Submitted:
17 February 2024
Posted:
20 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Germination and Growth Conditions
2.2. Treatment and Sampling
2.3. Elemental Analysis
2.4. Metabolites Extraction
2.5. GC-TOF-MS Analysis
2.6. Data Preprocessing and Annotation
2.7. Data Analysis
3. Results
3.1. Elemental Concentrations
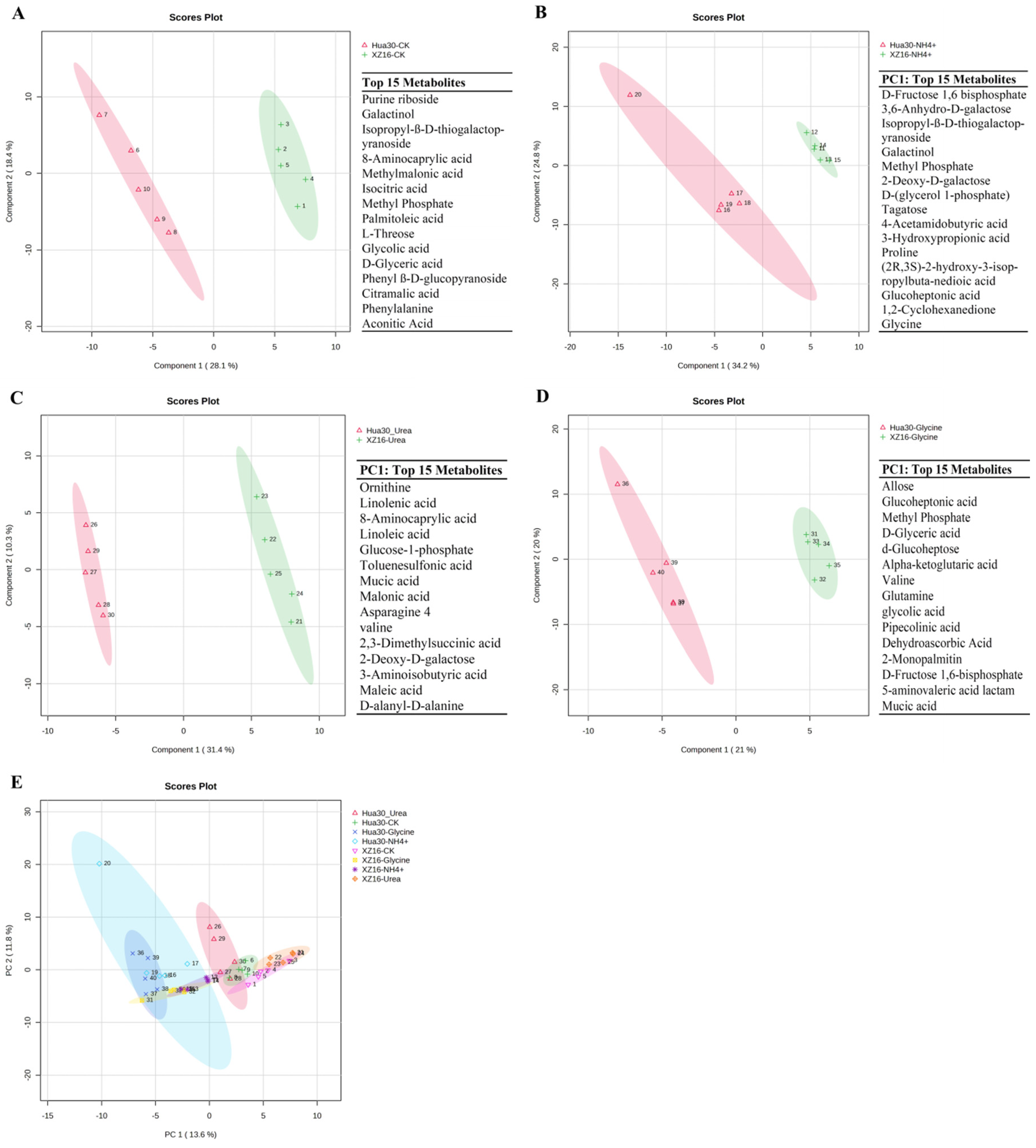
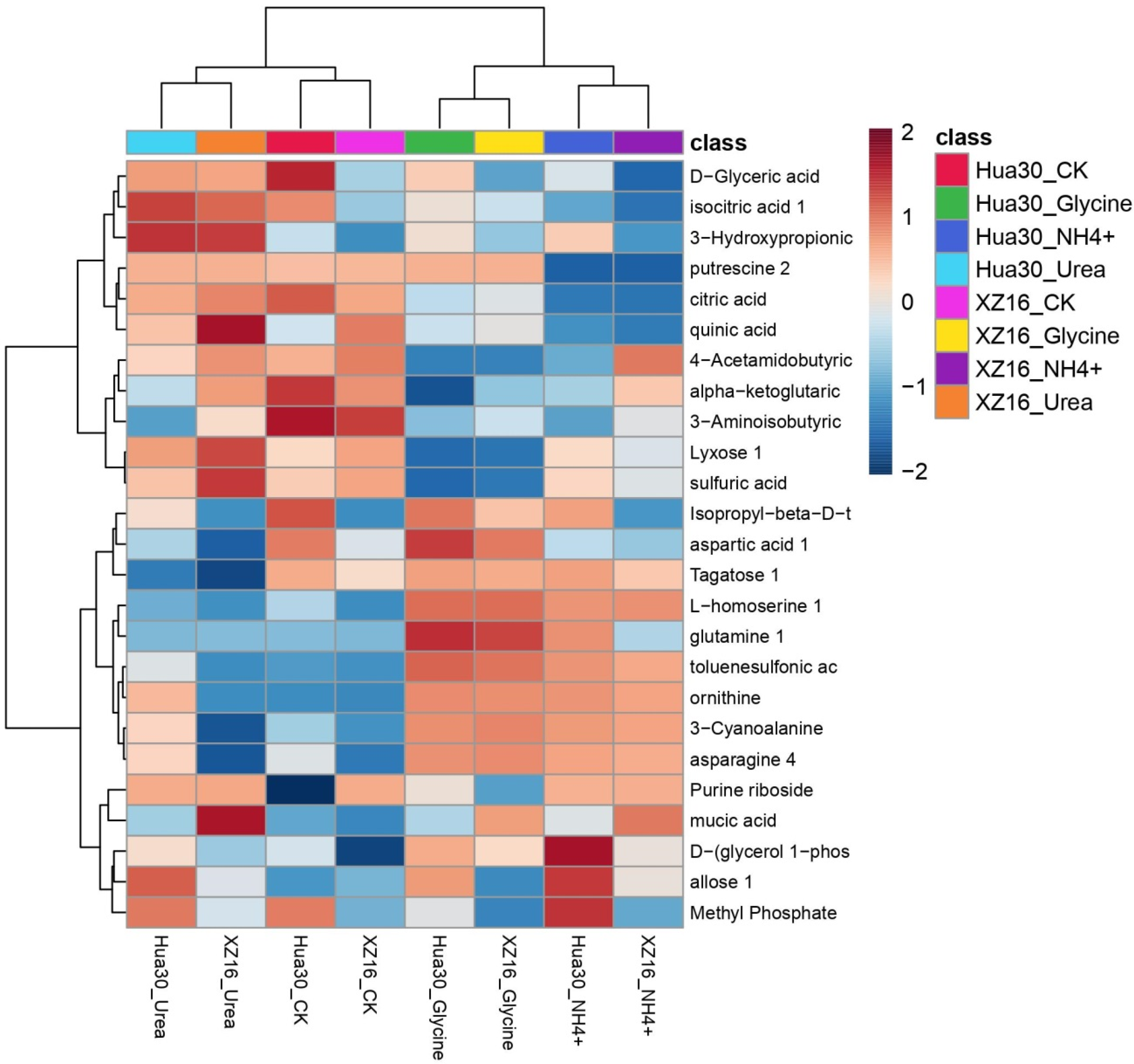
3.2. Metabolomics Profiles of XZ16 and Hua30
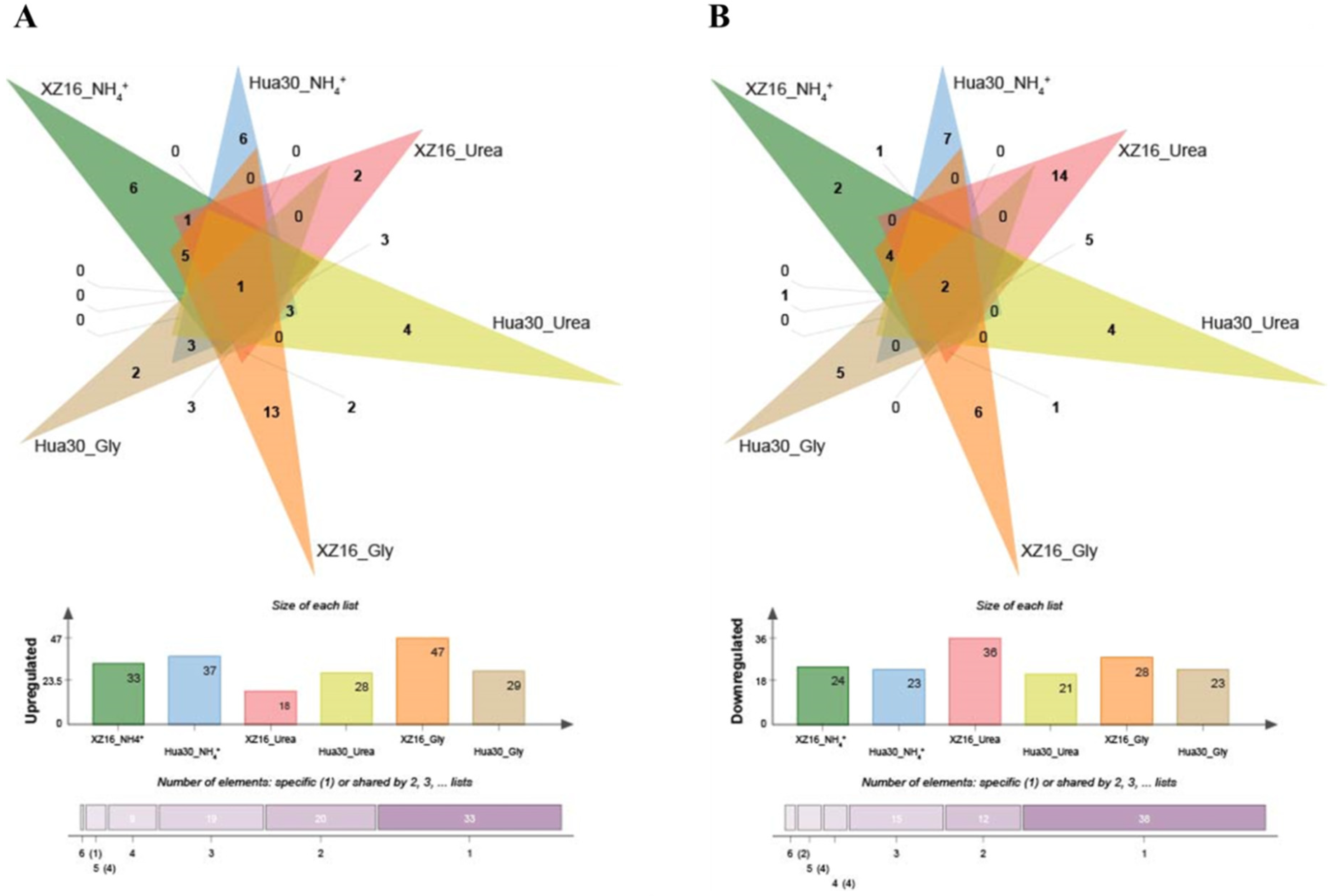
3.3. Differentially Expressed Metabolites
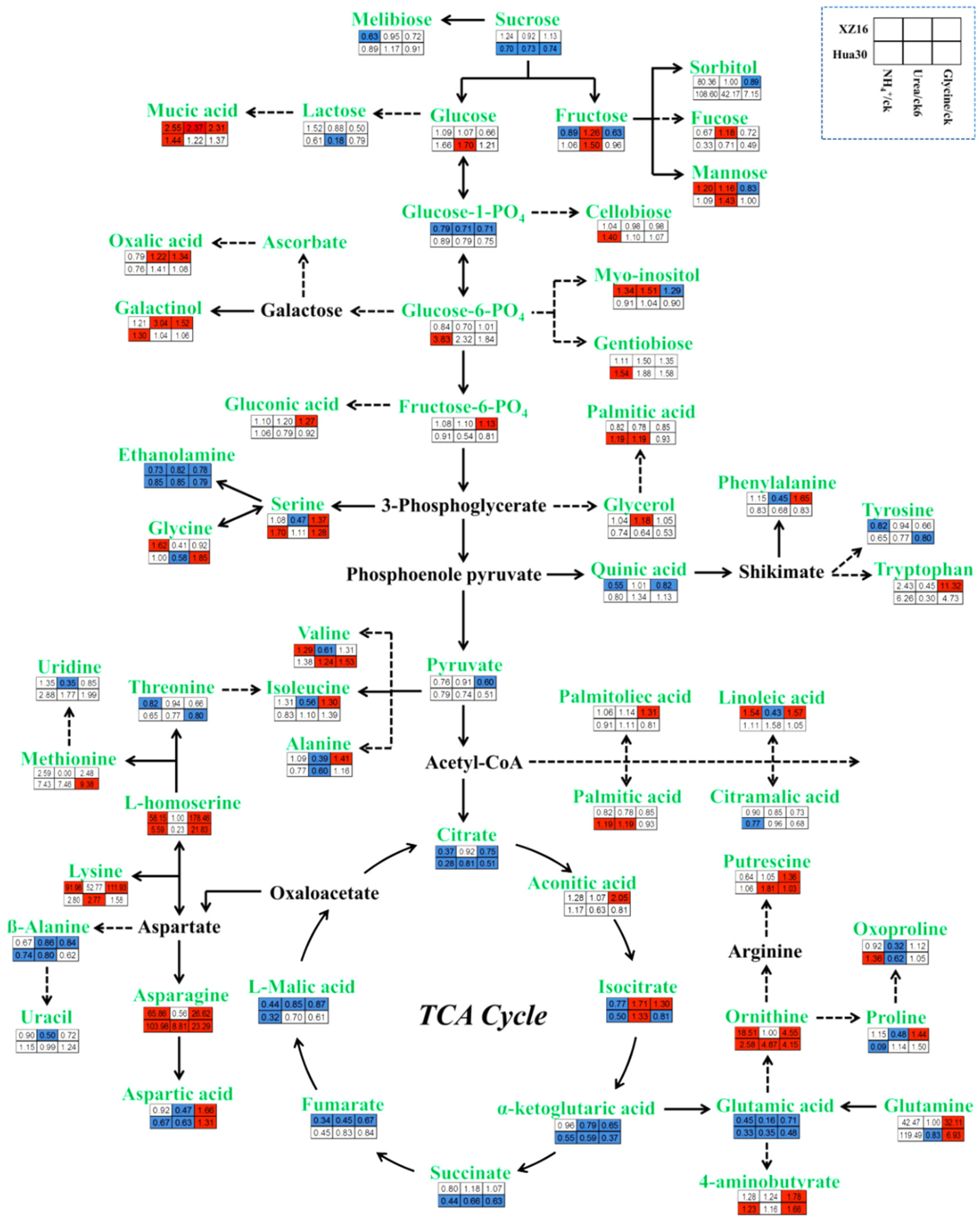
3.4. Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lemaire, G.; Sinclair, T.; Sadras, V.; Bélanger, G. Allometric Approach to Crop Nutrition and Implications for Crop Diagnosis and Phenotyping: A Review. Agron. Sustain. Dev. 2019, 39. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Higher Yield with Less Nitrogen Fertilizer. Nat. Plants 2020, 6, 1078–1079. [Google Scholar] [CrossRef]
- Zhao, B.; Ata-Ul-Karim, S.T.; Duan, A.; Liu, Z.; Wang, X.; Xiao, J.; Liu, Z.; Qin, A.; Ning, D.; Zhang, W.; Lian, Y. Determination of Critical Nitrogen Concentration and Dilution Curve Based on Leaf Area Index for Summer Maize. Field Crops Res. 2018, 228, 195–203. [Google Scholar] [CrossRef]
- Ge, J.; Tang, H.; Yang, N.; Hu, Y. Rapid Identification of Damaged Buildings Using Incremental Learning with Transferred Data from Historical Natural Disaster Cases. ISPRS J. Photogramm. Remote Sens. 2023, 195, 105–128. [Google Scholar] [CrossRef]
- Bloom, A.J. The Increasing Importance of Distinguishing among Plant Nitrogen Sources. Curr. Opin. Plant Biol. 2015, 25, 10–16. [Google Scholar] [CrossRef]
- Owen, A.G.; Jones, D.L. Competition for Amino Acids between Wheat Roots and Rhizosphere Microorganisms and the Role of Amino Acids in Plant N Acquisition. Soil biol. biochem. 2001, 33, 651–657. [Google Scholar] [CrossRef]
- Becker-Ritt, A.B.; Martinelli, A.H.S.; Mitidieri, S.; Feder, V.; Wassermann, G.E.; Santi, L.; Vainstein, M.H.; Oliveira, J.T.A.; Fiuza, L.M.; Pasquali, G. Antifungal Activity of Plant and Bacterial Ureases. Toxicon 2007, 50, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Perchlik, M.; Foster, J.; Tegeder, M. Different and Overlapping Functions of Arabidopsis LHT6 and AAP1 Transporters in Root Amino Acid Uptake. J. Exp. Bot. 2014, 65, 5193–5204. [Google Scholar] [CrossRef]
- Mérigout, P.; Lelandais, M.; Bitton, F.; Renou, J.-P.; Briand, X.; Meyer, C.; Daniel-Vedele, F. Physiological and Transcriptomic Aspects of Urea Uptake and Assimilation in Arabidopsis Plants. Plant Physiol. 2008, 147, 1225–1238. [Google Scholar] [CrossRef] [PubMed]
- Pinton, R.; Tomasi, N.; Zanin, L. Molecular and Physiological Interactions of Urea and Nitrate Uptake in Plants. Plant signal. behav. 2016, 11, e1076603. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Yu, M.; Xu, F.; Ouyang, S.; Xu, X.; Gao, Q.; Li, X. Uptake of Amino Acids and Inorganic Nitrogen by Two Dominant Temperate Grasses. Rhizosphere 2020, 14, 100199. [Google Scholar] [CrossRef]
- Näsholm, T.; Ekblad, A.; Nordin, A.; Giesler, R.; Högberg, M.; Högberg, P. Boreal Forest Plants Take up Organic Nitrogen. Nature 1998, 392, 914–916. [Google Scholar] [CrossRef]
- Henry, H.A.L.; Jefferies, R.L. Interactions in the Uptake of Amino Acids, Ammonium and Nitrate Ions in the Arctic Salt-marsh Grass, Puccinellia Phryganodes. Plant Cell Environ. 2003, 26, 419–428. [Google Scholar] [CrossRef]
- Hu, B.; Zhou, M.; Bilela, S.; Simon, J.; Dannenmann, M.; Liu, X.; Alfarraj, S.; Hou, L.; Chen, H.; Zhang, S.; et al. Nitrogen Nutrition of Native and Introduced Forest Tree Species in N-Limited Ecosystems of the Qinling Mountains, China. Trees 2017, 31, 1189–1202. [Google Scholar] [CrossRef]
- Ge, T.; Song, S.; Roberts, P.; Jones, D.L.; Huang, D.; Iwasaki, K. Amino Acids as a Nitrogen Source for Tomato Seedlings: The Use of Dual-Labeled (13C, 15N) Glycine to Test for Direct Uptake by Tomato Seedlings. Environ. Exp. Bot. 2009, 66, 357–361. [Google Scholar] [CrossRef]
- Gioseffi, E.; de Neergaard, A.; Schjørring, J.K. Interactions between Uptake of Amino Acids and Inorganic Nitrogen in Wheat Plants. Biogeosciences 2012, 9, 1509–1518. [Google Scholar] [CrossRef]
- Brackin, R.; Näsholm, T.; Robinson, N.; Guillou, S.; Vinall, K.; Lakshmanan, P.; Schmidt, S.; Inselsbacher, E. Nitrogen Fluxes at the Root-Soil Interface Show a Mismatch of Nitrogen Fertilizer Supply and Sugarcane Root Uptake Capacity. Sci. Rep. 2015, 5, 15727. [Google Scholar] [CrossRef]
- Wei, L.; Chen, C.; Yu, S. Uptake of Organic Nitrogen and Preference for Inorganic Nitrogen by Two Australian Native Araucariaceae Species. Plant Ecol. Divers. 2015, 8, 259–264. [Google Scholar] [CrossRef]
- Guo, S.; Zhou, Y.; Shen, Q.; Zhang, F. Effect of Ammonium and Nitrate Nutrition On Some Physiological Processes in Higher Plants - Growth, Photosynthesis, Photorespiration, and Water Relations. Plant Biol. 2007, 9, 21–29. [Google Scholar] [CrossRef]
- Szczerba, M.W.; Britto, D.T.; Ali, S.A.; Balkos, K.D.; Kronzucker, H.J. NH4+-Stimulated and -Inhibited Components of K+ Transport in Rice (Oryza Sativa L.). J. Exp. Bot. 2008, 59, 3415–3423. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; Ren, B.; Ding, L.; Gao, C.; Shen, Q.; Guo, S. Drought-Induced Root Aerenchyma Formation Restricts Water Uptake in Rice Seedlings Supplied with Nitrate. Plant Cell Physiol. 2012, 53, 495–504. [Google Scholar] [CrossRef]
- Ding, L.; Gao, C.; Li, Y.; Li, Y.; Zhu, Y.; Xu, G.; Shen, Q.; Kaldenhoff, R.; Kai, L.; Guo, S. The Enhanced Drought Tolerance of Rice Plants under Ammonium Is Related to Aquaporin (AQP). Plant Sci. 2015, 234, 14–21. [Google Scholar] [CrossRef]
- Cramer, M.D.; Lewis, O.A.M. The Influence of Nitrate and Ammonium Nutrition on the Growth of Wheat (Triticum Aestivum) and Maize (Zea Mays) Plants. Ann. Bot. 1993, 72, 359–365. [Google Scholar] [CrossRef]
- Raab, T.K.; Terry, N. Nitrogen Source Regulation of Growth and Photosynthesis in Beta Vulgaris L. Plant Physiol. 1994, 105, 1159–1166. [Google Scholar] [CrossRef]
- Guo, S.; Kaldenhoff, R.; Uehlein, N.; Sattelmacher, B.; Brueck, H. Relationship between Water and Nitrogen Uptake in Nitrate- and Ammonium-Supplied Phaseolus Vulgaris L. Plants. J. Plant Nutr. Soil Sci. 2007, 170, 73–80. [Google Scholar] [CrossRef]
- Lu, Y.X.; Li, C.J.; Zhang, F.S. Transpiration, Potassium Uptake and Flow in Tobacco as Affected by Nitrogen Forms and Nutrient Levels. Ann. Bot. 2005, 95, 991–998. [Google Scholar] [CrossRef]
- Gao, L.; Liu, M.; Wang, M.; Shen, Q.; Guo, S. Enhanced Salt Tolerance under Nitrate Nutrition Is Associated with Apoplast Na+ Content in Canola (Brassica. Napus L.) and Rice (Oryza Sativa L.) Plants. Plant Cell Physiol. 2016, 57, 2323–2333. [Google Scholar] [CrossRef]
- Li, Y.; Gao, Y.; Ding, L.; Shen, Q.; Guo, S. Ammonium Enhances the Tolerance of Rice Seedlings (Oryza Sativa L.) to Drought Condition. Agric. Water Manag. 2009, 96, 1746–1750. [Google Scholar] [CrossRef]
- Malagoli, M.; Dal Canal, A.; Quaggiotti, S.; Pegoraro, P.; Bottacin, A. Differences in Nitrate and Ammonium Uptake between Scots Pine and European Larch. Plant Soil 2000, 221, 1–3. [Google Scholar] [CrossRef]
- Nasir, C.; Malik, M.; Qadir, G.; Rafique, M.; Nawaz, N. Influence of Temperature and Osmotic Stress on Germination Induction of Different Castor Bean Cultivars. Pak. J. Bot. 2010, 42, 4035–4041. [Google Scholar]
- Khalilzadeh, R.; Tajbakhsh, M.; Jalilian, J. Growth Characteristics of Mung Bean (Vigna Radiata L.) Affected by Foliar Application of Urea and Bio-Organic Fertilizers. Int. J. Agric. Crop Sci. 2012, 4, 637–642. [Google Scholar]
- Forsum, O.; Svennerstam, H.; Ganeteg, U.; Näsholm, T. Capacities and Constraints of Amino Acid Utilization in Arabidopsis. New Phytol. 2008, 179, 1058–1069. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Yang, D.N.; Huang, D.F. Effects of different applied nitrogen forms on pakchoi (Brassica chinensis) growth and its carbon and nitrogen accumulation. Ying yong sheng tai xue bao = J. appl. ecol. 2012, 23, 1042–1048. [Google Scholar]
- Ti-Da, G.E.; Shi-Wei, S.; Wu, J.; Dong-Mei, T.; Dan-Feng, H. Influence of Glycine-N Concentration on the Growth and Nitrogen Metabolism of Tomato Seedlings under Sterile Hydroponics Cultivation. Shengtai Xuebao/ Acta Ecologica Sinica 2009, 29, 1994–2002. [Google Scholar]
- Wang, X.; Tang, D.; Huang, D. Proteomic Analysis of Pakchoi Leaves and Roots under Glycine–Nitrogen Conditions. Plant Physiol. Biochem. 2014, 75, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Tian, J.; Liao, H. Proteomics Dissection of Plant Responses to Mineral Nutrient Deficiency. Proteomics 2013, 13, 624–636. [Google Scholar] [CrossRef]
- Zhou, M.; Zhu, S.; Mo, X.; Guo, Q.; Li, Y.; Tian, J.; Liang, C. Proteomic Analysis Dissects Molecular Mechanisms Underlying Plant Responses to Phosphorus Deficiency. Cells 2022, 11, 651. [Google Scholar] [CrossRef]
- Luo, J. Metabolite-Based Genome-Wide Association Studies in Plants. Curr. Opin. Plant Biol. 2015, 24, 31–38. [Google Scholar] [CrossRef]
- Schlüter, U.; Mascher, M.; Colmsee, C.; Scholz, U.; Bräutigam, A.; Fahnenstich, H.; Sonnewald, U. Maize Source Leaf Adaptation to Nitrogen Deficiency Affects Not Only Nitrogen and Carbon Metabolism but Also Control of Phosphate Homeostasis. Plant Physiol. 2012, 160, 1384–1406. [Google Scholar] [CrossRef]
- Sung, J.; Lee, S.; Lee, Y.; Ha, S.; Song, B.; Kim, T.; Waters, B.M.; Krishnan, H.B. Metabolomic Profiling from Leaves and Roots of Tomato (Solanum Lycopersicum L.) Plants Grown under Nitrogen, Phosphorus or Potassium-Deficient Condition. Plant Sci. 2015, 241, 55–64. [Google Scholar] [CrossRef]
- Quan, X.; Qian, Q.; Ye, Z.; Zeng, J.; Han, Z.; Zhang, G. Metabolic Analysis of Two Contrasting Wild Barley Genotypes Grown Hydroponically Reveals Adaptive Strategies in Response to Low Nitrogen Stress. J. plant physiol. 2016, 206, 59–67. [Google Scholar] [CrossRef]
- Tawaraya, K.; Horie, R.; Wagatsuma, T.; Saito, K.; Oikawa, A. Metabolite Profiling of Shoot Extract, Root Extract, and Root Exudate of Rice under Nitrogen and Phosphorus Deficiency. Soil Sci. Plant Nutr. 2018, 64, 312–322. [Google Scholar] [CrossRef]
- Xin, W.; Zhang, L.; Zhang, W.; Gao, J.; Yi, J.; Zhen, X.; Li, Z.; Zhao, Y.; Peng, C.; Zhao, C. An Integrated Analysis of the Rice Transcriptome and Metabolome Reveals Differential Regulation of Carbon and Nitrogen Metabolism in Response to Nitrogen Availability. Int. J. Mol. Sci. 2019, 20, 2349. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xue, Y.; Xie, B.; Zhu, S.; Lu, X.; Liang, C.; Tian, J. Complex Gene Regulation between Young and Old Soybean Leaves in Responses to Manganese Toxicity. Plant Physiol. Biochem. 2020, 155, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, D.; Tao, Z.; Yang, Y.; Gao, Z.; Zhao, G.; Chang, X. Impacts of Nitrogen Deficiency on Wheat (Triticum Aestivum L.) Grain During the Medium Filling Stage: Transcriptomic and Metabolomic Comparisons. Front. Plant Sci. 2021, 12, 674433. [Google Scholar] [CrossRef]
- Shen, X.; Yang, L.; Han, P.; Gu, C.; Li, Y.; Liao, X.; Qin, L. Metabolic Profiles Reveal Changes in the Leaves and Roots of Rapeseed (Brassica Napus L.) Seedlings under Nitrogen Deficiency. Int. J. Mol. Sci. 2022, 23, 5784. [Google Scholar] [CrossRef]
- Tschoep, H.; Gibon, Y.; Carillo, P.; Armengaud, P.; Szecowka, M.; Nunes-Nesi, A.; Fernie, A.R.; Koehl, K.; Stitt, M. Adjustment of Growth and Central Metabolism to a Mild but Sustained Nitrogen-Limitation in Arabidopsis. Plant Cell Environ. 2009, 32, 300–318. [Google Scholar] [CrossRef]
- Schlüter, U.; Colmsee, C.; Scholz, U.; Bräutigam, A.; Weber, A.P.M.; Zellerhoff, N.; Bucher, M.; Fahnenstich, H.; Sonnewald, U. Adaptation of Maize Source Leaf Metabolism to Stress Related Disturbances in Carbon, Nitrogen and Phosphorus Balance. BMC genomics 2013, 14, 1–25. [Google Scholar] [CrossRef]
- Naz, S.; Shen, Q.; Wa Lwalaba, J.L.; Zhang, G. Genotypic Difference in the Responses to Nitrogen Fertilizer Form in Tibetan Wild and Cultivated Barley. Plants 2021, 10, 595. [Google Scholar] [CrossRef]
- Tsugawa, H.; Tsujimoto, Y.; Arita, M.; Bamba, T.; Fukusaki, E. GC/MS Based Metabolomics: Development of a Data Mining System for Metabolite Identification by Using Soft Independent Modeling of Class Analogy (SIMCA). BMC Bioinformatics 2011, 12, 131. [Google Scholar] [CrossRef]
- See, C.R.; Yanai, R.D.; Fisk, M.C.; Vadeboncoeur, M.A.; Quintero, B.A.; Fahey, T.J. Soil Nitrogen Affects Phosphorus Recycling: Foliar Resorption and Plant–Soil Feedbacks in a Northern Hardwood Forest. Ecology 2015, 96, 2488–2498. [Google Scholar] [CrossRef] [PubMed]
- Kobe, R.K.; Lepczyk, C.A.; Iyer, M. Resorption Efficiency Decreases With Increasing Green Leaf Nutrients in A Global Data Set. Ecology 2005, 86, 2780–2792. [Google Scholar] [CrossRef]
- Lü, X.T.; Reed, S.C.; Yu, Q.; Han, X.G. Nutrient Resorption Helps Drive Intra-Specific Coupling of Foliar Nitrogen and Phosphorus under Nutrient-Enriched Conditions. Plant Soil 2016, 398, 111–120. [Google Scholar] [CrossRef]
- Grime, J.P. Benefits of Plant Diversity to Ecosystems: Immediate, Filter and Founder Effects. J. Ecol. 1998, 86, 902–910. [Google Scholar] [CrossRef]
- Güsewell, S. Nutrient Resorption of Wetland Graminoids Is Related to the Type of Nutrient Limitation. Funct. Ecol. 2005, 19, 344–354. [Google Scholar] [CrossRef]
- Dubey, R.S.; Srivastava, R.K.; Pessarakli, M. Physiological mechanisms of nitrogen absorption and assimilation in plants under stressful conditions. In Handbook of plant and crop physiology; CRC Press, 2021; pp. 579–616.
- Bowman, W.D.; Cleveland, C.C.; Halada, Ĺ.; Hreško, J.; Baron, J.S. Negative Impact of Nitrogen Deposition on Soil Buffering Capacity. Nature Geoscience 2008, 1, 767–770. [Google Scholar] [CrossRef]
- Tian, D.; Niu, S. A Global Analysis of Soil Acidification Caused by Nitrogen Addition. Environ. Res. Lett. 2015, 10, 24019. [Google Scholar] [CrossRef]
- Wang, J.; Fu, Z.; Chen, G.; Zou, G.; Song, X.; Liu, F. Runoff Nitrogen (N) Losses and Related Metabolism Enzyme Activities in Paddy Field under Different Nitrogen Fertilizer Levels. Environ. Sci. Pollut. Res. Int. 2018, 25, 27583–27593. [Google Scholar] [CrossRef]
- Cao, Y.; Qu, R.; Tang, X.; Sun, L.; Chen, Q.; Miao, Y. UPLC-Triple TOF-MS/MS Based Metabolomics Approach to Reveal the Influence of Nitrogen Levels on Isatis Indigotica Seedling Leaf. Sci. Hortic. 2020, 266, 109280. [Google Scholar] [CrossRef]
- Wang, H.; Gong, M.; Xin, H.; Tang, L.; Dai, D.; Gao, Y.; Liu, C. Effects of Chilling Stress on the Accumulation of Soluble Sugars and Their Key Enzymes in Jatropha Curcas Seedlings. Physiol. Mol. Biol. Plants 2018, 24, 857–865. [Google Scholar] [CrossRef]
- Ramos, A.; Raven, N.; Sharp, R.J.; Bartolucci, S.; Rossi, M.; Cannio, R.; Lebbink, J.; J, V.D.O.; M, D.V.W.; Santos, H. Stabilization of Enzymes against Thermal Stress and Freeze-Drying by Mannosylglycerate. Appl. Environ. Microbiol. 1997, 63, 4020–4025. [Google Scholar] [CrossRef]
- Lin, Z.H.; Chen, C.S.; Zhong, Q.S.; Ruan, Q.C.; Chen, Z.H.; You, X.M.; Shan, R.Y.; Li, X.L. The GC-TOF/MS-Based Metabolomic Analysis Reveals Altered Metabolic Profiles in Nitrogen-Deficient Leaves and Roots of Tea Plants (Camellia Sinensis). BMC Plant Biol. 2021, 21, 506. [Google Scholar] [CrossRef]
- Wang, X.; Yu, W.; Zhou, Q.; Han, R.; Huang, D. Metabolic Response of Pakchoi Leaves to Amino Acid Nitrogen. J. Integr. Agric. 2014, 13, 778–788. [Google Scholar] [CrossRef]
- Liu, X.; Yang, X.; Wang, L.; Duan, Q.; Huang, D. Comparative Analysis of Metabolites Profile in Spinach (Spinacia Oleracea L.) Affected by Different Concentrations of Gly and Nitrate. Sci. Hortic. 2016, 204, 8–15. [Google Scholar] [CrossRef]
- Millward, D.J.; Layman, D.K.; Tomé, D.; Schaafsma, G. Protein Quality Assessment: Impact of Expanding Understanding of Protein and Amino Acid Needs for Optimal Health1. Am. J. Clin. Nutr. 2008, 87, 1576S–1581S. [Google Scholar] [CrossRef]
- Tessari, P.; Lante, A.; Mosca, G. Essential Amino Acids: Master Regulators of Nutrition and Environmental Footprint? Sci. Rep. 2016, 6, 26074. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sharma, A.; Kaur, R.; Thukral, A.K.; Bhardwaj, R.; Ahmad, P. Differential Distribution of Amino Acids in Plants. Amino Acids 2017, 49, 821–869. [Google Scholar] [CrossRef]
- Deng, Y.; Lu, S. Biosynthesis and Regulation of Phenylpropanoids in Plants. Crit. Rev. Plant Sci. 2017, 36, 257–290. [Google Scholar] [CrossRef]
- Anzano, A.; Bonanomi, G.; Mazzoleni, S.; Lanzotti, V. Plant Metabolomics in Biotic and Abiotic Stress: A Critical Overview. Phytochem. Rev. 2022, 21, 503–524. [Google Scholar] [CrossRef]
- Perchlik, M.; Tegeder, M. Leaf Amino Acid Supply Affects Photosynthetic and Plant Nitrogen Use Efficiency under Nitrogen Stress. Plant physiol. 2018, 178, 174–188. [Google Scholar] [CrossRef]
- Tan, Q.; Zhang, L.; Grant, J.; Cooper, P.; Tegeder, M. Increased Phloem Transport of S-Methylmethionine Positively Affects Sulfur and Nitrogen Metabolism and Seed Development in Pea Plants. Plant Physiol. 2010, 154, 1886–1896. [Google Scholar] [CrossRef]
- Zhang, L.; Garneau, M.G.; Majumdar, R.; Grant, J.; Tegeder, M. Improvement of Pea Biomass and Seed Productivity by Simultaneous Increase of Phloem and Embryo Loading with Amino Acids. The Plant J. 2015, 81, 134–146. [Google Scholar] [CrossRef]
- Santiago, J.P.; Tegeder, M. Connecting Source with Sink: The Role of Arabidopsis AAP8 in Phloem Loading of Amino Acids. Plant Physiol. 2016, 171, 508–521. [Google Scholar] [CrossRef]
- Perchlik, M.; Tegeder, M. Improving Plant Nitrogen Use Efficiency through Alteration of Amino Acid Transport Processes. Plant Physiol. 2017, 175, 235–247. [Google Scholar] [CrossRef]
- Oliveira, I.C.; Brenner, E.; Chiu, J.; Hsieh, M.H.; Kouranov, A.; Lam, H.M.; Shin, M.J.; Coruzzi, G. Metabolite and Light Regulation of Metabolism in Plants: Lessons from the Study of a Single Biochemical Pathway. Braz. J. Med. Biol. Res. 2001, 34, 567–575. [Google Scholar] [CrossRef]
- Kinnersley, A.M.; Turano, F.J. Gamma Aminobutyric Acid (GABA) and Plant Responses to Stress. Crit. Rev. Plant Sci. 2000, 19, 479–509. [Google Scholar] [CrossRef]
- Bown, A.W.; Shelp, B.J. Plant GABA: Not Just a Metabolite. Trends Plant Sci. 2016, 21, 811–813. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, R.; Planas, J.; Saxena, T.; Zarza, X.; Bortolotti, C.; Cuevas, J.; Bitrián, M.; Tiburcio, A.F.; Altabella, T. Putrescine Accumulation Confers Drought Tolerance in Transgenic Arabidopsis Plants Over-Expressing the Homologous Arginine Decarboxylase 2 Gene. Plant Physiol. Biochem. 2010, 48, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Feng, L.; Zhao, L.; Liu, X.; Hassani, D.; Huang, D. Effect of Glycine Nitrogen on Lettuce Growth under Soilless Culture: A Metabolomics Approach to Identify the Main Changes Occurred in Plant Primary and Secondary Metabolism. J. Sci. Food Agric. 2018, 98, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Luna, M.C.; Tudela, J.A.; Tomás-Barberán, F.A.; Gil, M.I. Modified Atmosphere (MA) Prevents Browning of Fresh-Cut Romaine Lettuce through Multi-Target Effects Related to Phenolic Metabolism. Postharvest Biol. Technol. 2016, 119, 84–93. [Google Scholar] [CrossRef]
- Beatty, P.H.; Klein, M.S.; Fischer, J.J.; Lewis, I.A.; Muench, D.G.; Good, A.G. Understanding Plant Nitrogen Metabolism through Metabolomics and Computational Approaches. Plants 2016, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Shen, Q.; Lwalaba, J.L.; Zhang, G. Genotypic Difference in the Responses to Nitrogen Fertilizer Form in Tibetan Wild and Cultivated Barley. Plants 2021, 10, 595. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zeng, X.; Wu, J.; Zhang, F.; Li, C.; Jiang, J.; Wang, Y.; Sun, W. ITRAQ-Based Quantitative Proteome Revealed Metabolic Changes in Winter Turnip Rape (Brassica Rapa L.) under Cold Stress. Int. J. Mol. Sci. 2018, 19, 3346. [Google Scholar] [CrossRef] [PubMed]
- Neto, J.C.R.; Vieira, L.R.; de Aquino Ribeiro, J.A.; de Sousa, C.A.F.; Júnior, M.T.S.; Abdelnur, P.V. Metabolic Effect of Drought Stress on the Leaves of Young Oil Palm (Elaeis Guineensis) Plants Using UHPLC–MS and Multivariate Analysis. Sci. Rep. 2021, 11, 18271. [Google Scholar] [CrossRef]
| Genotype | Treatments | K | Ca | Mg | P | S | Zn | Mn | Fe | Cu |
|---|---|---|---|---|---|---|---|---|---|---|
| mg g−1 DW | mg kg−1 DW | |||||||||
| XZ16 | NO3− | 55.75 b | 7.98 ab | 13.46 a | 13.10 a | 7.35 a | 2.30 a | 0.63 b | 2.84 a | 0.110 a |
| NH4+ | 46.82 d | 6.19 d | 10.18 de | 9.94 c | 6.09 cd | 1.79 bc | 0.42 de | 2.03 de | 0.087 b | |
| Urea | 58.48 a | 8.27 a | 12.51 b | 13.20 a | 7.11 ab | 2.03 ab | 0.73 a | 2.51 b | 0.093 ab | |
| Gly | 50.27 c | 6.42 cd | 11.03 c | 9.54 c | 6.55 bc | 2.16 ab | 0.53 c | 2.32 bc | 0.087 b | |
| Hua30 | NO3− | 46.27 de | 7.15 bc | 10.62 cd | 10.88 b | 6.51bcd | 1.33 cd | 0.43 de | 2.20 cd | 0.087 b |
| NH4+ | 40.68 f | 6.08 d | 8.44 f | 7.33 d | 5.87 d | 1.21 d | 0.34 ef | 1.83 ef | 0.073 bc | |
| Urea | 44.03 e | 6.58 cd | 9.47 e | 11.08 b | 6.77 ab | 1.46 cd | 0.46 cd | 1.84 ef | 0.083 bc | |
| Gly | 34.28 g | 4.55 e | 7.39 g | 5.48 e | 5.03 e | 1.17 d | 0.25 f | 1.57 f | 0.063 c | |
| Interaction (N×G) | *** | * | * | ** | * | ns | * | * | ns | |
| LSD(0.05) values | 2.6590 | 0.8377 | 0.7825 | 0.8229 | 0.6683 | 0.4676 | 0.095 | 0.2822 | 0.0223 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





