Preprint
Article
High HER2 intratumoral heterogeneity is a predictive factor for poor prognosis in Early-stage and Locally Advanced HER2-positive Breast Cancer
Altmetrics
Downloads
168
Views
64
Comments
0
A peer-reviewed article of this preprint also exists.
supplementary.zip (519.66KB )
This version is not peer-reviewed
Submitted:
20 February 2024
Posted:
20 February 2024
You are already at the latest version
Alerts
Abstract
PURPOSE: Breast cancer tumors frequently have intratumor heterogeneity. Tumors with high intratumor heterogeneity have caused therapeutic resistance and have human epidermal growth factor receptor 2 (HER2) heterogeneity in response to HER2-targeted therapies. This study aimed to investigate whether high HER2 heterogeneity levels were clinically related to poor prognosis for HER2-targeted adjuvant therapies resistance in primary breast cancers.
METHODS: This study included patients with primary breast cancer (n = 251) treated with adjuvant HER2-targeted therapies. HER2 heterogeneity was manifested by the shape of HER2 fluorescence in situ hybridization amplification (FISH) distributed histograms with HER2 gene copy number within a tumor sample. Those heterogeneity classified each tumor into the biphasic grade graph (high heterogeneity [HH]) group and the monophasic grade graph (low heterogeneity [LH]) group. Both groups were evaluated for disease-free survival (DFS) and overall survival (OS) for a median of ten years of annual follow-up.
RESULTS: Of 251 patients with HER2-positive breast cancer, 46 (18.3%) and 205 (81.7%) were classified into the HH and LH groups, respectively. The HH group had more distant metastases and a poorer prognosis than the LH group (DFS: P < 0.001 (HH:63% vs LH:91% at 10 years), and OS: P = 0.012 (HH:78% vs LH:95% at 10 years).
CONCLUSION: High HER2 heterogeneity is a poor prognostic factor in patients with HER2-positive breast cancer. A novel approach of heterogeneity, which is manifested by the shape of HER2 FISH distributions, might be clinically useful in the prognosis prediction of patients after HER2 adjuvant therapy.
Keywords:
Subject: Biology and Life Sciences - Biology and Biotechnology
1. Introduction
Cancer heterogeneity refers to the diversity of cancer cells within a tumor as well as the differences between tumors in different individuals [1,2]. It is a fundamental characteristic of cancer and can have important implications for diagnosis, treatment, and prognosis. Tumor heterogeneity includes genetic mutations, epigenetic changes, and cellular level signaling pathway differences [3,4]. This causes subpopulations of cancer cells within a tumor with different characteristics, such as different growth rates, treatment response, and metastasis potential [5]. The presence of heterogeneity can make developing effective cancer therapies challenging, as effective treatments against one cancer cell subpopulation may be ineffective against others. Breast cancer tumors have intratumor heterogeneity (ITH) and tumors with high ITH have demonstrated therapeutic resistance [6]. Also, Human epidermal growth factor receptor 2 (HER2) expression can be heterogeneous in breast cancer tumors. HER2 is a tyrosine kinase receptor which is encoded by the ERBB2 gene as a cancerogenic gene. HER2 is an orphan receptor characterized by a constitutively activated conformation and lacks specific ligands or ligand binding activity. The formation of HER2 homodimers leads to phosphorylation of the tyrosine kinase domain, thereby activating various downstream cancer signaling pathways, ultimately enhancing cancer cell proliferation, tumor formation, invasion, and related processes [7,8].
HER2-positive breast cancer is more aggressive than other breast cancer types, but it can be treated with targeted therapies such as trastuzumab and pertuzumab, which specifically target the HER2 protein [9,10]. Overall, the prognosis of patients with HER2-positive early breast cancer has significantly improved in recent years due to advances in targeted therapies. Currently, the primary application of HER2 assessment lies in the improvement for patient survival to anti-HER2 therapy in the neoadjuvant and adjuvant settings. HER2 heterogeneity is defined by the coexistence of at least two distinct cellular clones with differing HER2 statuses within the same tumor, and the heterogeneity is the coexistence of different tumor cell subpopulations with varying levels of HER2 protein expression [11]. The definition of HER2-positive cancer is typically determined primarily immunohistochemistry (IHC) and in situ hybridization (ISH). ISH involves the simultaneous detection of HER2 gene amplification and chromosome 17 (CEP17) to assess HER2 gene status in tumor tissue. If the IHC test reveals a score of 3+, indicating strong HER2 protein expression, or if the IHC test shows a score of 2+ along with confirmed HER2 gene amplification via ISH, the cancer is classified as HER2-positive [12]. HER2 heterogeneity is a well-established phenomenon in breast cancer, signifying the potential for distinct regions within the same tumor to exhibit varying levels of protein expression or genetic amplification [7,11]. Tumor heterogeneity refers to the diversity of cancer cells within a tumor or among tumors, encompassing variations in genetic mutations, gene expression patterns, and cellular characteristics. This heterogeneity can occur spatially, with different regions of a tumor exhibiting distinct features, and temporally, as cancer cells evolve and adapt over time [2,13,14]. Heterogeneous HER2 amplification was observed in 11%–40% of tumor cell populations in HER2-positive breast cancers [8,15,16,17]. On the geographic (spatial) distribution, HER2 heterogeneity has been described in a “clustered”form or “mosaic” pattern characterized by pockets of highly amplified cells [8,18,19,20]. HER2 heterogeneity may contribute to inaccurate HER2 status assessment and affect therapeutic decision-making, and validation of techniques to identify HER2 heterogeneity in the clinic and concurrent development of agents to effectively treat tumors with nonuniform HER2 expression is needed [21,22]. Dual-color in situ hybridization (DISH) is also a molecular diagnostic technique used to assess the status of the HER2 gene in breast cancer and other malignancies [23,24,25]. DISH probes are designed to bind to the HER2 gene and CEP17 and can be visualized under a microscope. DISH allows for the simultaneous detection of both the HER2 gene and CEP17, providing comprehensive information on gene amplification, and allows for quantitative analysis, enabling the precise measurement of gene amplification levels.
Recently, Seol et al. reported that the presence of HER2 heterogeneity associated with decreased disease-free survival (DFS) was treated with only 25% adjuvant trastuzumab therapy [26]. In contrast, Shen et al. revealed that HER2 heterogeneity detected with a HER2 gene protein assay (GPA) was associated with poor overall survival (OS) and increased distal metastasis in patients with HER2-positive breast cancer [21,27]. In the report, HER2 gene amplification was evaluated with HER2 GPA, which combines HER2 IHC and DISH for simultaneous evaluation at both the protein and gene at the tissue level. However, The DISH method frequently presents with non-specific background deposition, including debris as subtle silver deposits, which elude automated analysis. Furthermore, the DISH method yields manual counting dependent on the measurer, requiring training for signal counting and susceptible to inter-measurer variability in interpretation [28,23]. Recently, Laurence et.al. investigated the prevalence of a range of gene expression distributions in three different tumor types from the Cancer Genome Atlas (TCGA), and studied a genes distribution shape to model heterogeneity of transcriptomic data [29]. Also, their results indicate that prognostic genes identified based on consideration of the shape of the distribution were different from those identified through more standard assumptions. histogram analysis is the most common and popular method of characterization of ITH in imaging data [30]. Radziuviene et al. demonstrated automated image analysis of HER2 FISH histograms to refine their definition of genetic heterogeneity, and HER2 ITH could also be identified by histograms based on HER2 FISH amplification [31]. It is necessity to develop novel methods capable of analyzing HER2 ITH within cancer tissues automatically, and to make unbiased analyses, rather than relying on manual procedures. Also, to assess more clear-cut heterogeneity of HER2, the novel method is necessary to evaluate the HER2 copy number quantification, as HER2 gene copy numbers are present in each of those cancer cells. In additional, it is desirable that the method proves beneficial in predicting prognosis and anti-HER2 therapies sensitivity. We developed a novel method to confirm the shape of HER2 FISH distributions with HER2 gene copy number within a tumor sample, and analyze HER2 ITH automatically. Also, we conducted our method to investigate the association between HER2-targeted therapy and HER2 heterogeneity. This study defined HER2 ITH using Gaussian mixture modeling (GMM) analysis of histograms based on HER2 FISH distributions. Recently A GMM classifier demonstrated potential use for clinical validation of markers and determination of target populations to improve molecular stratification of patients with breast cancer [32].
We aimed to develop a novel method to analyze HER2 ITH automatically, and determine the clinical association of high HER2 heterogeneity with poor prognosis due to resistance to postoperative adjuvant therapy with HER2-targeted agents in primary breast cancer, through the application of GMM analysis.
2. Materials and Methods
2.1. Patients selection and breast tumor samples
This study consecutively recruited 251 patients with HER2-positive invasive breast carcinoma (mean age: 56.6 years; range: 28–97 years) treated with adjuvant or neoadjuvant HER2-targeted therapies. These patients underwent mastectomy or breast-conserving surgery from February 2009 to January 2018 at Osaka University Hospital, Osaka, Japan. All 251 patients received 12 months of adjuvant trastuzumab.
Surgery was performed on 185 patients, followed by anti-HER2 adjuvant chemotherapy, and 66 patients received neoadjuvant chemotherapy followed by surgery (anthracycline and taxane-based chemotherapy, n = 146; taxane-based chemotherapy, n = 87; anthracycline-chemotherapy, n = 5). Both anti-HER2 adjuvant therapy and hormonal therapy were given to 162 patients with hormone receptor-positive cancer (aromatase inhibitors, n = 99; tamoxifen, n = 45; or goserelin + tamoxifen, n = 18). We collected clinical–pathological characteristics from patients’ electronic medical records, and Table 1 summarized the details. Tumor tissues (surgical specimens) were obtained at surgery, and tumor tissues from patients who received neoadjuvant chemotherapy were obtained before the treatment by vacuum-assisted core needle biopsy. Solid intratumoral tissues were fixed in 10% buffered formalin and embedded in paraffin (FFPE). The Ethics Committee of Osaka University approved the study protocol.
2.2. HER2 FISH assay
All tumor tissues underwent HER2 FISH assay. HER2 status, including HER2 gene signals and ratios, was determined by FISH using the PathVysion HER-2 DNA probe Kit (Abbott Laboratories, Abbott Park, IL). Regarding the kits, the tumor FFPE tissue slides were dried and 20 µL DAPI counterstain solution was applied. Then, a fluorescence microscope was used to capture both orange-fluorescent signals corresponding to HER2/neu and green-fluorescent signals corresponding to CEP17. For the histograms of HER2 FISH, all of the fluorescence images were automatically identified by histograms based on HER2 FISH amplification for the protocol of Duet-3/Setup Station/SOLO2 system (BioView, Rehovot, Israel). Meanwhile, for counting the value of HER2 FISH, a fluorescence microscope was used to count the number of those fluorescent signals under the manual or semi-automated protocols analysis. First, samples were analyzed with semi-automated protocol and then samples failed semi-automated protocol analysis were analyzed with manual protocol. The number of cells counted was more than 60 in semi-automated protocols, and it was 20 in the manual. The ratio between the HER2/neu and the CEP17 signal count was calculated. HER2 was considered positive when a tumor contained more than two genes per cell for the FISH assay of HER2. Experienced molecular pathologists used the 2018 HER2 ASCO/CAP updated guidelines to analyze the HER2 status.
2.3. Assessment of HER2 intratumoral heterogeneity
We evaluated HER2 intratumoral heterogeneity (ITH), which was manifested by the shape of HER2 FISH distributions with HER2 gene copy number within a tumor sample, as shown in Figure 1A. We used scanned image data from FISH diagnostic reports. We extracted the FISH count information by reading the histogram and performing image processing analysis, such as OCR, because a few scanned FISH images were insufficient for image analysis. Then, the heterogeneity was analyzed by Gaussian mixture modeling (GMM) of histograms based on HER2 FISH. Parameters were estimated by fitting a mixed distribution with mean 𝜇 𝜎, and 𝜋 was the weight of each mixed component:
𝑓(𝑥) = 𝜋1𝑁(𝑥│𝜇1, 𝜎1) + 𝜋2𝑁(𝑥│𝜇2, 𝜎2)
Gaussian distribution f(x) to the distribution of signal counts, where N was the normal and standard deviation.
Here, the distributions of FISH signal counts 2 and those >2 are assumed to be normal (π , respectively). Then, 1/ 2 was used as a measure to calculate the diversity. This is because a large 1/ 2 means a FISH count of <2, but the average FISH count is >2. Figure S1 shows two representative cases where imaging for a diagnostic report was fitted with a mixture of Gaussian distributions. HER2 ITH, which was manifested by the shape of HER2 FISH distributions were classified into two groups: the biphasic grade graph (high heterogeneity [HH]) group and the monophasic grade graph (low heterogeneity [LH]) group for each tumor in Figure 1B. Both groups were evaluated in terms of DFS and OS. All patients had been receiving HER2 treatment. The median of 10 years after annual follow-up was obtained. We used Python (v.3.10.6) and the Python libraries “opencv” (v.4.7.0.72), “pillow” (v.9.0.1), “pyOCR” (v.0.8.3), and “pytesseract” (v.0.3.10) for diagnostic report image processing.
Figure 1 shows the representative histograms of HER2 FISH distributions.
2.4. Statistical analyzes
All patients was classified into the HH and LH groups, and the HH groups was compared to the LH groups In clinicopathological parameters. The chi-square test was used to assess the associations between HER2 heterogeneity and clinicopathological parameters. DFS was calculated from the date of diagnosis of primary breast cancer to the date of diagnosis of metastatic breast cancer, and OS was calculated from the date of diagnosis of metastatic breast cancer to the date of death from any cause, censoring at last known follow-up. The Kaplan–Meier method was used to calculate t disease-free survival (DFS) rates. Differences in DFS were evaluated by the log-rank test. The Cox proportional hazard model was used for univariate and multivariate analyses of various clinicopathological and biological factors for DFS. R (v.4.2.3; 14) was used for all statistical analyses, and statistical significance was set to P < 0.05 for two-sided tests.
2.5. Data availability
We obtained Medical Ethical Committee approval (ethical board approval number of Osaka University: 22080 (T1). This is an observational study conducted with patient enrollment based on the opt-out method for comprehensive informed consent. Informed consent was obtained in the form of opt-out on the web-site, and those who rejected were excluded.
3. Results
3.1. Association between HER2 intratumoral heterogeneity and patient Characteristics
HER2-positive breast cancer was classified into the HH and LH groups with high (18.3%) and low (81.7%) cases, respectively, of 251 tumors. The subsequent analysis considered HER2-positive breast cancers showing biphasic- or monophasic grade graph analysis of the HER2 FISH distributions as the HH or LH groups, respectively. The HER2-positive breast cancer tumors within the HH group had a tendency toward a higher proportion of HER2 IHC 2+ than 3+ expression, and the HER2 FISH ratio in the HH group was notably expressed higher than the LH group (Table 2). Their tumors within the HH group were significantly associated with estrogen receptor (ER) + (P = 0.002) and PgR + (P < 0.001). No significant association was observed between HER2 heterogeneity (high or low) and menopausal status, histological type, histological grade, lymph node status, or stage. Figure S2 shows the frequency of HH HER2-positive tumors following the ER status and progesterone estrogen receptor (PgR). The ER+ and PgR+ tumors demonstrated the highest frequency of HH HER2 heterogeneity (ER+ and PgR+: 28% [28/99], ER- and PgR-: 8% [6/75], ER+ and PgR-: 6% [4/70], and ER- and PgR+: 0% [0/7]).
3.2. HER2 intratumoral heterogeneity and patient prognosis
The DFS and OS rates of patients with tumors in the HH group were significantly longer and shorter, respectively (DFS: P < 0.001 (HH group:63% vs LH group:91% at 10 years), and OS: P = 0.012 (HH group:78% vs LH group:95% at 10 years)) (Figure 2A,B). Univariate analysis of various prognostic factors demonstrated that pretreatment lymph node metastasis and resected lymph node metastasis were significantly associated with patient prognosis (DFS, pretreatment: P = 0.029, resected: P = 0.009; OS, pretreatment: P = 0.014, resected: P = 0.009) (Table 3 and Table 4). Multivariate analysis revealed that HER2 heterogeneity was significantly associated with prognosis (DFS: P < 0.001, OS: P = 0.039). High levels of HER2 ITH were clinically significant for poor prognosis for resistance to adjuvant therapy with HER2-targeted therapies in primary breast cancers (Figures S3). The ratio of HER2 FISH (the mean number of HER2 FISH signals/cell) of the HH group (median: 2.3) was lower than that of the LH group (median: 5.4). However, the ratio of HER2 FISH was not significantly associated with prognosis. It is necessary to investigate whether the prognosis of LH is associated with the level of HER2 FISH amplification. LH tumors were divided into the ex-high amplification LH group (Ex-High LH) (>4.9%) and a high amplification group LH (High LH) (<4.9%) according to the median value of HER2 FISH as the cut-off, and we investigated whether the relationship of HER2 heterogeneity with prognosis was associated with the ratio of HER2 FISH (Figure 1B). LH tumors in the Ex-High LH group and High LH group were not associated with prognosis (DFS: P = 0.253, OS: P = 0.686) (Figure 2C,D). Figures S1 and S4 show the representative results between patient recurrence and the ratio of HER2 FISH. In particular, the proportion (%) of patient recurrence in the HH group was higher than that in the LH group.
Figure 2 shows Disease-free survival (DFS) and overall survival (OS) rates according to HER2 intratumoral heterogeneity.
4. Discussion
This study aimed to develop our novel method to analyze HER2 intratumoral heterogeneity (ITH) automatically, and to clarify the clinicopathological characteristics and prognosis of patients with breast cancer with HER2 ITH (Figure S1). We revealed an 18.3% frequency of HH tumors. HH tumors were significantly associated with high ER positivity, PgR positivity, and lower HER2 FISH ratio than LH tumors. This study aimed to develop our novel method to analyze HER2 intratumoral heterogeneity (ITH) automatically, and to clarify the clinicopathological characteristics and prognosis of patients with breast cancer with HER2 ITH (Figure S1). We revealed an 18.3% frequency of HH tumors. HH tumors were significantly associated with high ER positivity, PgR positivity, and lower HER2 FISH ratio than LH tumors. Thus, luminal HER2 (ER+ and PgR+) tumors have the highest frequency (28%) of HH tumors (Figure S3) in our study, and these observations are essentially consistent with the concept of tumor heterogeneity, where a biological diversity of cancer cells was found within a tumor [33]. In Table 2, HH tumors were not significantly associated in pretreatment LN metastasis (0.098). The reason is that the HER2 ITH may contribute to the therapeutic effect (suppression of metastatic recurrence) of HER2 treatment and may has little to do with cancer invasion, metastasis, and the progression to LN metastasis. High proliferative cancer cells in both HH and LH tumors may be susceptible to pretreatment LN metastasis.
Further, the present study demonstrated a difference in DFS and OS rates between patients with HH and LH tumors, indicating that the presence of high heterogeneity has prognostic significance (Table 3). High heterogeneity was an independent prognostic factor after adjustment for other factors, including the type of anti-HER2 adjuvant chemotherapy. Highly heterogeneous breast cancers may show poorer prognosis because of their association with different biological characteristics. On the other hand, the ratio of HER2 FISH, and HER2 IHC (3+, or 2+) could not predict prognosis in patients with HER2-positive breast cancer. In additional, we investigated whether the level of HER2 FISH amplification is associated with the prognosis of LH, and LH tumors were divided into the ex-high amplification LH group (Ex-High LH) and a high amplification group LH (High LH). As a result, in HER2-positive LH tumors, our study revealed that the ratio of HER2 FISH (mean number of HER2 FISH signals/cell) (Ex-High LH vs. High LH) was not associated with prognosis of DFS rates and OS rates (Figure 2C and 2D).
Similarly, Shen et al. reported a significantly poorer prognosis in patients with HH tumors than in those with LH tumors, to be evaluated with HER2 GPA, which combines HER2 IHC (the protein) and DISH (HER2 gene amplification) at the tissue level [27,21]. In addition, Hou et al. and Horii et al. revealed that HER2 ITH with HER2 GPA method is an independent factor predicting incomplete response to anti-HER2 neoadjuvant chemotherapy [34,35]. However, HER2 GPA method as well as DISH yields manual counting and demands advanced technical expertise and specialized equipment. Their methods lacks standardized staining and interpretation methods, making it susceptible to false positives influenced by background factors such as debris. Also, the HER2 GPA was not analyzed with the distribution of HER2 gene copy number within a tumor sample.
The strength of our method is able to analyze HER2 ITH within cancer tissues automatically and to make unbiased analyses of HER2 ITH, rather than relying on manual procedures. Also, our method can evaluate the HER2 copy number quantification within a tumor sample and assess more clear-cut heterogeneity of HER2, because HER2 gene copy numbers are present in each of those cancer cells within a tumor sample. Automated analysis of HER2 expression typically involves utilizing computer algorithms and image processing techniques to assess HER2 IHC status in tissue samples. Miglietta et.al. reported that the assessment of HER2 IHC status had significant observer variability of pathologist [18]. To accurately assess the state of HER2 IHC, alternative methods such as molecular examinations and digital pathology have been proposed [36]. Since the widespread adoption of whole slide imaging, Digital image analysis has emerged as a swift, cost-effective, objective, and reproducible scoring methodology for evaluating HER2 IHC status and the efficacy of anti-HER2 therapies through immunohistochemistry [37,38,39,40]. However, digital image analysis for evaluating the heterogeneity of HER2 IHC, which correlates with anti-HER2 treatment efficacy, is yet to be developed. Our study has the limitations not to compare HER2 IHC with HER2 gene copy number within a tumor sample, unlike HER2 dual-color imaging which combines HER2 IHC and HER2 gene amplification. However, we believe that the prognosis in patients of HER2-positive breast cancers can be estimated more accurately by adding our HER2 FISH distributions analysis of heterogeneity.
Besides, another strength of our analysis is an easy, low-cost method only to read histograms of FISH diagnostic reports and perform image processing analysis. The findings of this study provide valuable insights into the efficacy of our novel method of HER2 ITH analysis, and the highlighting is its potential as a clinically useful of predicting prognosis and anti-HER2 therapies sensitivity. And then, the clinical significance of identification of our analysis for the prediction of the prognosis seems to deserve further investigation of multicenter research.
Furthermore, our analysis has the potential to accurately assess ITH not only in HER2-positive breast cancers but also in HER2-low. It is hypothesized that HER2-low breast cancers commonly represent more heterogeneous population than HER2-positive breast cancers [41]. Recently, trastuzumab deruxtecan (T-DXd) treatment resulted in significantly longer relapse-free survival and OS than chemotherapy among patients with HER2-low metastatic breast cancer [42]. This study reported that HER2-low tumors constituted a heterogeneous population that varied in prognosis and sensitivity to systemic treatments. T-DXd, effectively targets tumor cells that express low levels of HER2 and can deliver a potent cytotoxic payload through the bystander effect to neighboring tumor cells heterogeneously expressing HER2, unlike many other approved HER2-targeted therapies [43,44]. Therefore, in the HER2-low as well as HER2-positive, highly heterogeneous breast cancers might be responsive to treatment of T-DXd that exhibits potent therapeutic efficacy through the delivery of cytotoxic payload. In addition, Mosele et. al. reported that HER2 is a determinant of sensitivity to T-DXd, although modest anti-tumor activity was also observed in a small subset of patients whose cancer did not express HER2 [45]. The study revealed that T-DXd anti-tumor activity increased when HER2 expression was high, modest anti-tumor activity was also observed in patients with HER2 IHC 0, and suggested that very low levels of HER2 could allow uptake of T-DXd and/or that partially mediated drug efficacy. That is to say, the level of HER2 IHC is extremely low or absent, which cannot have implications for T-DXd treatment decisions, and our HER2 FISH distributions analysis without HER2 IHC, might accurately identify the heterogeneity of HER2-low breast or absent, and predict T-DXd efficacy of breast cancers. Hence, our analysis may be useful for predicting the effectiveness of not only standard HER2-targeted therapies but also of the new drug against highly heterogeneous tumors. In the future, we are going to investigate whether our method our analysis can accurately identify the heterogeneity of HER2-low breast cancers or not.
Meanwhile, it has been confirmed that HER2-positive gastric cancer responds to HER2-targeted therapy as well [46]. The positivity rate of HER2 in gastric cancer is lower compared to breast cancer, resulting in lower effectiveness of HER2-targeted therapies [47,48]. Moreover, there is a higher probability of HER2 heterogeneity in gastric cancer compared to breast cancer [49,50]. Our study might have clinical practice impact that our novel analysis clinically t has the potential to become a new classification of HER2 ITH and predict prognosis in patients with HER2-positive gastric cancer as well as breast cancer, as a discriminative, inexpensive and easy-to perform method.
5. Conclusions
We have revealed the HH of HER2 as a poor prognostic factor in patients with HER2-positive breast cancer. Examining HER2 heterogeneity using HER2 FISH distributions is a discriminative, inexpensive, and easy-to-perform method. Our results indicate that our HER2 FISH distributions analysis of HER2 ITH might be clinically useful in the prognosis prediction of patients after HER2 adjuvant therapy.
Supplementary Materials
s1, Figure S1: Procedure for imaging a diagnostic report. Two representative cases in which imaging a diagnostic report of HER2 FISH signals (A), histogram of HER2 FISH signals (B), and fitting by a mixture of Gaussian distribution (C) classified the cases into the HH group (a) and LH group (b). HER2 FISH signals and ratio of mixtures (1 2) in the total sample of patients with HER2-positive breast cancer (D). s2, Figure S2: Relationship between the population of patients with highly heterogeneous/total HER2-positive breast cancer (%) and ER/PgR status (n = 251). The ER+ and PgR+ tumors demonstrated the highest frequency of HER2 heterogeneity (ER+ and PgR+: 28% [28/99], ER and PgR-: 8% [6/75], ER+ and PgR-: 6% [4/70], and ER- and PgR+: 0% [0/7]). S3, Figure S3: Schematic diagram of the effect of HER2-targeted therapies in HER2 intratu- moral heterogeneity. High levels of heterogeneity of HER2 were clinically significant for poor prognosis for resistance to adjuvant therapy with HER2-targeted therapies in primary breast cancers. S4, Figure S4: Relationship between the HER2 ratio (the mean HER2 signals/cell) and HER2 intratumoral heterogeneity. The HER2 ratio was compared between tumors with high and low heterogeneity (P-value, Mann–Whitney U-test; bars, median; patients experiencing dis- ease recurrence: white triangles; and patients with no recurrence: black dots).
Author Contributions
Tomonori Tanei: Conceptualization, Visualization, Formal analysis, Investigation, Methodology, Project administration, Writing original draft; Shigeto Seno: Investigation, Methodology, Software, Visualization, Writing; Review & Editing; Yoshiaki Sota: Data curation, Methodology, Validation, Visualization, Writing - Review & Editing, Project administration; Kaori Abe: Writing - Review & Editing; Nanae Masunaga: Writing - Review & Editing; Masami Tsukabe: Writing - Review & Editing; Tetsuhiro Yoshinami: Writing - Review & Editing; Tomohiro Miyake: Writing - Review & Editing; Masafumi Shimoda: Writing - Review & Editing; Hideo Matsuda: Writing - Review & Editing; Kenzo Shimazu: Writing - Review & Editing, Supervision, fouding acuistration.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Osaka University 22080 (T1) on September 20th, 2022.
Informed Consent Statement
Our opt-out comprehensive Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author (accurately indicate status).
Conflicts of Interest
The authors declare no conflict of interest.
References
- IJzerman, M.J.; Berghuis, A.S.; Bono, J.S. de; Terstappen, L.W. Health Economic Im- pact of Liquid Biopsies in Cancer Management. Expert review of pharmacoeconomics & outcomes research 2018, 18, 593–599. [Google Scholar]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Ther- apies. Nature reviews Clinical oncology 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Ramón y Cajal, S.; Sesé, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz- Cano, S.J.; Hernández-Losa, J.; Castellvı́, J. Clinical Implications of Intratumor Het- erogeneity: Challenges and Opportunities. Journal of Molecular Medicine 2020, 98, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Beca, F.; Polyak, K. Intratumor Heterogeneity in Breast Cancer. Novel biomarkers in the continuum of breast cancer 2016, 169–189. [Google Scholar]
- Marusyk, A.; Polyak, K. Tumor Heterogeneity: Causes and Consequences. Biochim- ica et Biophysica Acta (BBA)-Reviews on Cancer 2010, 1805, 105–117. [Google Scholar] [CrossRef]
- Marusyk, A.; Janiszewska, M.; Polyak, K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer cell 2020, 37, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F. HER2 and Breast Cancer—a Phenomenal Success Story. New England Journal of Medicine 2019, 381, 1284–1286. [Google Scholar] [CrossRef]
- Hanna, W.M.; Rüschoff, J.; Bilous, M.; Coudry, R.A.; Dowsett, M.; Osamura, R.Y.; Penault-Llorca, F.; Van De Vijver, M.; Viale, G. HER2 in Situ Hybridization in Breast Cancer: Clinical Implications of Polysomy 17 and Genetic Heterogeneity. Modern Pathology 2014, 27, 4–18. [Google Scholar] [CrossRef]
- Von Minckwitz, G.; Procter, M.; Azambuja, E. de; Zardavas, D.; Benyunes, M.; Viale, G.; Suter, T.; Arahmani, A.; Rouchet, N.; Clark, E.; et al. Adjuvant Pertuzumab and Trastuzumab in Early HER2-Positive Breast Cancer. New England Journal of Medicine 2017, 377, 122–131. [Google Scholar] [CrossRef]
- Cameron, D.; Piccart-Gebhart, M.J.; Gelber, R.D.; Procter, M.; Goldhirsch, A.; Azambuja, E. de; Castro, G.; Untch, M.; Smith, I.; Gianni, L.; et al. 11 Years’ Follow-up of Trastuzumab After Adjuvant Chemotherapy in HER2-Positive Early Breast Cancer: Final Analysis of the HERceptin Adjuvant (HERA) Trial. The Lancet 2017, 389, 1195–1205. [Google Scholar] [CrossRef]
- Marchiò, C.; Annaratone, L.; Marques, A.; Casorzo, L.; Berrino, E.; Sapino, A. Evolving Concepts in HER2 Evaluation in Breast Cancer: Heterogeneity, HER2-Low Carcinomas and Beyond. In Proceedings of the Seminars in cancer biology; Elsevier, 2021; Vol. 72; pp. 123–135. [Google Scholar]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical On- cology/College of American Pathologists Clinical Practice Guideline Focused Update. Archives of pathology & laboratory medicine 2018, 142, 1364–1382. [Google Scholar]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical Management of Breast Cancer Heterogeneity. Nature reviews Clinical oncology 2015, 12, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Brocks, D.; Gerhäuser, C. Intratumor Heterogeneity in Epigenetic Pat- terns. In Proceedings of the Seminars in cancer biology; Elsevier, 2018; Vol. 51, pp. 12–21. [Google Scholar]
- Wolff, A.C.; Hammond, M.E.H.; Schwartz, J.N.; Hagerty, K.L.; Allred, D.C.; Cote, R.J.; Dowsett, M.; Fitzgibbons, P.L.; Hanna, W.M.; Langer, A.; et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommen- dations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. Archives of pathology & laboratory medicine 2007, 131, 18–43. [Google Scholar]
- Allison, K.H.; Dintzis, S.M.; Schmidt, R.A. Frequency of HER2 Heterogeneity by Flu- orescence in Situ Hybridization According to CAP Expert Panel Recommendations: Time for a New Look at How to Report Heterogeneity. American journal of clinical pathology 2011, 136, 864–871. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.C.; Malowany, J.I.; Mazurkiewicz, J.; Wood, M. ‘Genetic Heterogeneity’in HER2/Neu Testing by Fluorescence in Situ Hybridization: A Study of 2522 Cases. Modern pathology 2012, 25, 683–688. [Google Scholar] [CrossRef]
- Miglietta, F.; Griguolo, G.; Bottosso, M.; Giarratano, T.; Lo Mele, M.; Fassan, M.; Cacciatore, M.; Genovesi, E.; De Bartolo, D.; Vernaci, G.; et al. Evolution of HER2- Low Expression from Primary to Recurrent Breast Cancer. NPJ Breast Cancer 2021, 7, 137. [Google Scholar] [CrossRef] [PubMed]
- Sapino, A.; Goia, M.; Recupero, D.; Marchiò, C. Current Challenges for HER2 Testing in Diagnostic Pathology: State of the Art and Controversial Issues. Frontiers in oncology 2013, 3, 129. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Seither, F.; Schneeweiss, A.; Link, T.; Blohmer, J.-U.; Just, M.; Wim- berger, P.; Forberger, A.; Tesch, H.; Jackisch, C.; et al. Clinical and Molecular Characteristics of HER2-Low-Positive Breast Cancer: Pooled Analysis of Individual Patient Data from Four Prospective, Neoadjuvant Clinical Trials. The Lancet Oncol- ogy 2021, 22, 1151–1161. [Google Scholar] [CrossRef]
- Hou, Y.; Nitta, H.; Li, Z. HER2 Intratumoral Heterogeneity in Breast Cancer, an Evolving Concept. Cancers 2023, 15, 2664. [Google Scholar] [CrossRef]
- Hamilton, E.; Shastry, M.; Shiller, S.M.; Ren, R. Targeting HER2 Heterogeneity in Breast Cancer. Cancer Treatment Reviews 2021, 100, 102286. [Google Scholar] [CrossRef]
- Mansfield, A.S.; Sukov, W.R.; Eckel-Passow, J.E.; Sakai, Y.; Walsh, F.J.; Lonzo, M.; Wiktor, A.E.; Dogan, A.; Jenkins, R.B. Comparison of Fluorescence in Situ Hybridization (FISH) and Dual-ISH (DISH) in the Determination of HER2 Status in Breast Cancer. American journal of clinical pathology 2013, 139, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Schiavon, B.N.; Jasani, B.; De Brot, L.; Vassallo, J.; Damascena, A.; Cirullo-Neto, J.; Neves, J.I.; Soares, F.A.; Gobbi, H.; Rocha, R.M. Evaluation of Reliability of FISH Versus Brightfield Dual-Probe in Situ Hy-Bridization (BDISH) for Frontline Assess- ment of HER2 Status in Breast Cancer Samples in a Community Setting: Influence of Poor Tissue Preservation. The American journal of surgical pathology 2012, 36, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Babic, A.; Loftin, I.R.; Stanislaw, S.; Wang, M.; Miller, R.; Warren, S.M.; Zhang, W.; Lau, A.; Miller, M.; Wu, P.; et al. The Impact of Pre-Analytical Processing on Staining Quality for h&e, Dual Hapten, Dual Color in Situ Hybridization and Fluorescent in Situ Hybridization Assays. Methods 2010, 52, 287–300. [Google Scholar] [PubMed]
- Seol, H.; Lee, H.J.; Choi, Y.; Lee, H.E.; Kim, Y.J.; Kim, J.H.; Kang, E.; Kim, S.-W.; Park, S.Y. Intratumoral Heterogeneity of HER2 Gene Amplification in Breast Cancer: Its Clinicopathological Significance. Modern pathology 2012, 25, 938–948. [Google Scholar] [CrossRef]
- Shen, T.; Nitta, H.; Wei, L.; Parwani, A.V.; Li, Z. HER2 Intratumoral Heterogeneity Is Independently Associated with Distal Metastasis and Overall Survival in HER2- Positive Breast Carcinomas. Breast cancer research and treatment 2020, 181, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Furrer, D.; Sanschagrin, F.; Jacob, S.; Diorio, C. Advantages and Disadvantages of Technologies for HER2 Testing in Breast Cancer Specimens. American journal of clinical pathology 2015, 144, 686–703. [Google Scholar] [CrossRef] [PubMed]
- Torrenté, L. de; Zimmerman, S.; Suzuki, M.; Christopeit, M.; Greally, J.M.; Mar, J.C. The Shape of Gene Expression Distributions Matter: How Incorporating Dis- tribution Shape Improves the Interpretation of Cancer Transcriptomic Data. BMC bioinformatics 2020, 21, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Patkulkar, P.A.; Subbalakshmi, A.R.; Jolly, M.K.; Sinharay, S. Mapping Spatiotem- poral Heterogeneity in Tumor Profiles by Integrating High-Throughput Imaging and Omics Analysis. ACS omega 2023, 8, 6126–6138. [Google Scholar] [CrossRef]
- Radziuviene, G.; Rasmusson, A.; Augulis, R.; Lesciute-Krilaviciene, D.; Laurinavi- ciene, A.; Clim, E.; Laurinavicius, A. Automated Image Analysis of HER2 Fluores- cence in Situ Hybridization to Refine Definitions of Genetic Heterogeneity in Breast Cancer Tissue. BioMed research international 2017, 2017. [Google Scholar] [CrossRef]
- Prabakaran, I.; Wu, Z.; Lee, C.; Tong, B.; Steeman, S.; Koo, G.; Zhang, P.J.; Gu- vakova, M.A. Gaussian Mixture Models for Probabilistic Classification of Breast Can- cer. Cancer research 2019, 79, 3492–3502. [Google Scholar] [CrossRef]
- Skibinski, A.; Kuperwasser, C. The Origin of Breast Tumor Heterogeneity. Oncogene 2015, 34, 5309–5316. [Google Scholar] [CrossRef] [PubMed]
- Horii, R.; Nitta, H.; Nojima, M.; Maruyama, R.; Ueno, T.; Ito, Y.; Ohno, S.; Banks, P.; Kanda, H.; Akiyama, F. Predictive Significance of HER2 Intratumoral Hetero- geneity, Determined by Simultaneous Gene and Protein Analysis, for Resistance to Trastuzumab-Based Treatments for HER2-Positive Breast Cancer. Virchows Archiv 2021, 479, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Nitta, H.; Wei, L.; Banks, P.M.; Portier, B.; Parwani, A.V.; Li, Z. HER2 Intratumoral Heterogeneity Is Independently Associated with Incomplete Response to Anti-HER2 Neoadjuvant Chemotherapy in HER2-Positive Breast Carcinoma. Breast cancer research and treatment 2017, 166, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Zhang, S.; Yazdanparast, A.; Li, M.; Pawar, A.V.; Liu, Y.; Inavolu, S.M.; Cheng, L. Comprehensive Comparison of Molecular Portraits Between Cell Lines and Tumors in Breast Cancer. BMC genomics 2016, 17, 281–301. [Google Scholar] [CrossRef]
- Bui, M.M.; Riben, M.W.; Allison, K.H.; Chlipala, E.; Colasacco, C.; Kahn, A.G.; Lac- chetti, C.; Madabhushi, A.; Pantanowitz, L.; Salama, M.E.; et al. Quantitative Image Analysis of Human Epidermal Growth Factor Receptor 2 Immunohistochemistry for Breast Cancer: Guideline from the College of American Pathologists. Archives of pathology & laboratory medicine 2019, 143, 1180–1195. [Google Scholar]
- Yousif, M.; Huang, Y.; Sciallis, A.; Kleer, C.G.; Pang, J.; Smola, B.; Naik, K.; McClintock, D.S.; Zhao, L.; Kunju, L.P.; et al. Quantitative Image Analysis as an Adjunct to Manual Scoring of ER, PgR, and HER2 in Invasive Breast Carcinoma. American Journal of Clinical Pathology 2022, 157, 899–907. [Google Scholar] [CrossRef]
- Farahmand, S.; Fernandez, A.I.; Ahmed, F.S.; Rimm, D.L.; Chuang, J.H.; Reisenbich- ler, E.; Zarringhalam, K. Deep Learning Trained on Hematoxylin and Eosin Tumor Region of Interest Predicts HER2 Status and Trastuzumab Treatment Response in HER2+ Breast Cancer. Modern Pathology 2022, 35, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Helin, H.O.; Tuominen, V.J.; Ylinen, O.; Helin, H.J.; Isola, J. Free Digital Image Analysis Software Helps to Resolve Equivocal Scores in HER2 Immunohistochemistry. Virchows Archiv 2016, 468, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Tarantino, P.; Hamilton, E.; Tolaney, S.M.; Cortes, J.; Morganti, S.; Ferraro, E.; Marra, A.; Viale, G.; Trapani, D.; Cardoso, F.; et al. HER2-Low Breast Cancer: Pathological and Clinical Landscape. Journal of Clinical Oncology 2020, 38, 1951–1962. [Google Scholar] [CrossRef]
- Modi, S.; Jacot, W.; Yamashita, T.; Sohn, J.; Vidal, M.; Tokunaga, E.; Tsurutani, J.; Ueno, N.T.; Prat, A.; Chae, Y.S.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. New England Journal of Medicine 2022, 387, 9–20. [Google Scholar] [CrossRef]
- Ogitani, Y.; Aida, T.; Hagihara, K.; Yamaguchi, J.; Ishii, C.; Harada, N.; Soma, M.; Okamoto, H.; Oitate, M.; Arakawa, S.; et al. DS-8201a, a Novel HER2-Targeting ADC with a Novel DNA Topoisomerase i Inhibitor, Demonstrates a Promising An- titumor Efficacy with Differentiation from t-DM1Preclinical Efficacy of DS-8201a, a Novel HER2-Targeting ADC. Clinical Cancer Research 2016, 22, 5097–5108. [Google Scholar] [CrossRef] [PubMed]
- Ogitani, Y.; Hagihara, K.; Oitate, M.; Naito, H.; Agatsuma, T. Bystander Killing Effect of DS-8201a, a Novel Anti-Human Epidermal Growth Factor Receptor 2 Antibody–Drug Conjugate, in Tumors with Human Epidermal Growth Factor Re- ceptor 2 Heterogeneity. Cancer science 2016, 107, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Deluche, E.; Lusque, A.; Le Bescond, L.; Filleron, T.; Pradat, Y.; Ducoulombier, A.; Pistilli, B.; Bachelot, T.; Viret, F.; et al. Trastuzumab Derux- tecan in Metastatic Breast Cancer with Variable HER2 Expression: The Phase 2 DAISY Trial. Nature medicine 2023, 29, 2110–2120. [Google Scholar] [CrossRef]
- Bang, Y.-J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in Combination with Chemotherapy Versus Chemotherapy Alone for Treatment of HER2-Positive Advanced Gastric or Gastro-Oesophageal Junction Cancer (ToGA): A Phase 3, Open- Label, Randomised Controlled Trial. The Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Johnson, A.M.; Dumbrava, E.E.I.; Raghav, K.; Balaji, K.; Bhatt, M.; Murthy, R.K.; Rodon, J.; Piha-Paul, S.A. Advances in HER2-Targeted Therapy: Novel Agents and Opportunities Beyond Breast and Gastric Cancer. Clinical Cancer Research 2019, 25, 2033–2041. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, J.T.; Winther, H.; Askaa, J.; Andresen, L.; Olsen, D.; Mollerup, J. A Companion Diagnostic with Significant Clinical Impact in Treatment of Breast and Gastric Cancer. Frontiers in Oncology 2021, 11, 676939. [Google Scholar] [CrossRef]
- Grillo, F.; Fassan, M.; Sarocchi, F.; Fiocca, R.; Mastracci, L. HER2 Heterogeneity in Gastric/Gastroesophageal Cancers: From Benchside to Practice. World journal of gastroenterology 2016, 22, 5879. [Google Scholar] [CrossRef]
- Bang, K.; Cheon, J.; Park, Y.S.; Kim, H.-D.; Ryu, M.-H.; Park, Y.; Moon, M.; Lee, H.; Kang, Y.-K. Association Between HER2 Heterogeneity and Clinical Outcomes of HER2-Positive Gastric Cancer Patients Treated with Trastuzumab. Gastric cancer 2022, 25, 794–803. [Google Scholar] [CrossRef]
Figure 1.
HER2 intratumoral heterogeneity was identified by the shape of HER2 FISH amplification distributed histograms with HER2 gene copy number within a tumor sample (A). Those histograms were classified into two groups: the biphasic grade graph (high heterogeneous [HH] group) and the monophasic biphasic grade graph (low heterogeneous [LH] group) (B). Additionally, the LH group was divided into the ex-high amplification LH (Ex-High LH) group and the high amplification LH (High LH) group by the ratio of HER2 FISH (mean number of HER2 FISH signals/cell). Representative images on the left were HER2 FISH imaging with a HER2 (red signals)/CEP17 (green signals) in tumor cell nuclei (DAPI: blue).
Figure 1.
HER2 intratumoral heterogeneity was identified by the shape of HER2 FISH amplification distributed histograms with HER2 gene copy number within a tumor sample (A). Those histograms were classified into two groups: the biphasic grade graph (high heterogeneous [HH] group) and the monophasic biphasic grade graph (low heterogeneous [LH] group) (B). Additionally, the LH group was divided into the ex-high amplification LH (Ex-High LH) group and the high amplification LH (High LH) group by the ratio of HER2 FISH (mean number of HER2 FISH signals/cell). Representative images on the left were HER2 FISH imaging with a HER2 (red signals)/CEP17 (green signals) in tumor cell nuclei (DAPI: blue).
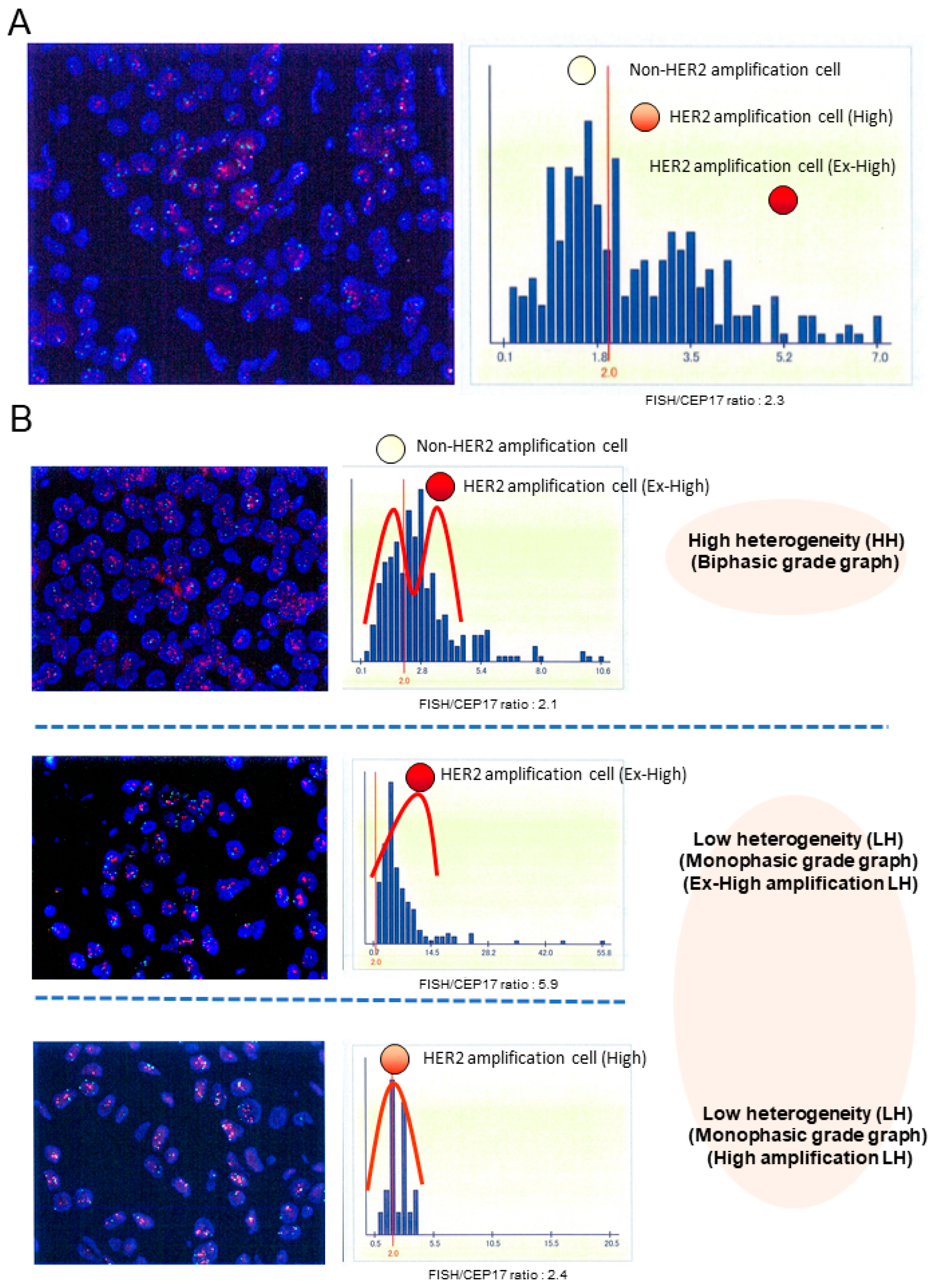
Figure 2.
Disease-free survival (DFS) and overall survival (OS) rates according to HER2 intratumoral heterogeneity. DFS (A) and OS rates (B) of patients with breast cancer com- paring the high heterogeneous (HH) group to the low heterogeneous (LH) group (DFS: P <. 0.001 (HH group:63% vs LH group:91% at 10 years), and OS: P = 0.012 (HH group:78% vs LH group:95% at 10 years). DFS rates (C) and OS rates (D) of patients with breast cancer were compared between the HH, Ex-High LH, and High LH groups.
Figure 2.
Disease-free survival (DFS) and overall survival (OS) rates according to HER2 intratumoral heterogeneity. DFS (A) and OS rates (B) of patients with breast cancer com- paring the high heterogeneous (HH) group to the low heterogeneous (LH) group (DFS: P <. 0.001 (HH group:63% vs LH group:91% at 10 years), and OS: P = 0.012 (HH group:78% vs LH group:95% at 10 years). DFS rates (C) and OS rates (D) of patients with breast cancer were compared between the HH, Ex-High LH, and High LH groups.
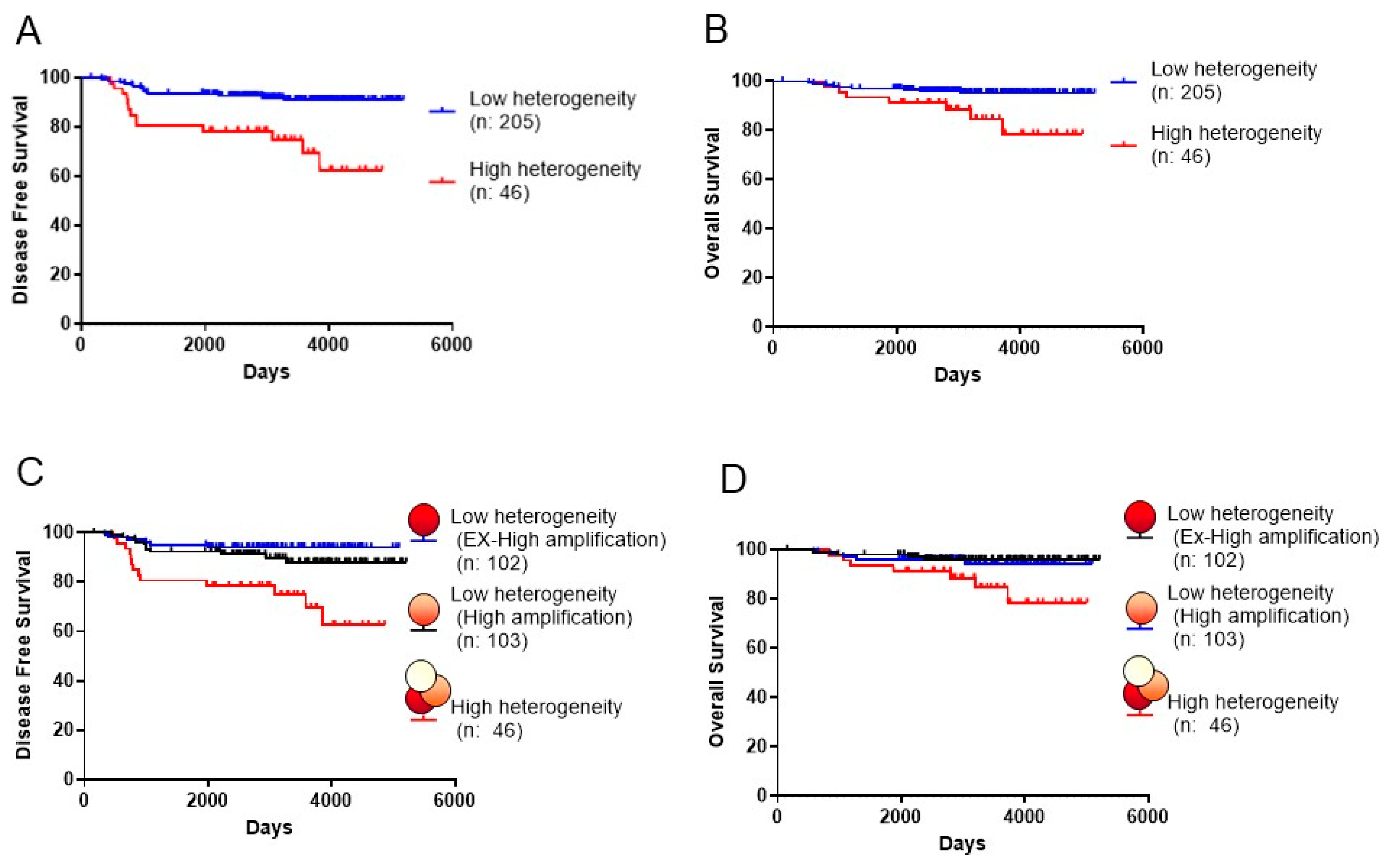
Table 1.
Patient Characteristics.
| Number | (%) | Number | (%) | ||
| Menopause | ER | ||||
| pre | 96 | (38.2) | Positive | 167 | (66.5) |
| post | 155 | (61.8) | Negative | 84 | (33.5) |
| Histological type | PgR | ||||
| IDC | 244 | (97.2) | Positive | 99 | (39.4) |
| ILC | 7 | (2.8) | Negative | 152 | (60.6) |
| Histological type | HER2 IHC | ||||
| 1 | 26 | (10.4) | 3+ | 232 | (92.4) |
| 2 | 91 | (36.3) | 2+ | 19 | (7.6) |
| 3 | 134 | (53.4) | HER2 ratio | ||
| Tumor size | Mean | 4.8 ± 2.5 | |||
| T1 | 86 | (34.3) | HER2 heterogeneity | ||
| T2 | 139 | (55.4) | High (HH) | 46 | (18.3) |
| T3 or T4 | 26 | (10.4) | Low (LH) | 205 | (81.7) |
| Pretreatment LN metastasis | (Ex-High-Amplification LH | 102 | (49.8)) | ||
| Negative | 134 | (53.4) | (High-Amplification LH | 103 | (50.2)) |
| Positive | 117 | (46.6) | Stage | ||
| Resected LN metastasis | I | 68 | (27.1) | ||
| Yes | 116 | (46.2) | II | 161 | (64.1) |
| No | 135 | (53.8) | III | 22 | (8.8) |
| Neoadjuvant chemotherapy | |||||
| Negative | 185 | (73.7) | |||
| Positive | 66 | (26.3) | |||
Table 2.
Association between Patient Characteristics and HER2 heterogeneity.
| HER-2 heterogeneity | ||||
| Total | High(HH) | Low(LH) | P value | |
| n = 251, % | n = 46, % | n = 205, % | ||
| Menopause | ||||
| pre | 96 (38.2%) | 23 (50.0%) | 73 (35.6%) | 0.1 |
| post | 155 (61.8%) | 23 (50.0%) | 132 (64.4%) | |
| Histological type | ||||
| IDC | 244 (97.2%) | 43 (93.5%) | 201 (98.0%) | 0.118 |
| ILC | 7 ( 2.8%) | 3 ( 6.5%) | 4 ( 2.0%) | |
| Histological grade | ||||
| 1 | 26 (10.4%) | 3 ( 6.5%) | 23 (11.2%) | 0.421 |
| 2 | 91 (36.3%) | 20 (43.5%) | 71 (34.6%) | |
| 3 | 134 (53.4%) | 23 (50.0%) | 111 (54.1%) | |
| Tumor size | ||||
| T1 | 86 (34.3%) | 14 (30.4%) | 72 (35.1%) | 0.665 |
| T2-T4 | 165 (65.7%) | 32 (69.6%) | 133 (64.9%) | |
| Pretreatment LN metastasis | ||||
| Negative | 134 (53.4%) | 19 (41.3%) | 115 (56.1%) | 0.098 |
| Positive | 117 (46.6%) | 27 (58.7%) | 90 (43.9%) | |
| Resected LN metastasis | ||||
| Negative | 185 (73.7%) | 31 (67.4%) | 154 (75.1%) | 0.373 |
| Positive | 66 (26.3%) | 15 (32.6%) | 51 (24.9%) | |
| ER | ||||
| Positive | 167 (66.5%) | 40 (87.0%) | 127 (62.0%) | 0.002 |
| Negative | 84 (33.5%) | 6 (13.0%) | 78 (38.0%) | |
| PgR | ||||
| Positive | 99 (39.4%) | 29 (63.0%) | 70 (34.1%) | 5.46E-04 |
| Negative | 152 (60.6%) | 17 (37.0%) | 135 (65.9%) | |
| HER2 IHC | ||||
| 3+ | 232 (92.4%) | 40 (87.0%) | 192 (93.7%) | 0.128 |
| 2+ | 19 ( 7.6%) | 6 (13.0%) | 13 ( 6.3%) | |
| HER2 ratio | ||||
| Mean | 4.8 ± 2.5 | 2.3 ± 0.4 | 5.4 ± 2.4 | 1.03E-43 |
| Neoadjuvant chemotherapy | ||||
| Yes | 116 (46.2%) | 27 (58.7%) | 89 (43.4%) | 0.086 |
| No | 135 (53.8%) | 19 (41.3%) | 116 (56.6%) | |
| Stage | ||||
| I | 68 (27.1%) | 8 (17.4%) | 60 (29.3%) | 0.094 |
| II | 161 (64.1%) | 31 (67.4%) | 130 (63.4%) | |
| III | 22 ( 8.8%) | 7 (15.2%) | 15 ( 7.3%) | |
Table 3.
Univariate and Multivariate Analysis of Various Factors on Disease Free Survival in HER2-Positive Patients.
Table 3.
Univariate and Multivariate Analysis of Various Factors on Disease Free Survival in HER2-Positive Patients.
| Univariate analysis | Multivariate analysis | |||||||
| Characteristic | N | Event(N) | HR | 95% CI | P value | HR | 95% CI | P value |
| cT | ||||||||
| T1 | 86 | 8 | 1 | 1 | ||||
| T2-T4 | 165 | 22 | 1.41 | 0.63 - 3.16 | 0.41 | 1.04 | 0.45 - 2.40 | 0.932 |
| ER | ||||||||
| Positive | 167 | 20 | 1 | 1 | ||||
| Negative | 84 | 10 | 0.99 | 0.46 - 2.12 | 0.985 | 1.63 | 0.62 - 4.29 | 0.326 |
| PgR | ||||||||
| Positive | 99 | 14 | 1 | 1 | ||||
| Negative | 152 | 16 | 0.76 | 0.37 - 1.56 | 0.452 | 0.95 | 0.39 - 2.33 | 0.918 |
| Pretreatment LN metastasis | ||||||||
| Negative | 134 | 10 | 1 | 1 | ||||
| Positive | 117 | 20 | 2.33 | 1.09 - 4.98 | 0.029 | 1.23 | 0.43 - 3.52 | 0.696 |
| Resected LN metastasis | ||||||||
| Negative | 185 | 16 | 1 | 1 | ||||
| Positive | 66 | 14 | 2.6 | 1.27 - 5.32 | 0.009 | 2.26 | 0.81 - 6.30 | 0.12 |
| HG | ||||||||
| 1 or 2 | 117 | 14 | 1 | 1 | ||||
| 3 | 134 | 16 | 1.04 | 0.51 - 2.13 | 0.919 | 1.24 | 0.59 - 2.63 | 0.57 |
| HER2 heterogeneity | ||||||||
| Low (LH) | 205 | 17 | 1 | 1 | ||||
| High (HH) | 46 | 13 | 3.8 | 1.84 - 7.83 | 2.95E-04 | 4.03 | 1.83 - 8.87 | 5.51E-04 |
Table 4.
Univariate and Multivariate Analysis of Various Factors on Overall Survival in HER2-Positive Patients.
Table 4.
Univariate and Multivariate Analysis of Various Factors on Overall Survival in HER2-Positive Patients.
| Univariate analysis | Multivariate analysis | |||||||
| Characteristic | N | Event(N) | HR | 95% CI | P value | HR | 95% CI | P value |
| cT | ||||||||
| T1 | 86 | 8 | 1 | 1 | ||||
| T2-T4 | 165 | 22 | 2.22 | 0.63 - 7.78 | 0.214 | 1.44 | 0.39 - 5.24 | 0.584 |
| ER | ||||||||
| Positive | 167 | 20 | 1 | 1 | ||||
| Negative | 84 | 10 | 0.67 | 0.22 - 2.07 | 0.485 | 0.78 | 0.21 - 2.94 | 0.715 |
| PgR | ||||||||
| Positive | 99 | 14 | 1 | 1 | ||||
| Negative | 152 | 16 | 0.88 | 0.33 - 2.36 | 0.797 | 1.67 | 0.53 - 5.26 | 0.382 |
| Pretreatment LN metastasis | ||||||||
| Negative | 134 | 10 | 1 | 1 | ||||
| Positive | 117 | 20 | 4.84 | 1.38 - 16.99 | 0.014 | 2.79 | 0.61 - 12.90 | 0.188 |
| Resected LN metastasis | ||||||||
| Negative | 185 | 16 | 1 | 1 | ||||
| Positive | 66 | 14 | 3.71 | 1.38 - 9.96 | 0.009 | 1.83 | 0.53 - 6.26 | 0.336 |
| HG | ||||||||
| 1 or 2 | 117 | 14 | 1 | 1 | ||||
| 3 | 134 | 16 | 0.7 | 0.26 - 1.88 | 0.482 | 0.84 | 0.31 - 2.33 | 0.745 |
| HER2 heterogeneity | ||||||||
| Low (LH) | 205 | 17 | 1 | 1 | ||||
| High (HH) | 46 | 13 | 3.55 | 1.32 - 9.54 | 0.012 | 3.1 | 1.06 - 9.08 | 0.039 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Copyright: This open access article is published under a Creative Commons CC BY 4.0 license, which permit the free download, distribution, and reuse, provided that the author and preprint are cited in any reuse.
MDPI Initiatives
Important Links
© 2024 MDPI (Basel, Switzerland) unless otherwise stated






