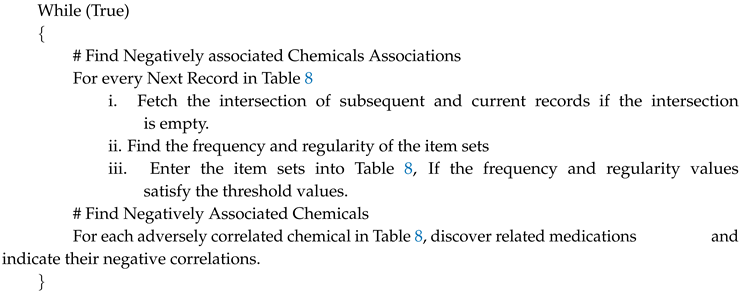
Table 10 displays the number of negative Associations with frequency 1.75 and regularity 2–3 for 20,000 and 10,000 data stream instances. Negative associations respond to regularity modifications in
Figure 1, with frequency fixed at 1.75.
Figure 2 demonstrates the variation in negative correlations with regularity varying and Frequency set at 1.75.
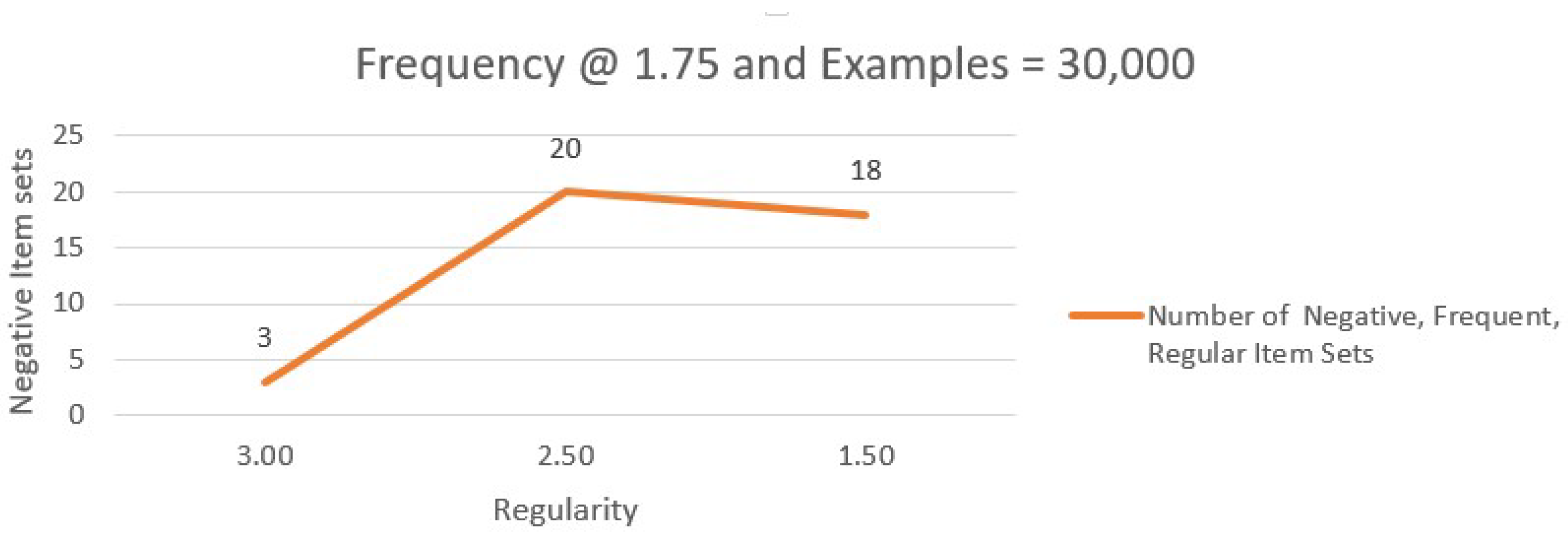
Table 11 illustrates the number of negative connections with Frequency 1.5 and Regularity 2–3 in 30,000 cases. The line graph of negative associations with 1.5 frequency and variable regularity is shown in
Figure 2.
Figure 3 shows the variation in negative associations with regularity varying and frequency set at 1.50.
The larger example set was analysed further.
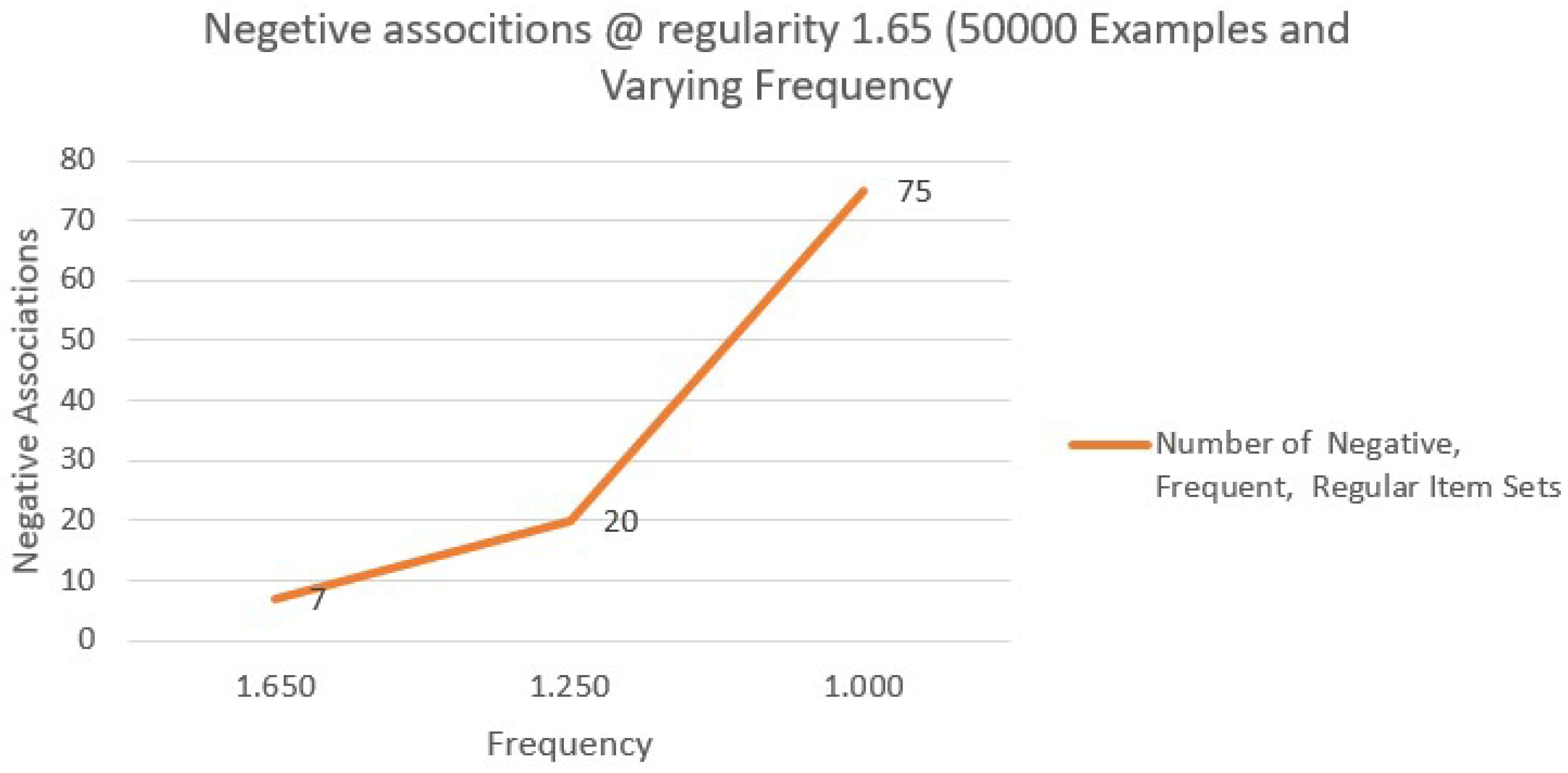
Table 12 illustrates the percentage of negative correlations per 1000 Examples for sample sizes (20,000, 50,000, 80,000), including 10,000 DataStream instances. Frequency varies.
From the above Table, it can be inferred that Considering a sample size of 50,000 examples, fixing the regularity 1.65 and frequency 1.25 gives the least and most effective negative associations 20 out of 50,000 could be identified.
Further analysis examines the effects of fixing regularity, altering frequency, and different sample sizes.
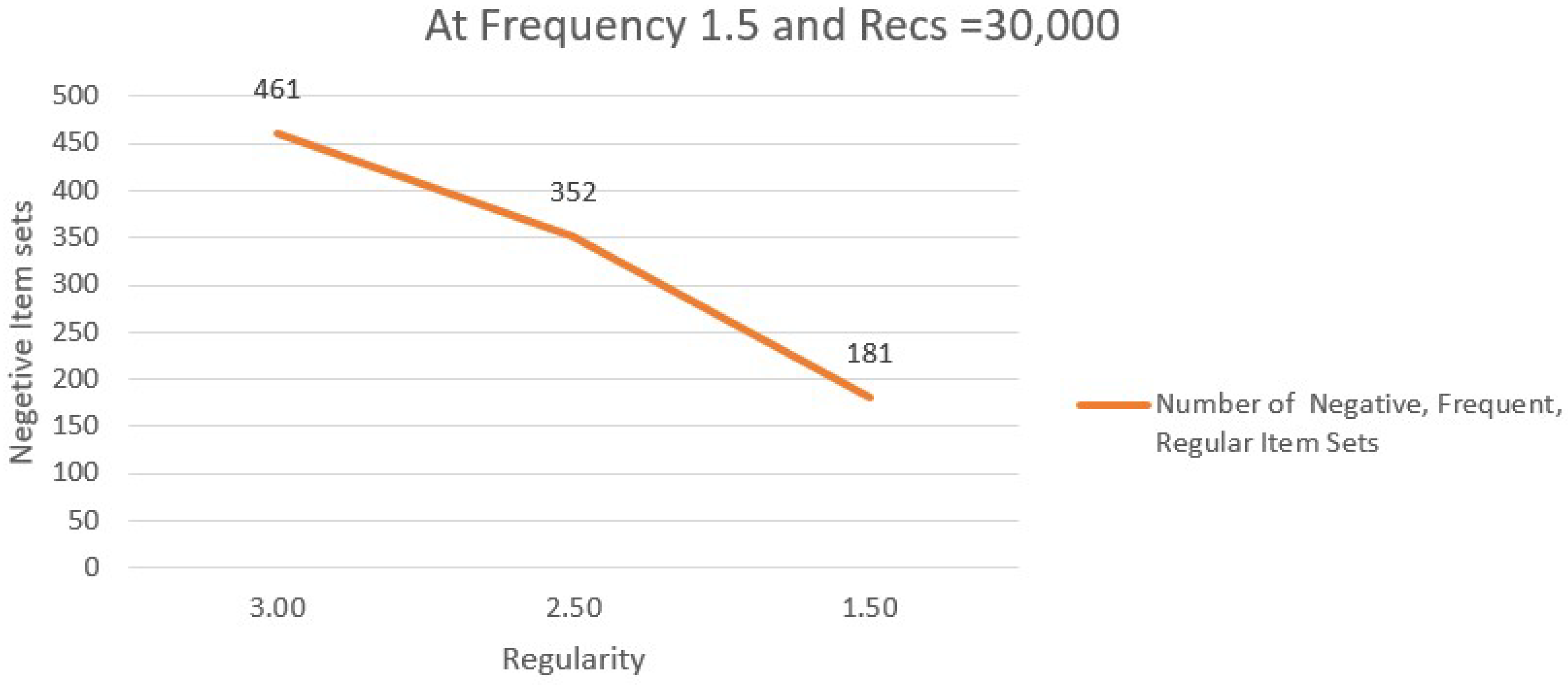
Table 13 lists unfavourable relationships with Regularity = 1.50 and Sample Size = 30,000.
Figure 4 exhibits negative association variation.
Table lists negative relationships with a regularity 1.65 and a sample size of 50,000. Negative connections dropped 73%.
Figure 5 shows variances.
7. Conclusions and Future Scope
Medical data moves in streams, and doctors must be warned soon after prescription if any negative associations exist among drugs prescribed by them, or else there could be a devastating effect on the patients. Many patients died during the coronavirus time as no system existed at that time that warned doctors not to use some drugs in combination. The use of humidifier drugs killed many due to many side reactions that affected the human organs.
Finding minimum and effective negative correlations is critical when medical data is rapidly shared online. Critical items are emphasised. Minifying regular and common item sets from historical databases and data streams yields the most effective negative associations. Since the complete data set is unavailable for study, finding the maximal and closed data sets is impossible. Regularity at 1.5 and frequency at 1.75 yields the last and most effective negative associations.
This research identifies all negative correlations with a collection of medications given to patients. Finding positive associations can be challenging due to many negative ones. Every medicine is chemically made. Chemical mixtures can be harmful. Consider negative correlations among chemicals and decode them into medications to determine the most important ones. This research is mainly limited by the chemical composition of medications doctors give to treat disorders.
Research medication administration limits and large trends. Rare and new medicinal item sets must be investigated. Further research may reveal intriguing elements beyond frequency and support that directly fit to uncover effective negative associations. The method may find negative links in remote or incremental medical databases and streaming data.
References
- Aggarwal, C.C.; Yu, P.S. Mining associations with the collective strength approach. IEEE Trans. Knowl. Data Eng. 2001, 13, 863–873. [CrossRef]
- Aggarwal, C.C.; Yu, P.S. A new framework for item-set generation. In Proceedings of the Seventeenth ACM SIGACTSIGMODSIGART Symposium on Principles of Database Systems, PODS’98, Seattle, WA, USA, 1–4 June 1998; pp. 18–24.
- Agrawal R, Imielinski T, Swami A. Mining association rules between sets of items in large databases. In: ACM SIGMOD conference. New York City: ACM Press; 1993. p. 207–216.
- Agrawal R, Srikant R. Fast algorithms for mining association rules. In: VLDB 1994 proceedings of the 20th international conference on large databases. 1994. p. 487–499.
- Mahmood S, Shahbaz M, Guergachi A. Negative and positive association rules mining from text using frequent and infrequent itemsets. Sci World J. 2014;2014:1–11. [CrossRef]
- Ashok SAVASERE, A., OMIECINSKI, E., AND NAVATHE, S. 1998. Mining for strong negative associations in a large database of customer transactions. In Proceedings of the Fourteenth International Conference on Data Engineering. IEEE Computer Society, Orlando, Florida, 494–502.
- Zaki, M.J. Fast Vertical Mining Using Diffsets. In Proceedings of the KDD03: The Ninth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Washington, DC, USA, 24–27 August 2003.
- Chuan Yao Yang1 Yuqin Li1 Chenghong Zhang2 Yunfa Hu1 “A Novel Algorithm of Mining Maximal Frequent Pattern Based on Projection Sum Tree,” IEEE Transactions. 2007.
- Chih-Hsiang Lin, Ding-Ying Chiu, Yi-Hung Wu, Arbee L. P. Chen, “Mining Frequent Itemsets from Data Streams with a Time-Sensitive Sliding Window,” in Society for industrial and applied mathematics, 2006.
- X. Dong, F. Sun, X. Han, R. Hou, “Study of positive and negative association rules based on multiconfidence and chi-squared test,” in ADMA06, in LNCS, vol.4093, Springer-Verlag, Berlin–Heidelberg, 2006, pp.100–109.
- Fahad Anwar, Ilias Petrounias, Tim Morris, Vassilis Kodogiannis, “ Discovery of events with negative behavior against given sequential patterns” -1-4244- 5164-7.
- Syed Khairuzzaman Tanbeer, Chowdhury Farhan Ahmed, and Byeong-Soo Jeong,” Mining Regular Patterns in Data Streams,” DASFAA 2010, Part I, LNCS 5981, pp. 399–413, 2010, Springer-Verlag Berlin Heidelberg 2010.
- Shirin Mirabedini, Mahdi Ahmadi Panah, Maryam Darbanian, “Frequent Pattern Mining in Data Streams,” Journal of Engineering and Applied Sciences 12(4); 857-863, 2014.
- VE Lee, R. Jin, G. Agarwal, Frequent Pattern Mining in Data Streams, Frequent Pattern Mining, DOI 10.1007/978-3-319-07821-2 9, © Springer International Publishing Switzerland 2014.
- Juni Yang, Yanjun Wei, Fenfen Zhou, “An efficient algorithm for mining maximal frequent patterns over data streams,” 7th International Conference on Intelligent Human-Machine Systems and Cybernetics IEEE 2015.
- Daly, O.; Taniar, D. Exception Rules Mining Based On Negative Association Rules. Lect. Notes Comput. Sci. 2004, 3046, 543–552.
- Thiruvady, D.R.;Webb, G.I. Mining Negative Association Rules Using GRD. In Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining, Sydney, Australia, 26–28 May 2004; pp. 161–165.
- Antonie, M.; Zaiane, O.R. Mining Positive and Negative Association Rules: An Approach for Confined Rules.. In Proceedings of the 8th European Conference on Principles and Practice of Knowledge Discovery in Databases (PKDD04), Pisa, Italy, 20–24 September 2004; pp. 27–38.
- Cornelis, C.; Yan, P.; Zhang, X.; Chen, G. Mining Positive and Negative Association Rules from Large Databases. In Proceedings of the 2006 IEEE Conference on Cybernetics and Intelligent Systems, Bangkok, Thailand, 7–9 June 2006.
- Chongqing, China, 29–31 May 2012. 23. Kumar, N.V.S.P.; Rao, K.R. Mining Positive and Negative Regular Item-Sets using Vertical Databases. Int. J. Simul. Syst. Sci. Technol. 2016, 17, 33.1–33.4.
- Bagui, S.; Dhar, P.C. Positive and negative association rule mining in Hadoop’s MapReduce environment. J. Big Data 2019, 6, 75. [CrossRef]
- Kishor, P.; Porika, S. An efficient approach for mining positive and negative association rules from large transactional databases. In Proceedings of the 2016 International Conference on Inventive Computation Technologies (ICICT), Coimbatore, India, 26–27 August 2016.
- Ramasubbareddy, B.; Govardhan, A.; Ramamohanreddy, A. Mining positive and negative association rules. In Proceedings of the 5th International Conference on Computer Science and Education, Hefei, China, 24–27 August 2018; pp. 1403–1406.
- Kaur, S.; Singla, J.; Nkenyereye, L.; Jha, S.; Prashar, D.; Joshi, G.P.; El-Sappagh, S.; Islam, M.S.; Islam, S.M. Medical Diagnostic Systems Using Artificial Intelligence (AI) Algorithms: Principles and Perspectives. IEEE Access 2020, 8, 228049–228069. [CrossRef]
- Wei, J.; Lu, Z.; Qiu, K.; Li, P.; Sun, A.H. Predicting Drug Risk Level from Adverse Drug Reactions Using SMOTE and Machine, Learning Approaches. IEEE Access 2020, 8, 185761–185775. [CrossRef]
- Lu, Y.; Chen, S.; Zhang, H. Detecting Potential Serious Adverse Drug Reactions using Sequential Pattern Mining Method. In Proceedings of the 2018 IEEE 9th International Conference on Software Engineering and Service Science (ICSESS), Beijing, China, 23–25 November 2018; pp. 56–59.
- Lu, Y.; Seidl, T. Towards Efficient, Closed Infrequent Item set Mining using Bi-directional Traversing. In Proceedings of the 2018, IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA), Turin, Italy, 1–3 October 2018; pp. 140–149.
- Zhang, J.; Liu, W.; Wang, P. Drug-Drug Interaction Extraction from Chinese Biomedical Literature, using distant supervision. In Proceedings of the 2020 IEEE International Conference on Knowledge Graph (ICKG), Nanjing, China, 9–11 August 2020; pp. 593–598.
- Kim, Y.; Cho, Y. Predicting Drug–Gene–Disease Associations by Tensor Decomposition for Network-Based Computational Drug Repositioning. Biomedicines 2023, 11, 1998. [CrossRef]
- Aan Ahmed Toor, Muhammad Usman, Farah Younas, Alvis Cheuk M. Fong, Sajid Ali Khan 3 and Simon Fong, Mining Massive E-Health Data Streams for IoMT Enabled Healthcare Systems, sensors, 2020, 20, 2131.
- U. H. W. A. Hewage• R. Sinha • M. Asif Naeem, Privacy-preserving data (stream) mining techniques and their impact on data mining accuracy: a systematic literature review, Artificial Intelligence Review (2023) 56:10427–10464. [CrossRef]
- Aref Faisal Mourtada, Mining Association Rules Events over Data Streams, A Thesis submitted to Concordia Institute for Information Systems Engineering (CIISE), 2017.
- Nannan zhang, Xiaoqiang ren, and Xiangjun dong , An Effective Method for Mining Negative Sequential Patterns From Data Streams, IEEE Access, Vol 11, 2023, pp 31842 – 31854. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).