Submitted:
18 February 2024
Posted:
19 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
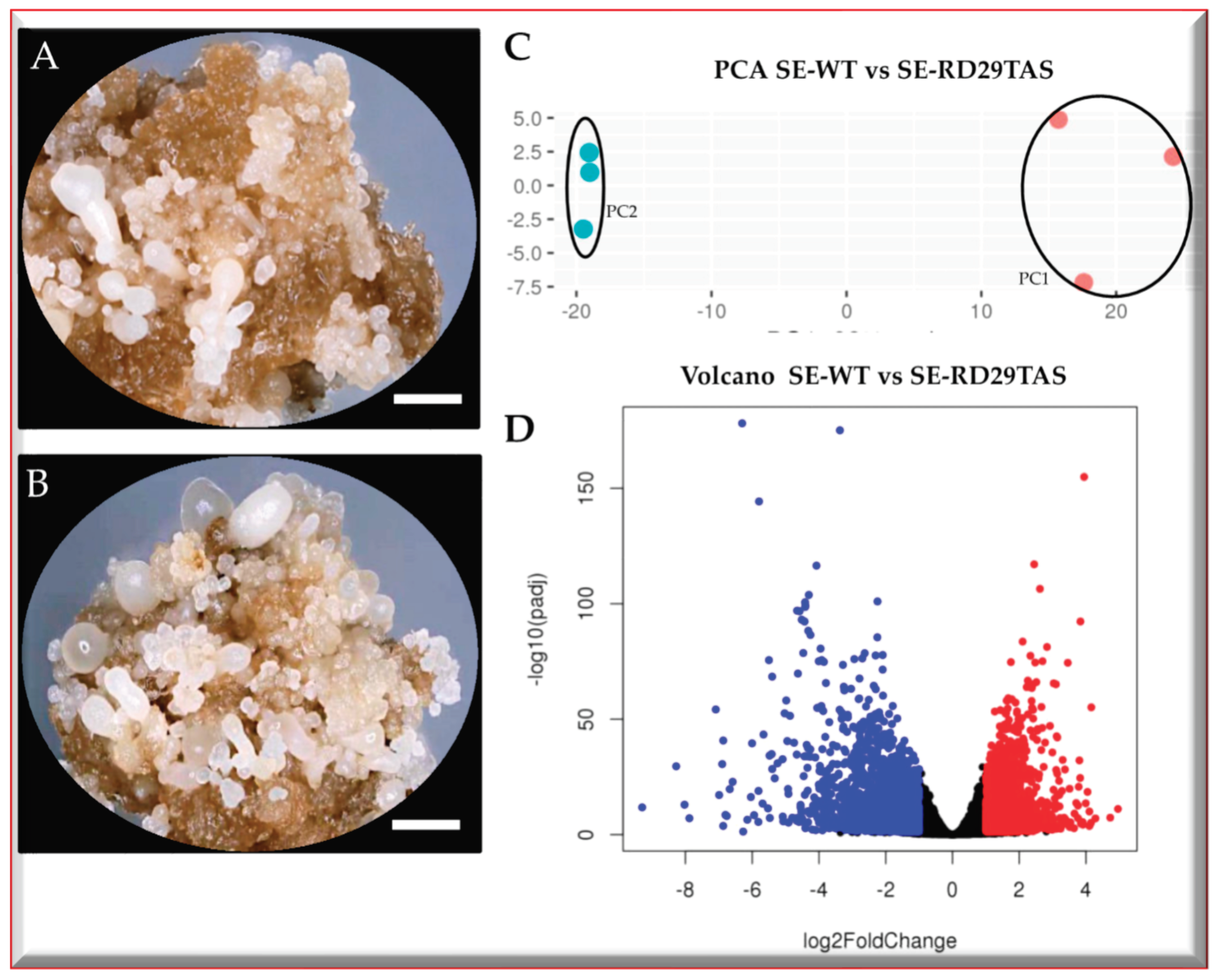
2.1. SE-RD29TAS transcriptomic-wide analysis
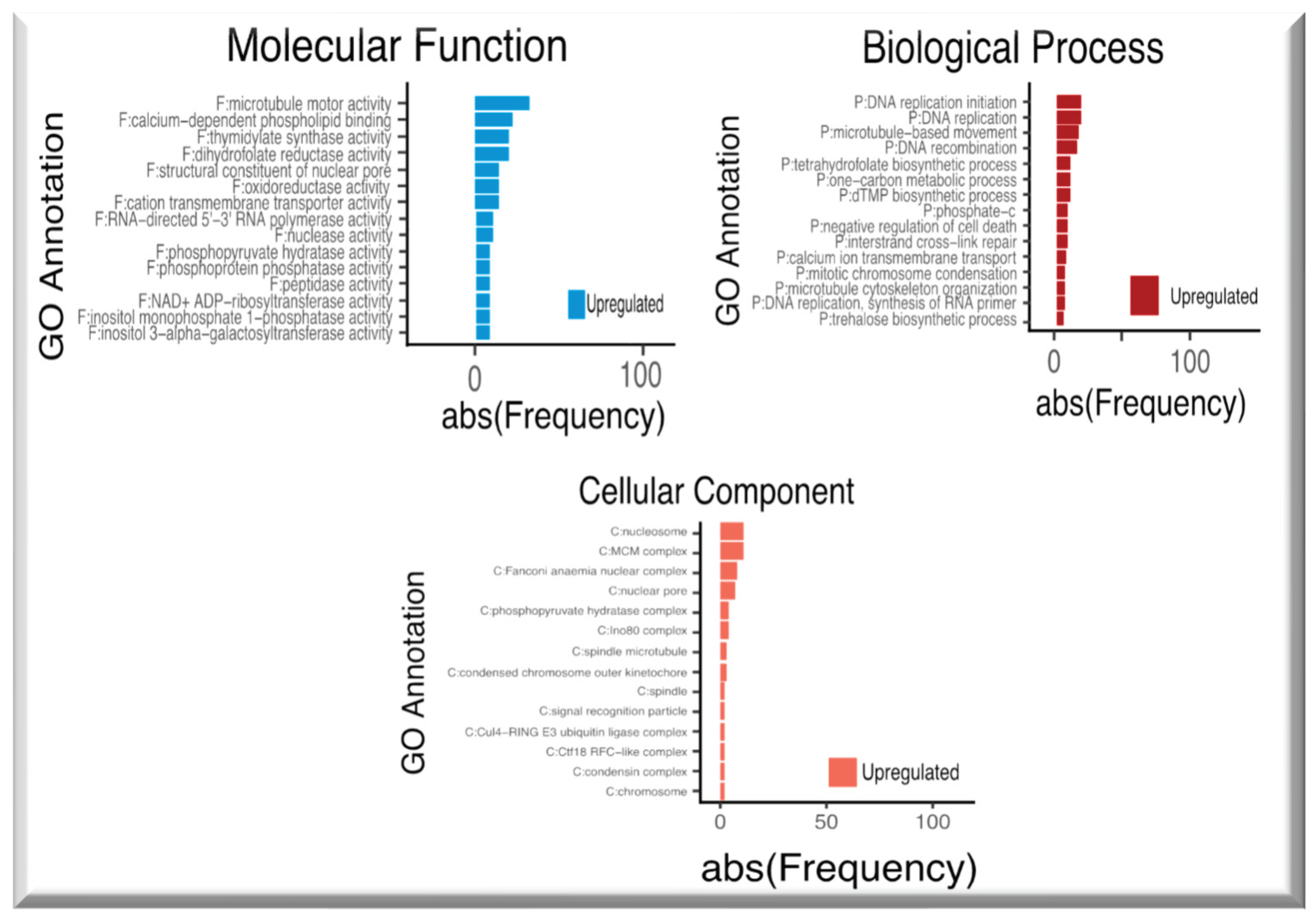
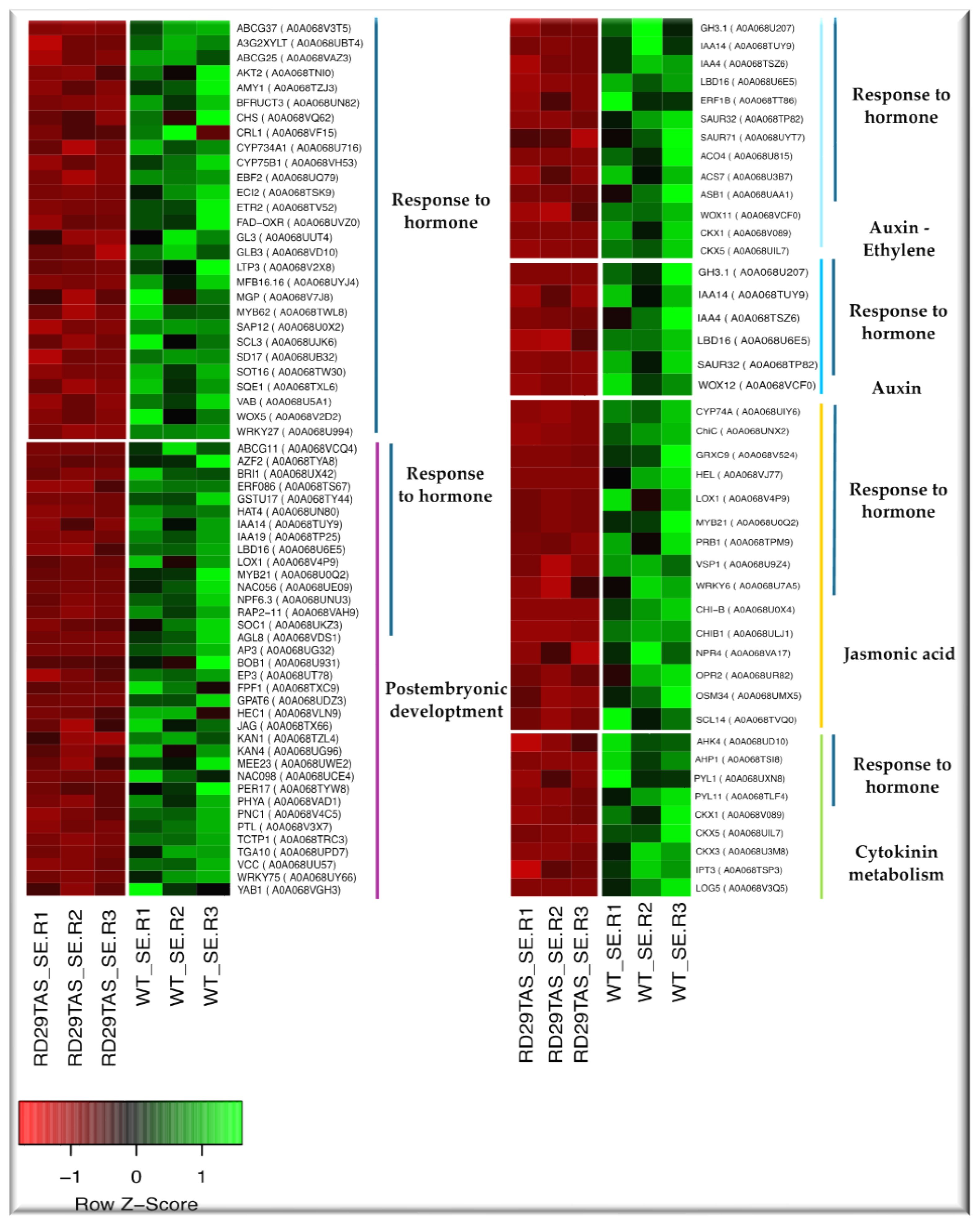
2.2. Up-regulated genes in SE-RD29TAS
2.3. Down-regulated genes in SE-RD29TAS
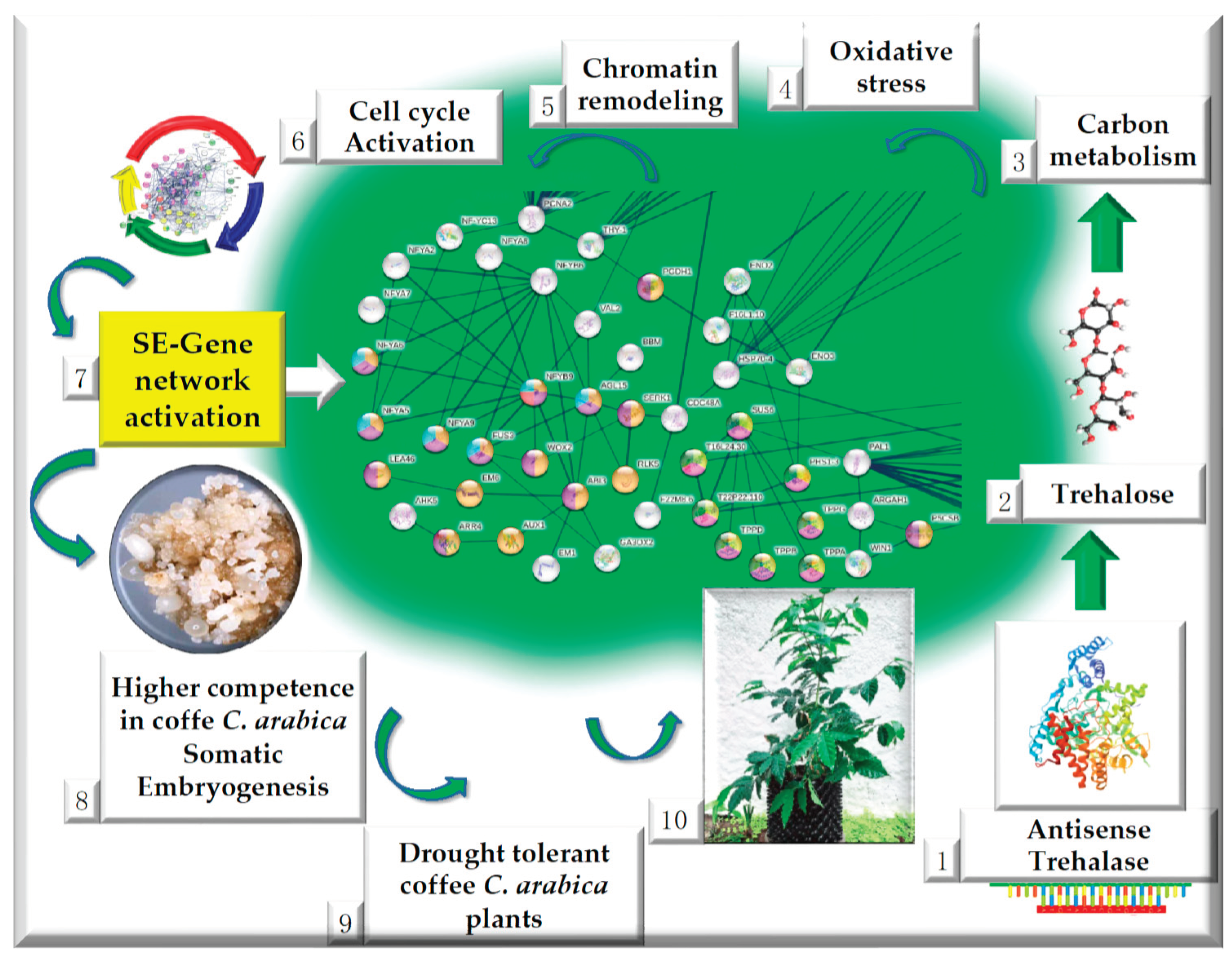
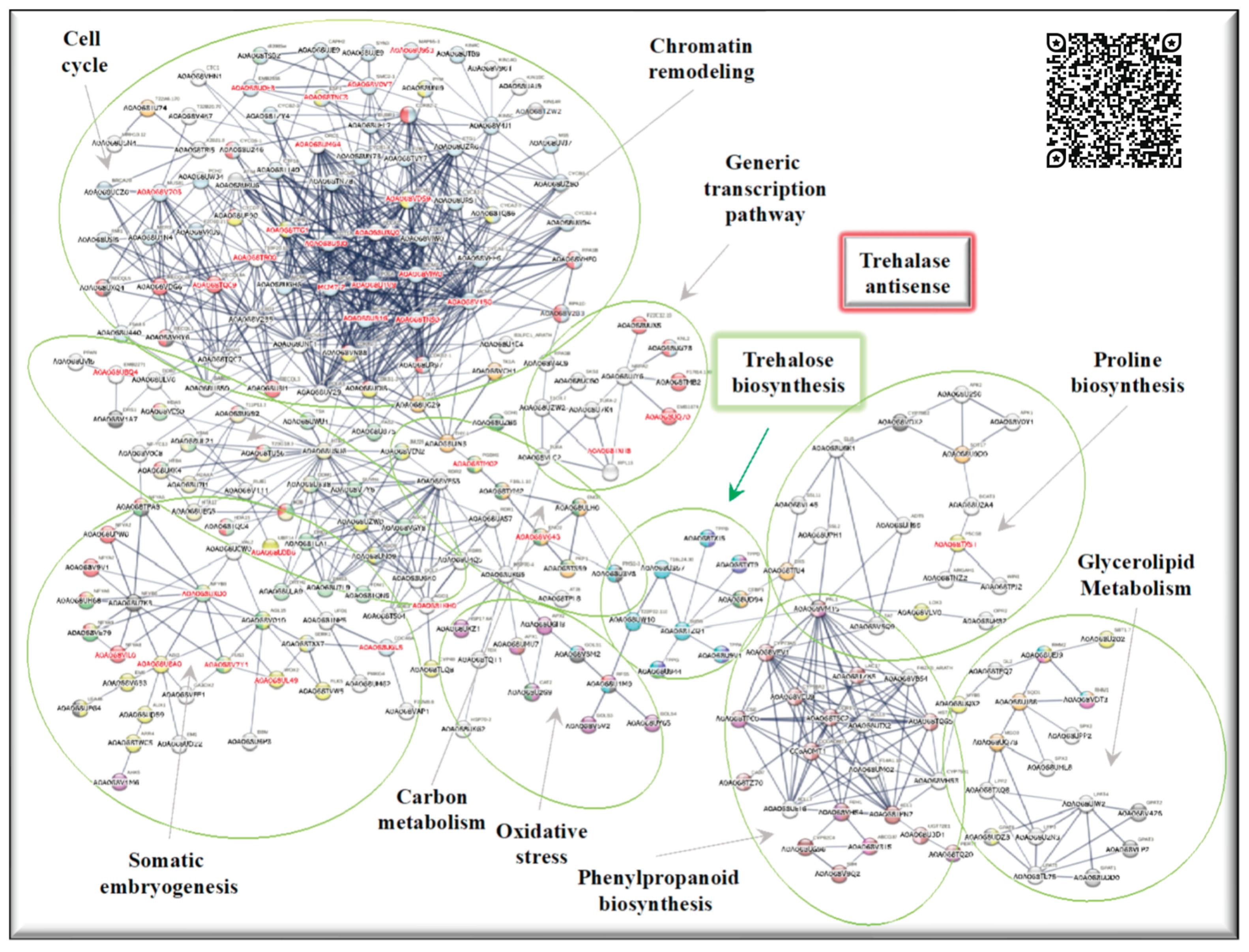
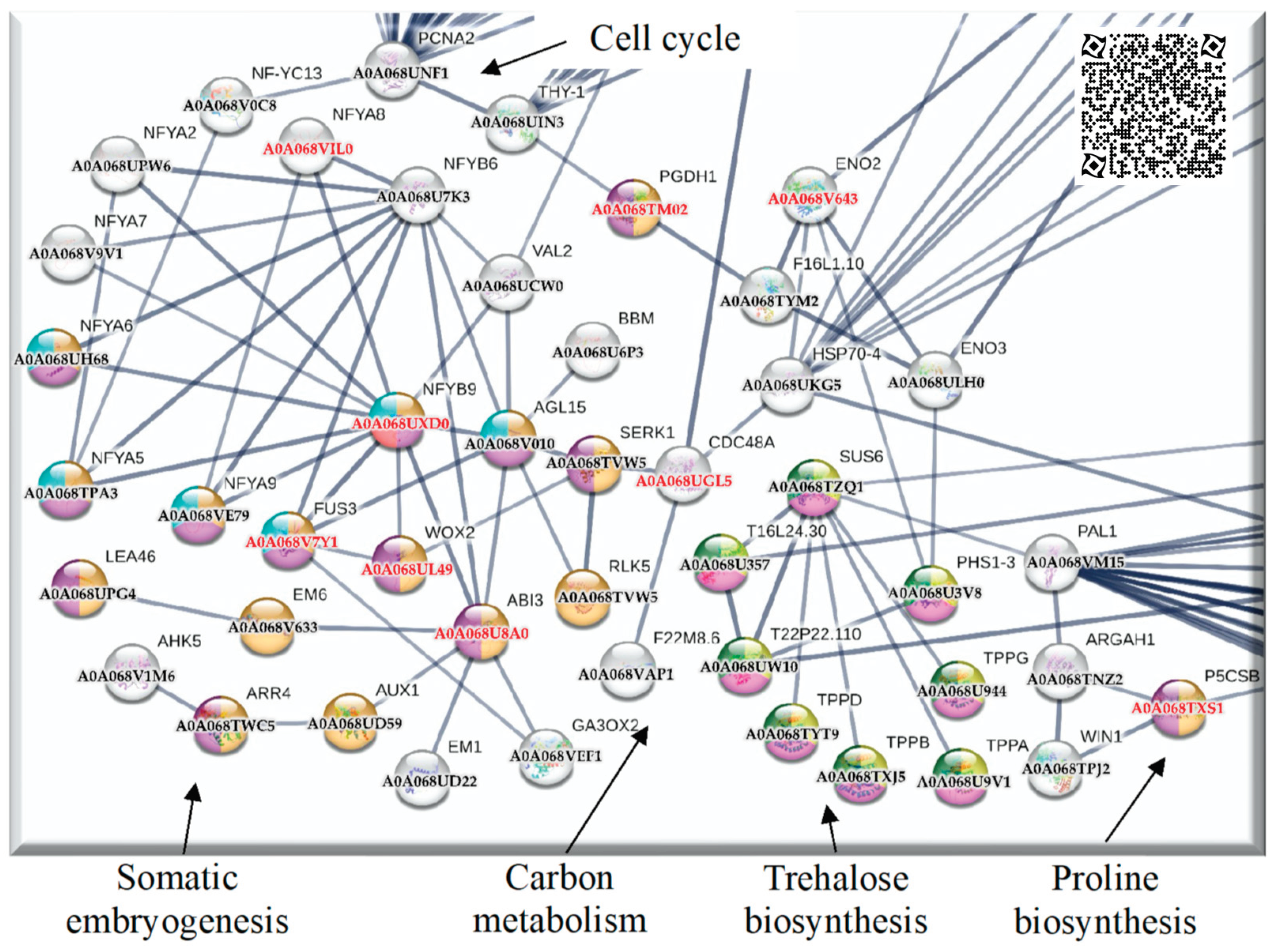
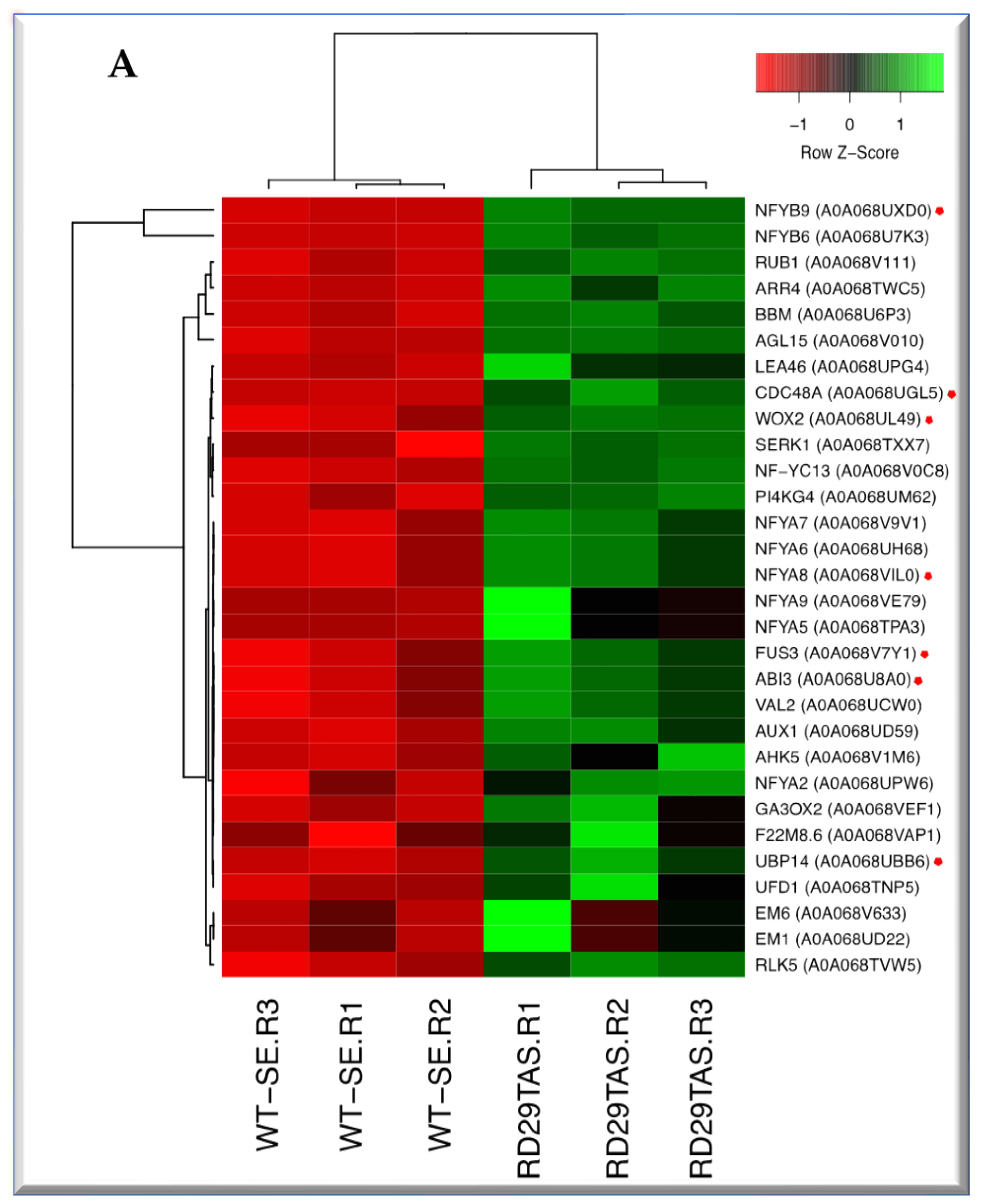
2.4. Up-regulated genes related to SE in SE-RD29TAS
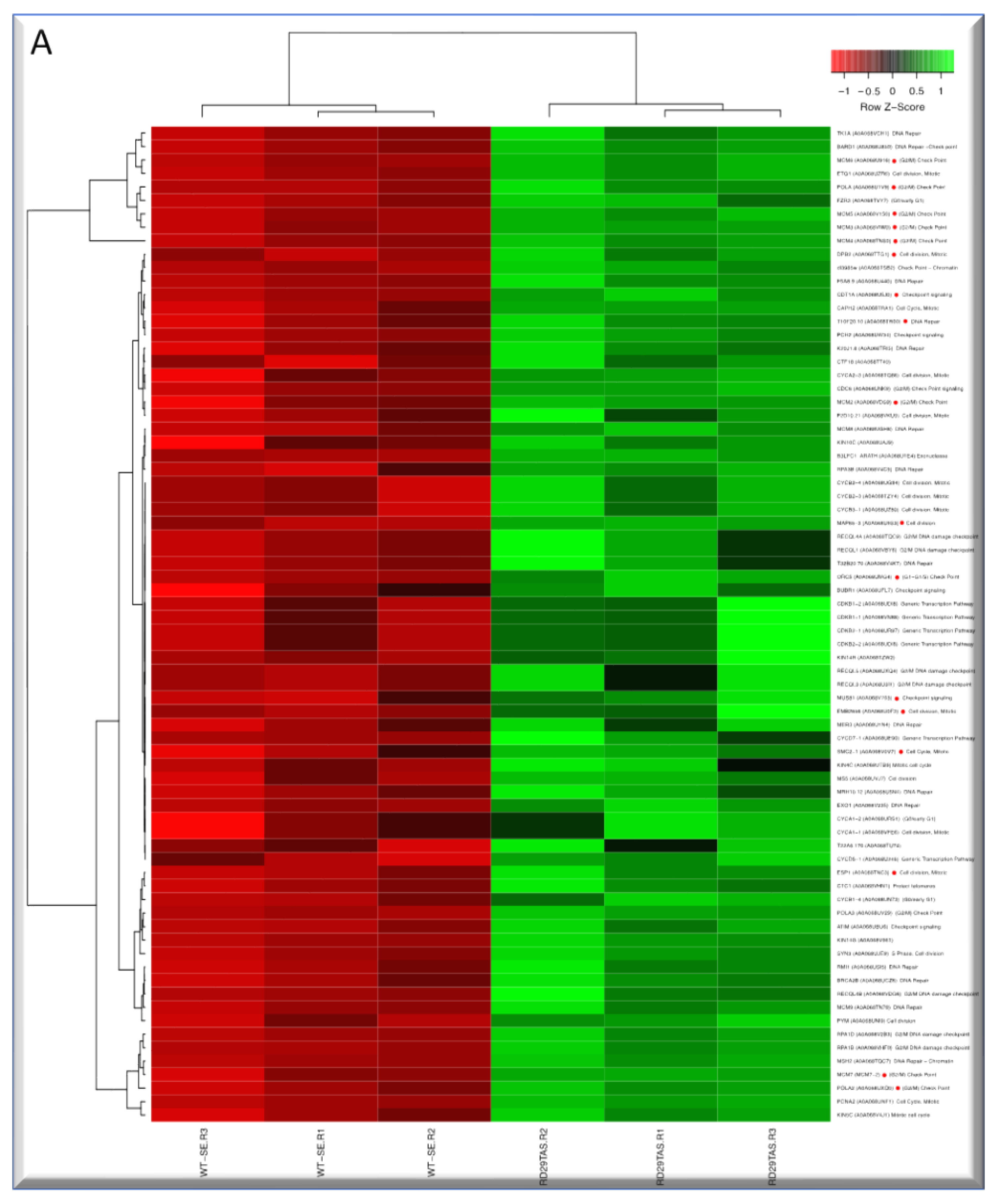
2.5. Up-regulated genes related to CC in SE-RD29TAS
2.6. Up-regulated genes related to chromatin remodeling in SE-RD29TAS
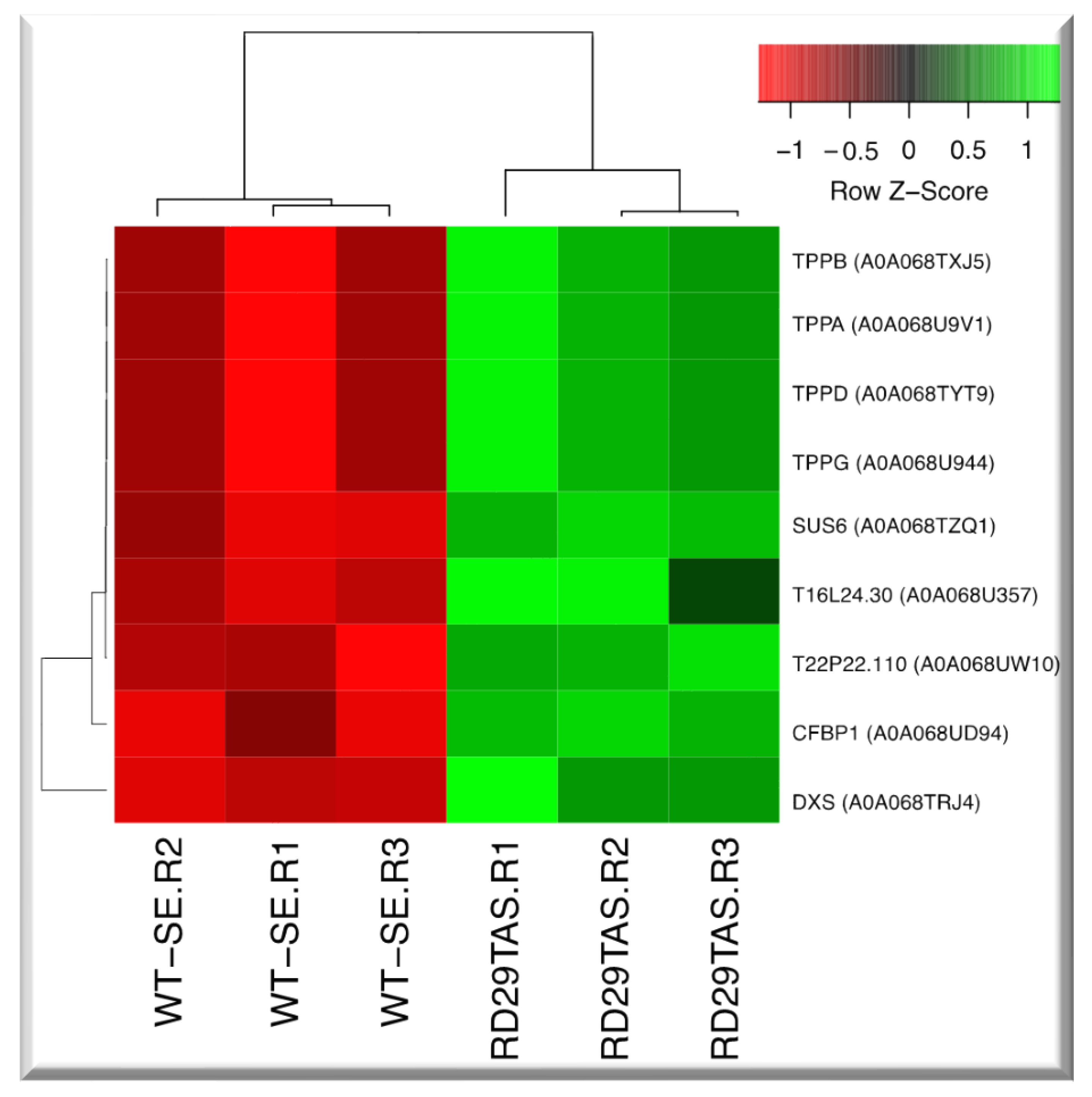
2.7. Up-regulated genes related to trehalose biosynthesis in SE-RD29TAS
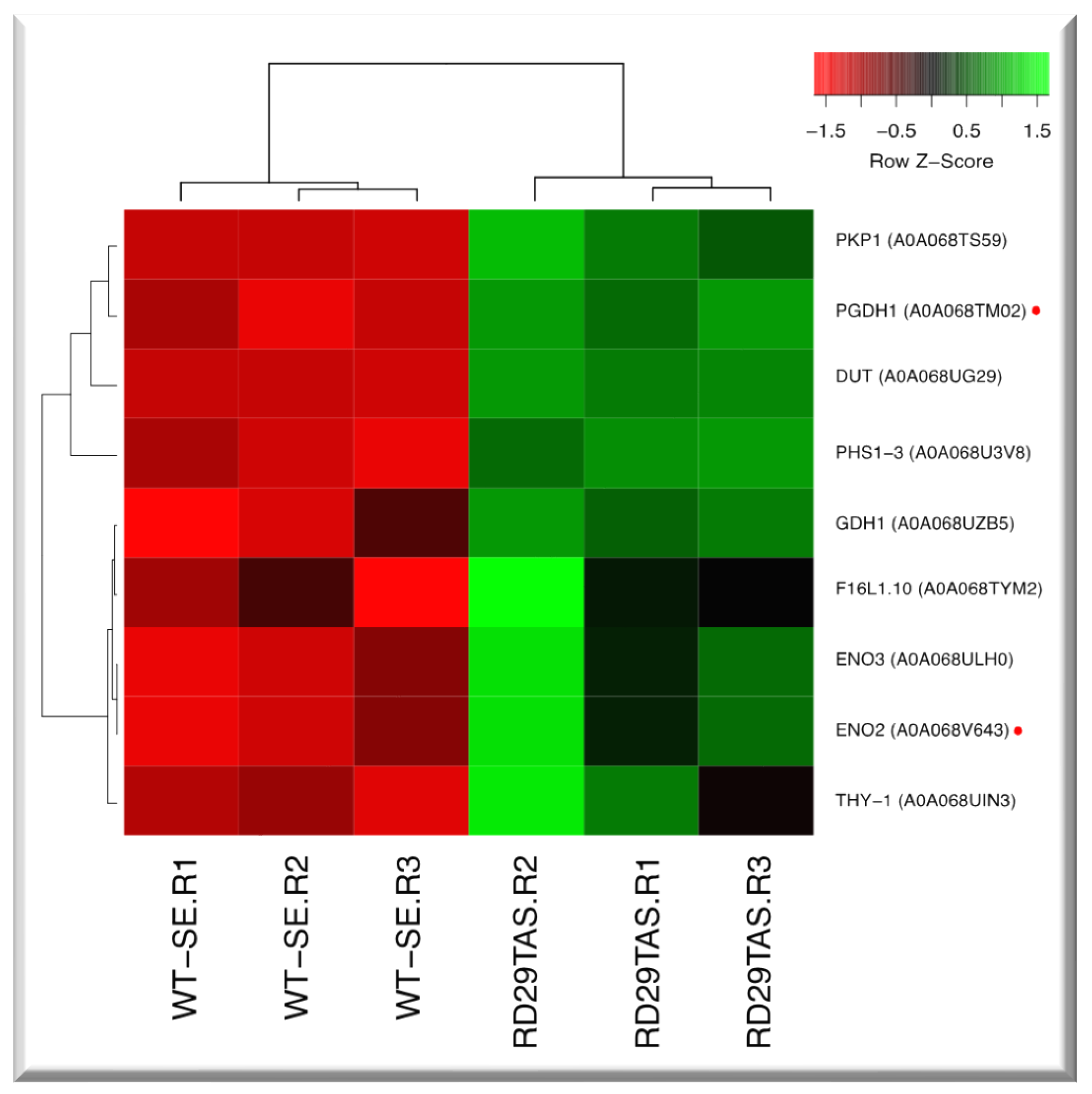
2.8. Up-regulated genes related to carbon metabolism in SE-RD29TAS
2.9. Up-regulated genes related to oxidative stress in SE-RD29TAS
2.10. Up-regulated genes related to phenylpropanoid biosynthesis, generic transcription pathway, glycerol-lipid metabolism, proline module in SE-RD29TAS.
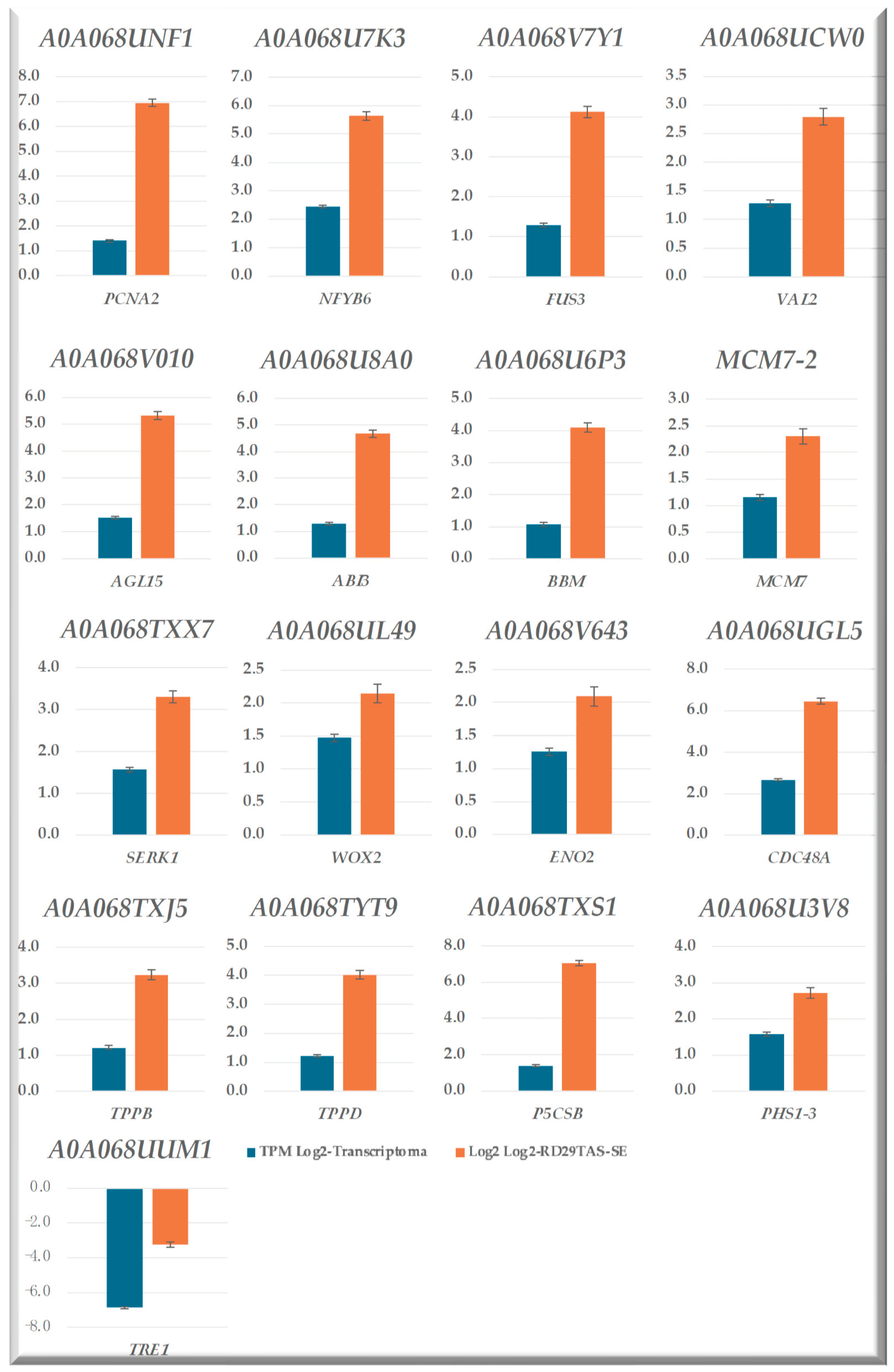
2.11. Validation of the Transcriptome-Wide Analysis
3. Discussion
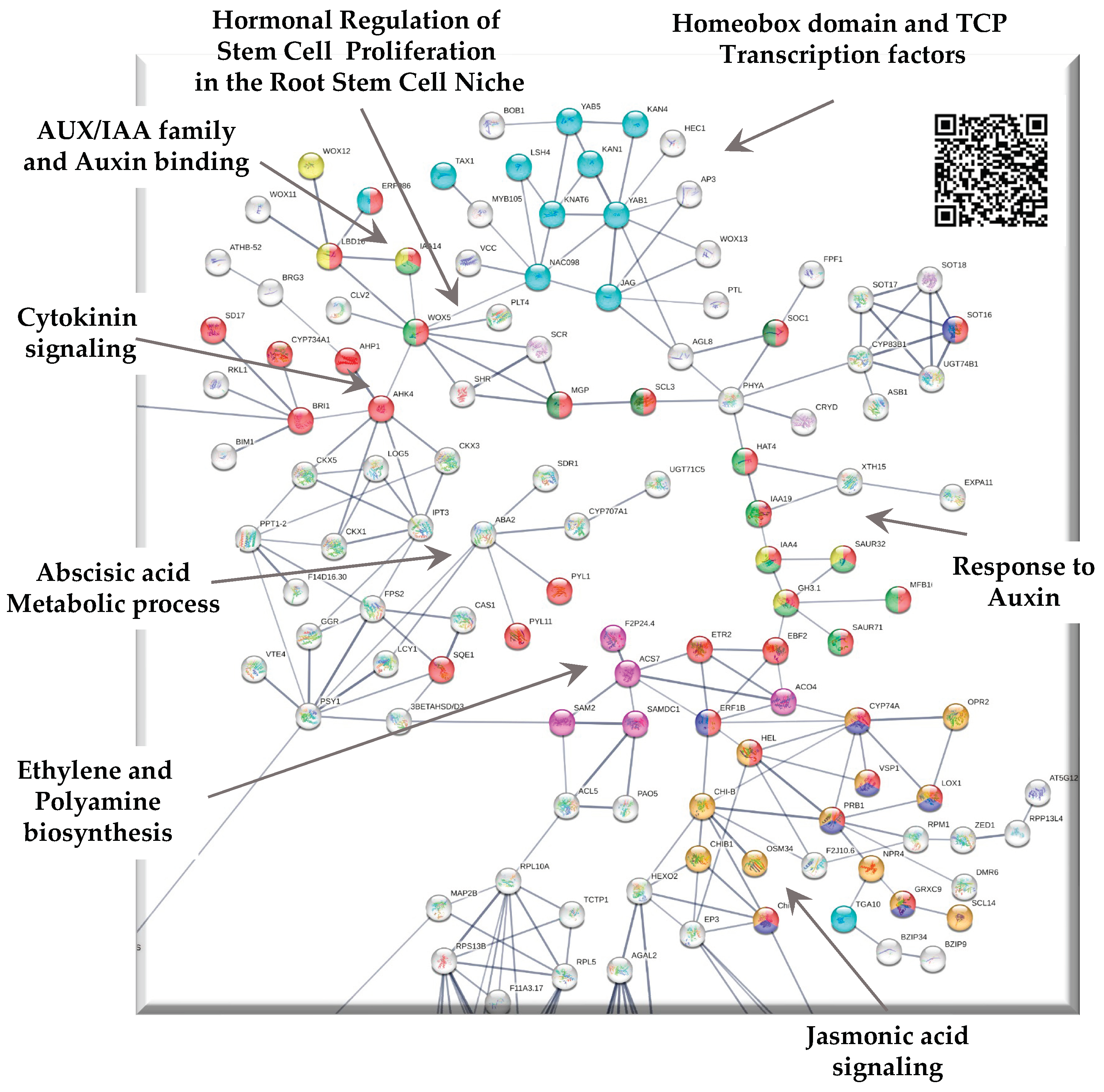
3.1. Up-regulated genes in SE-RD29TAS
3.1.1. Interaction with cell cycle
3.1.2. PCNA2 interacts with the somatic embryogenesis module
3.1.3. Proline biosynthesis module
3.2. Down-regulated genes in SE-RDTAS
4. Materials and Methods
4.1. Generation of marker-free SE-RD29TAS of coffee C. arabica L. var Typica.
4.2. RNA isolation and qPCR Analysis
4.3. Aligned and Analysis of DEG and GO in the SE-RD29TAS line of coffee C. arabica
4.4. PPI analysis of SE-RD29TAS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinheiro, H.A.; Da MATTA, F.M.; Chaves, A.R.; Loureiro, M.E.; Ducatti, C. Drought tolerance is associated with rooting depth and stomatal control of water use in clones of Coffea canephora. Ann. Bot. 2005, 96, 101–108. [Google Scholar] [CrossRef]
- Bunn, C.; Läderach, P.; Pérez Jimenez, J.G.; Montagnon, C.; Schilling, T. Multiclass classification of agroecological zones for Arabica coffee: an improved understanding of the impacts of climate change. PLoS ONE 2015, 10, e0140490. [Google Scholar] [CrossRef] [PubMed]
- ICO. International Coffee Organization. Coffee price continues in November reaching a 10-year high. Coffee Market Report. https://www.ico.org/documents/cy2021-22/cmr-1121-e.pdf/ (2021).
- Assad, E.D.; Pinto, H.; Zullo, J.; Junior, A.M.H. Impacto das mudanças climáticas no zoneamento agroclimático do café no Brasil. Pesqui Agropecu. Bras. 2004, 39, 1057–1064. [Google Scholar] [CrossRef]
- DaMatta, F. M, Ramalho, J.D.C. Impacts of drought and temperature stress on coffee physiology and production: a review. Braz. J. Plant Physiol. 2006, 18, 55–81. [Google Scholar] [CrossRef]
- Bertrand, B.; Marraccini, P.; Villain, L.; Breitler, J.C.; Etienne, H. Healthy tropical plants to mitigate the impact of climate change—As exemplified in coffee. Climate change and agriculture worldwide. 2016, 83–95. [Google Scholar] [CrossRef]
- Mofatto, L.S.; Carneiro, F.D.A.; Vieira, N.G.; Duarte, K.E.; Vidal, R.O.; Alekcevetch, J. C.; ... & Marraccini, P. Identification of candidate genes for drought tolerance in coffee by high-throughput sequencing in the shoot apex of different Coffea arabica cultivars. BMC Plant Biol. 2016, (1), 1-18. [CrossRef]
- DaMatta, F.M.; Avila, R.T.; Cardoso, A.A.; Martins, S.C.; Ramalho, J.C. Physiological and agronomic performance of the coffee crop in the context of climate change and global warming: A review. J. Agric. Food Chem. 2018, 66, 5264–5274. [Google Scholar] [CrossRef] [PubMed]
- Pham, Y.; Reardon-Smith, K.; Mushtaq, S.; Cockfield, G. The impact of climate change and variability on coffee production: a systematic review. Clim. Change 2019, 156, 609–630. [Google Scholar] [CrossRef]
- Ahmed, S.; Brinkley, S.; Smith, E.; Sela, A.; Theisen, M.; Thibodeau, C.; ... & Cash, S.B. Climate change and coffee quality: systematic review on the effects of environmental and management variation on secondary metabolites and sensory attributes of Coffea arabica and Coffea canephora. Front. Plant Sci. 2021, 12, 708013. [CrossRef]
- Marques, I.; Gouveia, D.; Gaillard, J.C.; Martins, S.; Semedo, M.C.; Lidon, F.C.; ... & Ramalho, J.C. Next-generation proteomics reveals a greater antioxidative response to drought in Coffea arabica than in Coffea canephora. Agronomy 2022, 12, 148. [CrossRef]
- Mofatto, L.S.; Carneiro, F.D.A.; Vieira, N.G.; Duarte, K.E.; Vidal, R.O.; Alekcevetch, J. C.; ... & Marraccini, P. Identification of candidate genes for drought tolerance in coffee by high-throughput sequencing in the shoot apex of different Coffea arabica cultivars. BMC Plant Biol. 2016, 16, 1–18. [CrossRef]
- Zeng, F.; Zhang, X.; Cheng, L.; Hu, L.; Zhu, L.; Cao, J.; Guo, X. A draft gene regulatory network for cellular totipotency reprogramming during plant somatic embryogenesis. Genomics 2007, 90, 620–628. [Google Scholar] [CrossRef]
- Karami, O.; Saidi, A. The molecular basis for stress-induced acquisition of somatic embryogenesis. Mol. Biol. Rep. 2010, 37, 2493–2507. [Google Scholar] [CrossRef]
- Elhiti, M.; Stasolla, C.; Wang, A. Molecular regulation of plant somatic embryogenesis. In Vitro Cell. Dev. Biol. Plant. 2013, 49, 631–642. [Google Scholar] [CrossRef]
- Horstman, A.; Bemer, M.; Boutilier, K. A transcriptional view on somatic embryogenesis. Regeneration 2017, 4, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Hernández, H.A.; Ledezma-Rodríguez, M.; Avilez-Montalvo, R.N.; Juárez-Gómez, Y.L.; Skeete, A.; Avilez-Montalvo, J.; ... & Loyola-Vargas, V.M. Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 2019, 10, 77. [CrossRef]
- Gulzar, B.; Mujib, A.; Malik, M.Q.; Sayeed, R.; Mamgain, J.; Ejaz, B. Genes, proteins and other networks regulating somatic embryogenesis in plants. J. Genet. Eng. Biotechnol. 2020, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Salaün, C.; Lepiniec, L.; Dubreucq, B. Genetic and molecular control of somatic embryogenesis. Plants 2021, 10, 1467. [Google Scholar] [CrossRef]
- de Silva, K.K.; Dunwell, J.M.; Wickramasuriya, A.M. Weighted Gene Correlation Network Analysis (WGCNA) of Arabidopsis Somatic Embryogenesis (SE) and Identification of Key Gene Modules to Uncover SE-Associated Hub Genes. Int. J. Genom. 2022, 4, 7471063. [Google Scholar] [CrossRef]
- Yuan, H.Y.; Kagale, S.; Ferrie, A.M.R. Multifaceted roles of transcription factors during plant embryogenesis. Front. Plant Sci. 2024, 14, 1322728–1322728. [Google Scholar] [CrossRef]
- Tonietto, Â.; Sato, J.H.; Teixeira, J.B.; de Souza, E.M.; Pedrosa, F.O.; Franco, O.L.; Mehta, A. 2012. Proteomic analysis of developing somatic embryos of Coffea arabica. Plant Mol. Biol. Rep. 2012, 30, 1393–1399. [Google Scholar] [CrossRef]
- Pinto, R.T.; Freitas, N.C.; Máximo, W.P.F.; Cardoso, T.B.; Prudente, D.D.O.; Paiva, L.V. 2019. Genome-wide analysis, transcription factor network approach and gene expression profile of GH3 genes over early somatic embryogenesis in Coffea spp. BMC genom. 2019, 20, 1–15. [Google Scholar] [CrossRef]
- Quintana-Escobar, A.O.; Nic-Can, G.I.; Avalos, R.M.G.; Loyola-Vargas, V.M.; Gongora-Castillo, E. Transcriptome analysis of the induction of somatic embryogenesis in Coffea canephora and the participation of ARF and Aux/IAA genes. PeerJ 2019, 7, e7752. [Google Scholar] [CrossRef]
- Quintana-Escobar, A.O.; Bojórquez-Velázquez, E.; Ruiz-May, E.; Loyola-Vargas, V.M. Proteomic Approach during the Induction of Somatic Embryogenesis in Coffea canephora. Plants 2023, 12, 4095. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Hernández, H.A.; Quintana-Escobar, A.O.; Uc-Chuc, M.A.; Loyola-Vargas, V.M. Genome-wide analysis, modeling, and identification of amino acid binding motifs suggest the involvement of GH3 genes during somatic embryogenesis of Coffea canephora. Plants 2021, 10, 2034. [Google Scholar] [CrossRef]
- Awada, R.; Lepelley, M.; Breton, D.; Charpagne, A.; Campa, C.; Berry, V.; ... & Etienne, H. Global transcriptome profiling reveals differential regulatory, metabolic and hormonal networks during somatic embryogenesis in Coffea arabica. BMC genom. 2023, 24, 41.
- Birch, G.G. Trehaloses. Adv. Carbohydr. Chem. 1963, 18, 201–225. [Google Scholar] [CrossRef] [PubMed]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 1–15. [Google Scholar] [CrossRef]
- Müller, J.; Aeschbacher, R.A.; Wingler, A.; Boller, T.; Wiemken, A. Trehalose and trehalase in Arabidopsis. Plant Physiol. 2001, 125, 1086–1093. [Google Scholar] [CrossRef]
- Gámez Escobedo, I.A.; Cabrera Ponce, J.L.; Herrera Estrella, L.R.; Hernández Luna, C. E.; Montes de Oca Luna, R. Mejora del crecimiento de plantas de tabaco mediante la inhibición del gen de la trehalasa. Ciencia UANL 2004, 7, 4. [Google Scholar]
- Kenny Alejandra AGREDA LAGUNA, José Luis (MX). CABRERA PONCE, Analí (MX). GAMEZ ESCOBEDO, Luis Rafael (MX). HERRERA ESTRELLA, Roberto (MX). MONTES DE OCA LUNA, Roberto (MX). RUÍZ MEDRANO, Beatriz (MX). XOCONOSTLE CÁZARES, Luis Antonio (MX) Agent: CARREÑO SÁNCHEZ, Av. Instituto: (EN) METHODS TO OBTAIN DROUGHT RESISTANT PLANTS (FR) PROCÉDÉS POUR OBTENIR DES PLANTES RÉSISTANTES À LA SÉCHERESSE. Year: 01/2012. (WO2012085806) METHODS TO OBTAIN DROUGHT RESISTANT PLANTS.
- Agreda-Laguna, K.A.; Cabrera-Ponce, J.L.; Medrano, R.R.; Garzon-Tiznado, J.A.; Xoconostle-Cazares, B. TREHALOSE ACCUMULATION PROVIDES DROUGHT TOLERANCE TO GENETICALLY-MODIFIED MAIZE IN OPEN FIELD TRIALS. Pak. J. Agric. Sci. 2018, 55. [Google Scholar]
- Valencia-Lozano, E.; Cabrera-Ponce, J.L.; Gómez-Lim, M.A.; Ibarra, J.E. Development of an efficient protocol to obtain transgenic coffee, Coffea arabica L.; expressing the Cry10Aa toxin of Bacillus thuringiensis. Int. J. Mol. Sci. 2019, 20, 5334. [Google Scholar] [CrossRef]
- Meinke, D.W. Genome-wide identification of EMBRYO-DEFECTIVE EMB genes required for growth and development in Arabidopsis. New Phytol. 2020, 226, 306–325. [Google Scholar] [CrossRef]
- Byrareddy, V.; Kouadio, L.; Mushtaq, S.; Kath, J.; Stone, R. Coping with drought: Lessons learned from robusta coffee growers in Vietnam. Clim. Serv. 2021, 22, 100229. [Google Scholar] [CrossRef]
- de Freitas Guedes, F.A., Nobres, P., Ferreira, D.C.R., Menezes-Silva, P.E., Ribeiro-Alves, M., Correa, R.L., ... & Alves-Ferreira, M. (2018). Transcriptional memory contributes to drought tolerance in coffee (Coffea canephora) plants. Environ. Exp. Bot. 2018, 147, 220–233. [CrossRef]
- Fernandes, I.; Marques, I.; Paulo, O. S.; Batista, D.; Partelli, F.L.; Lidon, F.C.; ... & Ribeiro-Barros, A.I. Understanding the impact of drought in Coffea genotypes: Transcriptomic analysis supports a common high resilience to moderate water deficit but a genotype dependent sensitivity to severe water deficit. Agronomy 2021, 11, 2255. [CrossRef]
- Zrenner, R.; Salanoubat, M.; Willmitzer, L.; Sonnewald, U. Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants Solanum tuberosum L. Plant J. 1995, 7, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Craig, J.; Barratt, P.; Tatge, H.; Déjardin, A.; Handley, L.; Gardner, C.D.; ... & Smith, A. M. Mutations at the rug4 locus alter the carbon and nitrogen metabolism of pea plants through an effect on sucrose synthase. Plant J. 1999, 17, 353–362. [CrossRef]
- Zeeman, S.C.; Thorneycroft, D.; Schupp, N.; Chapple, A.; Weck, M.; Dunstan, H.; ... & Smith, S.M. Plastidial α-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol. 2004, 135, 849–858. [CrossRef]
- Geshi, N.; Johansen, J.N.; Dilokpimol, A.; Rolland, A.; Belcram, K.; Verger, S.; ... & Mouille, G. A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J. 2013, 76, 128–137. [CrossRef]
- Eremina, M.; Rozhon, W.; Yang, S.; Poppenberger, B. ENO 2 activity is required for the development and reproductive success of plants and is feedback-repressed by A t MBP-1. Plant J. 2015, 81, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, H.; Bing, J.; Zhang, G. Integrative transcriptomic and TMT-based proteomic analysis reveals the mechanism by which AtENO2 affects seed germination under salt stress. Front. Plant Sci. 2022, 13, 1035750. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Tan, H.; Zheng, Q.; Fu, F.; Liang, Y.; Zhang, J.; ... & Zuo, J. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 1042–1054. [CrossRef]
- Zhao, B.T.; Ge, L.F.; Liang, R.Q.; Li, W.; Ruan, K.C.; Lin, H.X.; Jin, Y.X. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 2009, 10, 29–41. [Google Scholar] [CrossRef]
- Mu, J.; Tan, H.; Hong, S.; Liang, Y.; Zuo, J. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol. Plant. 2013, 6, 188–201. [Google Scholar] [CrossRef]
- West, M.A.; Yee, K.M.; Danao, J.; Zimmerman, J.L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 1994, 6, 1731–1745. [Google Scholar] [CrossRef]
- Lotan, T.; Ohto, M.A.; Yee, K.M.; West, M.A.; Lo, R.; Kwong, R.W.; ... & Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [CrossRef]
- Gaj, M.D.; Zhang, S.; Harada, J.J.; Lemaux, P.G. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 2005, 222, 977–988. [Google Scholar] [CrossRef]
- Fornari, M.; Calvenzani, V.; Masiero, S.; Tonelli, C.; Petroni, K. The Arabidopsis NF-YA3 and NF-YA8 genes are functionally redundant and are required in early embryogenesis. PLoS ONE 2013, 8, e82043. [Google Scholar] [CrossRef] [PubMed]
- Thakare, D.; Tang, W.; Hill, K.; Perry, S.E. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiol. 2008, 146, 1663–1672. [Google Scholar] [CrossRef] [PubMed]
- Nowak, K.; Morończyk, J.; Wójcik, A.; Gaj, M.D. AGL15 controls the embryogenic reprogramming of somatic cells in Arabidopsis through the histone acetylation-mediated repression of the miRNA biogenesis genes. Int. J. Mol. Sci. 2020, 21 18, 6733. [Google Scholar] [CrossRef]
- Suzuki, M.; Wang, H.H.Y.; McCarty, D.R. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 2007, 143, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef]
- Jia, H.; McCarty, D.R.; Suzuki, M. Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol. 2013, 163, 1293–1305. [Google Scholar] [CrossRef]
- Freitas, N.C.; Barreto, H.G.; Torres, L.F.; Freire, L.L.; Rodrigues, L.A.Z.; Diniz, L.E.C.; ... & Paiva, L.V. In silico and in vivo analysis of ABI3 and VAL2 genes during somatic embryogenesis of Coffea arabica: competence acquisition and developmental marker genes. Plant Cell Tissue Organ Cult. PCTOC 2019, 137, 599–611. [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L., ... & van Lookeren Campagne, M.M. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [CrossRef]
- Hecht, V.; Vielle-Calzada, J.P.; Hartog, M.V.; Schmidt, E.D.L.; Boutilier, K.; Grossniklaus, U.; De Vries, S.C. The Arabidopsis somatic embryogenesis receptor kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 2001, 127, 803–816. [Google Scholar] [CrossRef]
- Albrecht, C.; Russinova, E.; Hecht, V.; Baaijens, E.; de Vries, S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 2005, 17, 3337–3349. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi-Darvari, F.; Noor, N.M.; Ismanizan, I. Epigenetic regulation and gene markers as signals of early somatic embryogenesis. Plant Cell, Tissue Organ Cult. PCTOC 2015, 120, 407–422. [Google Scholar] [CrossRef]
- Harada, J.J.; Belmonte, M.F.; Kwong, R.W. Plant embryogenesis (zygotic and somatic). eLS. John Wiley & Sons Ltd, Chichester. 2010. [CrossRef]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; ... & Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811. [CrossRef]
- Nambara, E.; Keith, K.; McCourt, P.; Naito, S. A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 1995, 121, 629–636. [Google Scholar] [CrossRef]
- Keith, K.; Kraml, M.; Dengler, N.G.; McCourt, P. fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 1994, 6, 589–600. [Google Scholar] [CrossRef]
- Roberts, J.K.; DeSimone, N.A.; Lingle, W.L.; Dure 3rd, L. Cellular concentrations and uniformity of cell-type accumulation of two lea proteins in cotton embryos. Plant Cell 1993, 5, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Stasolla, C.; Bozhkov, P.V.; Chu, T. M.; Van Zyl, L.; Egertsdotter, U.; Suarez, M. F.; ... & Sederoff, R. R. Variation in transcript abundance during somatic embryogenesis in gymnosperms. Tree Physiol. 2004, 24, 1073–1085. [CrossRef]
- Müller, B.; Sheen, J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 2008, 453, 1094–1097. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, J.M.; Cortizo, M.; Ordás, R.J. Characterization of a type-A response regulator differentially expressed during adventitious caulogenesis in Pinus pinaster. J. Plant. Physiol. 2012, 169, 1807–1814. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, W.; Toyomasu, T.; Masui, H.; Katho, T.; Nakaminami, K.; Kashiwagi, Y.; ... & Kamada, H. Gibberellin is essentially required for carrot Daucus carota L. somatic embryogenesis: dynamic regulation of gibberellin 3-oxidase gene expressions. Biosci. Biotechnol. Biochem. 2003, 67, 2438–2447. [CrossRef]
- Curaba, J.; Moritz, T.; Blervaque, R.; Parcy, F.; Raz, V.; Herzog, M.; Vachon, G. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004, 136, 3660–3669. [Google Scholar] [CrossRef] [PubMed]
- Haecker, A.; Groß-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef]
- Park, S.; Rancour, D.M.; Bednarek, S.Y. In planta analysis of the cell cycle-dependent localization of AtCDC48A and its critical roles in cell division, expansion, and differentiation. Plant Physiol. 2008, 148, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Wehmeyer, N.; Hernandez, L.D.; Finkelstein, R.R.; Vierling, E. Synthesis of small heat-shock proteins is part of the developmental program of late seed maturation. Plant Physiol. 1996, 112, 747–757. [Google Scholar] [CrossRef]
- Leng, L.; Liang, Q.; Jiang, J.; Zhang, C.; Hao, Y.; Wang, X.; Su, W. A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana. J. Plant Res. 2017, 130, 349–363. [Google Scholar] [CrossRef]
- Sun, W.; Bernard, C.; Van De Cotte, B.; Van Montagu, M.; Verbruggen, N. At-HSP17. 6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J. 2001, 27, 407–415. [Google Scholar] [CrossRef]
- Sung, D.Y.; Kaplan, F.; Guy, C.L. Plant Hsp70 molecular chaperones: protein structure, gene family, expression and function. Physiol. Plant 2001, 113, 443–451. [Google Scholar] [CrossRef]
- Mattioli, R.; Marchese, D.; D'Angeli, S.; Altamura, M.M.; Costantino, P.; Trovato, M. Modulation of intracellular proline levels affects flowering time and inflorescence architecture in Arabidopsis. Plant Mol. Biol. 2008, 66, 277–288. [Google Scholar] [CrossRef]
- Funck, D.; Winter, G.; Baumgarten, L.; Forlani, G. Requirement of proline synthesis during Arabidopsis reproductive development. BMC Plant Biol. 2012, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Biancucci, M.; Mattioli, R.; Forlani, G.; Funck, D.; Costantino, P.; Trovato, M. Role of proline and GABA in sexual reproduction of angiosperms. Front. Plant Sci. 2015, 6, 680. [Google Scholar] [CrossRef] [PubMed]
- Capron, A.; Chatfield, S., Provart, N.; Berleth, T. (2009). Embryogenesis: pattern formation from a single cell. The Arabidopsis Book/American Society of Plant Biologists, 7. [CrossRef]
- Valencia-Lozano, E.; Barraza, A.; Ibarra,J.; Cabrera-Ponce, J.L. Marker-free Coffea arabica plants mediated by antisense trehalase: Activation of somatic embryogenesis and drought tolerance. In preparation. Special issue: Molecular mechanisms involved in somatic embryogenesis and organogenesis. Plants 2024.
- Valencia-Lozano, E.; Cabrera-Ponce, J.L.; Noa-Carrazana, J.C.; Ibarra, J.E. Coffea arabica L. resistant to coffee berry borer Hypothenemus hampei mediated by expression of the Bacillus thuringiensis Cry10Aa protein. Front. Plant Sci. 2021, 12, 765292. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Von Mering, C. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




