Submitted:
21 February 2024
Posted:
22 February 2024
You are already at the latest version
Abstract
Keywords:
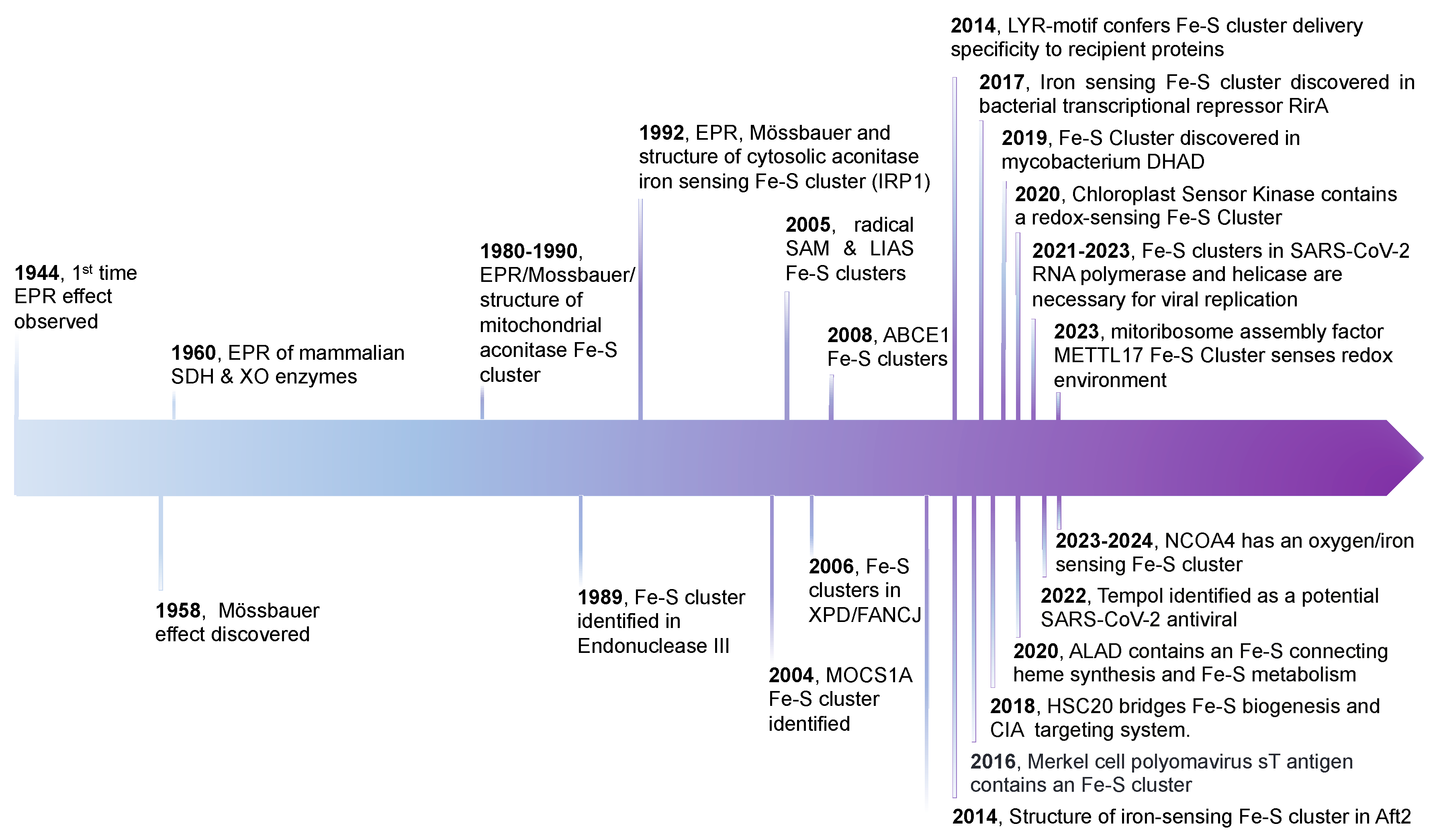
1. Introduction
2. FeS Cluster Structure, Geometries, and Biogenesis
3. Human Iron Metabolism
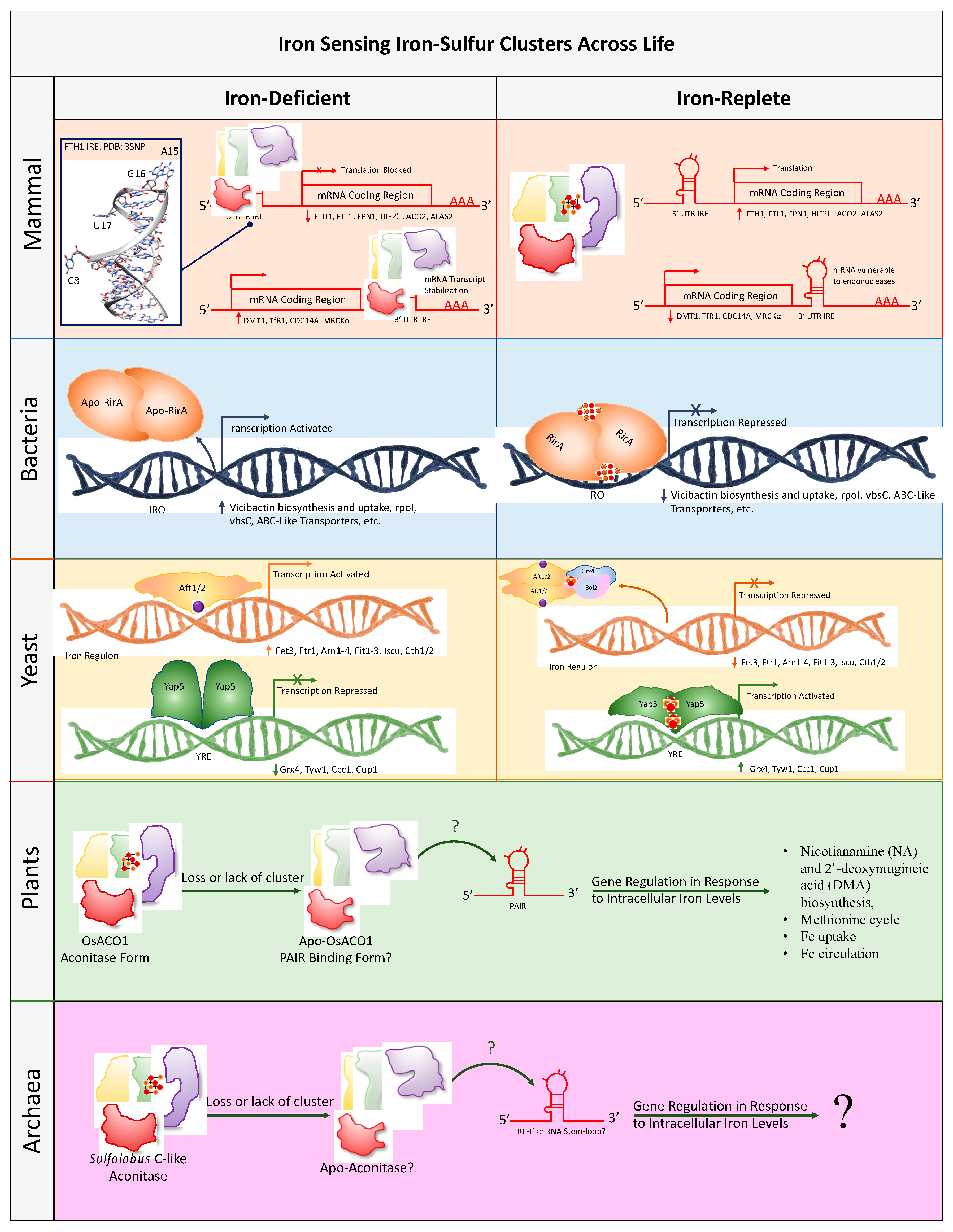
4. Intracellular Iron and Oxygen Sensing
4.1. Mammalian Iron Sensing Iron-Sulfur Clusters
4.2. Bacterial Iron Sensing Iron-Sulfur Clusters
4.3. Yeast Iron Sensing Iron-Sulfur Clusters
4.4. Plant Iron Sensing Iron-Sulfur Clusters
4.5. Archaea Iron Sensing Iron-Sulfur Clusters
5. Conclusions, Future Perspectives and Remaining Questions
References
- Rouault, T.A. The indispensable role of mammalian iron sulfur proteins in function and regulation of multiple diverse metabolic pathways. Biometals 2019, 32, 343–353. [CrossRef]
- Beinert, H. Iron-sulfur proteins: ancient structures, still full of surprises. J Biol Inorg Chem 2000, 5, 2–15. [CrossRef]
- Beinert, H.; Holm, R.H.; Münck, E. sIron-sulfur clusters: Nature's modular, multipurpose structures. Science 1997, 277, 653–659. [CrossRef]
- Koonin, E.V.; Martin, W. On the origin of genomes and cells within inorganic compartments. Trends in Genetics 2005, 21, 647–654. [CrossRef]
- Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol 2006, 59, 1073–1082. [CrossRef]
- Barton, J.K.; Silva, R.M.B.; O'Brien, E. Redox Chemistry in the Genome: Emergence of the [4Fe4S] Cofactor in Repair and Replication. Annual Review of Biochemistry 2019, 88, 163–190. [CrossRef]
- Maio, N.; Zhang, D.-L.; Ghosh, M.C.; Jain, A.; SantaMaria, A.M.; Rouault, T.A. Mechanisms of cellular iron sensing, regulation of erythropoiesis and mitochondrial iron utilization. Seminars in Hematology 2021, 58, 161–174. [CrossRef]
- Pellicer Martinez, M.T.; Crack, J.C.; Stewart, M.Y.; Bradley, J.M.; Svistunenko, D.A.; Johnston, A.W.; Cheesman, M.R.; Todd, J.D.; Le Brun, N.E. Mechanisms of iron- and O(2)-sensing by the [4Fe-4S] cluster of the global iron regulator RirA. Elife 2019, 8. [CrossRef]
- Mettert, E.L.; Kiley, P.J. Fe-S proteins that regulate gene expression. Biochimica et Biophysica Acta - Molecular Cell Research 2014, 1853, 1284–1293. [CrossRef]
- Rouault, T.A.; Maio, N. Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. Journal of Biological Chemistry 2017, 292, 12744–12753. [CrossRef]
- Rouault, T.A. Biogenesis of iron-sulfur clusters in mammalian cells: New insights and relevance to human disease. DMM Disease Models and Mechanisms 2012, 5, 155–164. [CrossRef]
- Beinert, H.; Sands, R.H. Studies on succinic and DPNH dehydrogenase preparations by paramagnetic resonance (EPR) spectroscopy. 1960.
- Rouault, T.A. Iron-sulfur proteins hiding in plain sight. Nature Chemical Biology 2015, 11, 442–445. [CrossRef]
- Oliveira, M.T.; Garesse, R.; Kaguni, L.S. Animal models of mitochondrial DNA transactions in disease and ageing. Experimental Gerontology 2010, 10.1016/j.exger.2010.01.019. [CrossRef]
- Prakash, A.; Doublié, S. Base Excision Repair in the Mitochondria. Journal of Cellular Biochemistry 2015, 116, 1490–1499. [CrossRef]
- Fuss, J.O.; Tsai, C.L.; Ishida, J.P.; Tainer, J.A. Emerging critical roles of Fe-S clusters in DNA replication and repair. In Biochimica et Biophysica Acta - Molecular Cell Research, Elsevier: 2015; Vol. 1853, pp 1253-1271.
- Puig, S.; Ramos-Alonso, L.; Romero, A.M.; Martinez-Pastor, M.T. The elemental role of iron in DNA synthesis and repair. Metallomics 2017, 9, 1483–1500. [CrossRef]
- Baranovskiy, A.G.; Siebler, H.M.; Pavlov, Y.I.; Tahirov, T.H. Iron–Sulfur Clusters in DNA Polymerases and Primases of Eukaryotes, 1 ed.; Methods in Enzymology. Elsevier Inc.: 2018; Vol. 599, pp. 1–20.
- Holt, M.E.; Salay, L.E.; Chazin, W.J. A Polymerase With Potential: The Fe–S Cluster in Human DNA Primase, 1 ed.; Methods in Enzymology. Elsevier Inc.: 2017; Vol. 595, pp. 361–390.
- Netz, D.J.A.; Stith, C.M.; Stümpfig, M.; Köpf, G.; Vogel, D.; Genau, H.M.; Stodola, J.L.; Lill, R.; Burgers, P.M.J.; Pierik, A.J. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nature Chemical Biology 2012, 8, 125–132. [CrossRef]
- Troadec, M.B.; Loréal, O.; Brissot, P. The interaction of iron and the genome: For better and for worse. Mutation Research - Reviews in Mutation Research 2017, 774, 25–32. [CrossRef]
- O'Brien, E.; Barton, J.K.; Holt, M.E.; Thompson, M.K.; Salay, L.E.; Ehlinger, A.C.; Chazin, W.J. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 2017, 355. [CrossRef]
- Lukianova, O.A.; David, S.S. A role for iron-sulfur clusters in DNA repair. Current Opinion in Chemical Biology 2005, 9, 145–151. [CrossRef]
- ter Beek, J.; Parkash, V.; Bylund, G.O.; Osterman, P.; Sauer-Eriksson, A.E.; Johansson, E. Structural evidence for an essential Fe–S cluster in the catalytic core domain of DNA polymerase ϵ. Nucleic Acids Research 2019, 47, 5712–5722. [CrossRef]
- Veatch, J.R.; McMurray, M.A.; Nelson, Z.W.; Gottschling, D.E. Mitochondrial Dysfunction Leads to Nuclear Genome Instability via an Iron-Sulfur Cluster Defect. Cell 2009, 137, 1247–1258. [CrossRef]
- Yien, Y.Y.; Shi, J.; Chen, C.; Cheung, J.T.M.; Grillo, A.S.; Shrestha, R.; Li, L.; Zhang, X.; Kafina, M.D.; Kingsley, P.D.; et al. FAM210B is an erythropoietin target and regulates erythroid heme synthesis by controlling mitochondrial iron import and ferrochelatase activity. Journal of Biological Chemistry 2018, 293, 19797–19811. [CrossRef]
- Todd, J.D.; Wexler, M.; Sawers, G.; Yeoman, K.H.; Poole, P.S.; Johnston, A.W.B. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology (Reading) 2002, 148 (Pt 12), 4059–4071. [CrossRef]
- Mancias, J.D.; Vaites, L.P.; Nissim, S.; Biancur, D.E.; Kim, A.J.; Wang, X.; Liu, Y.; Goessling, W.; Kimmelman, A.C.; Harper, J.W. Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. eLife 2015, 4, 1–19. [CrossRef]
- Zhang, D.L.; Ghosh, M.C.; Rouault, T.A. The physiological functions of iron regulatory proteins in iron homeostasis - an update. Front Pharmacol 2014, 5, 124. [CrossRef]
- Couturier, J.; Przybyla-Toscano, J.; Roret, T.; Didierjean, C.; Rouhier, N. The roles of glutaredoxins ligating Fe-S clusters: Sensing, transfer or repair functions? Biochimica et Biophysica Acta - Molecular Cell Research 2015, 1853, 1513–1527. [CrossRef]
- Jain, R.; Vanamee, E.S.; Dzikovski, B.G.; Buku, A.; Johnson, R.E.; Prakash, L.; Prakash, S.; Aggarwal, A.K. An iron-sulfur cluster in the polymerase domain of yeast DNA polymerase epsilon. J Mol Biol 2014, 426, 301–308. [CrossRef]
- Maio, N.; Lafont, B.A.P.; Sil, D.; Li, Y.; Bollinger, J.M., Jr.; Krebs, C.; Pierson, T.C.; Linehan, W.M.; Rouault, T.A. Fe-S cofactors in the SARS-CoV-2 RNA-dependent RNA polymerase are potential antiviral targets. Science 2021, 373, 236–241. [CrossRef]
- Maio, N.; Raza, M.K.; Li, Y.; Zhang, D.L.; Bollinger, J.M., Jr.; Krebs, C.; Rouault, T.A. An iron-sulfur cluster in the zinc-binding domain of the SARS-CoV-2 helicase modulates its RNA-binding and -unwinding activities. Proc Natl Acad Sci U S A 2023, 120, e2303860120. [CrossRef]
- Maio, N.; Cherry, S.; Schultz, D.C.; Hurst, B.L.; Linehan, W.M.; Rouault, T.A. TEMPOL inhibits SARS-CoV-2 replication and development of lung disease in the Syrian hamster model. iScience 2022, 25, 105074. [CrossRef]
- Rouault, T.A.; Klausner, R.D. Iron-sulfur clusters as biosensors of oxidants and iron. Trends Biochem Sci 1996, 21, 174–177. [CrossRef]
- Bak, D.W.; Elliott, S.J. Alternative FeS cluster ligands: tuning redox potentials and chemistry. Curr Opin Chem Biol 2014, 19, 50–58. [CrossRef]
- Gray, E.; Stewart, M.Y.Y.; Hanwell, L.; Crack, J.C.; Devine, R.; Stevenson, C.E.M.; Volbeda, A.; Johnston, A.W.B.; Fontecilla-Camps, J.C.; Hutchings, M.I.; et al. Stabilisation of the RirA [4Fe-4S] cluster results in loss of iron-sensing function. Chem Sci 2023, 14, 9744–9758. [CrossRef]
- Rouault, T.A. Mammalian iron-sulphur proteins: Novel insights into biogenesis and function. Nature Reviews Molecular Cell Biology 2015, 16, 45–55. [CrossRef]
- Maio, N.; Kim, K.S.; Singh, A.; Rouault, T.A. A Single Adaptable Cochaperone-Scaffold Complex Delivers Nascent Iron-Sulfur Clusters to Mammalian Respiratory Chain Complexes I–III. Cell Metabolism 2017, 25, 945-953.e946. [CrossRef]
- Ibrahim, I.M.; Wu, H.; Ezhov, R.; Kayanja, G.E.; Zakharov, S.D.; Du, Y.; Tao, W.A.; Pushkar, Y.; Cramer, W.A.; Puthiyaveetil, S. An evolutionarily conserved iron-sulfur cluster underlies redox sensory function of the Chloroplast Sensor Kinase. Commun Biol 2020, 3, 13. [CrossRef]
- Lu, Y. Assembly and Transfer of Iron-Sulfur Clusters in the Plastid. Front Plant Sci 2018, 9, 336. [CrossRef]
- Kim, K.S.; Maio, N.; Singh, A.; Rouault, T.A. Cytosolic HSC20 integrates de novo iron-sulfur cluster biogenesis with the CIAO1-mediated transfer to recipients. Human Molecular Genetics 2018, 27, 837–852. [CrossRef]
- Sharma, A.K.; Pallesen, L.J.; Spang, R.J.; Walden, W.E. Cytosolic iron-sulfur cluster assembly (CIA) system: Factors, mechanism, and relevance to cellular iron regulation. Journal of Biological Chemistry 2010, 285, 26745–26751. [CrossRef]
- Shi, R.; Hou, W.; Wang, Z.Q.; Xu, X. Biogenesis of Iron-Sulfur Clusters and Their Role in DNA Metabolism. Front Cell Dev Biol 2021, 9, 735678. [CrossRef]
- Maio, N.; Rouault, T.A. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochimica et Biophysica Acta - Molecular Cell Research 2015, 1853, 1493–1512. [CrossRef]
- Maio, N.; Rouault, T.A. Outlining the Complex Pathway of Mammalian Fe-S Cluster Biogenesis. Trends Biochem Sci 2020, 45, 411–426. [CrossRef]
- Rouault, T.A.; Tong, W.H. Iron-sulfur cluster biogenesis and human disease. Trends in Genetics 2008, 24, 398–407. [CrossRef]
- Uhrigshardt, H.; Singh, A.; Kovtunovych, G.; Ghosh, M.; Rouault, T.A. Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis. Hum Mol Genet 2010, 19, 3816–3834. [CrossRef]
- Esquilin-Lebron, K.; Dubrac, S.; Barras, F.; Boyd, J.M. Bacterial Approaches for Assembling Iron-Sulfur Proteins. mBio 2021, 12, e0242521. [CrossRef]
- Jain, A.; Singh, A.; Maio, N.; Rouault, T.A. Assembly of the [4Fe-4S] cluster of NFU1 requires the coordinated donation of two [2Fe-2S] clusters from the scaffold proteins, ISCU2 and ISCA1. Hum Mol Genet 2020, 29, 3165–3182. [CrossRef]
- Legati, A.; Reyes, A.; Ceccatelli Berti, C.; Stehling, O.; Marchet, S.; Lamperti, C.; Ferrari, A.; Robinson, A.J.; Muhlenhoff, U.; Lill, R.; et al. A novel de novo dominant mutation in ISCU associated with mitochondrial myopathy. J Med Genet 2017, 54, 815–824. [CrossRef]
- Srour, B.; Gervason, S.; Hoock, M.H.; Monfort, B.; Want, K.; Larkem, D.; Trabelsi, N.; Landrot, G.; Zitolo, A.; Fonda, E.; et al. Iron Insertion at the Assembly Site of the ISCU Scaffold Protein Is a Conserved Process Initiating Fe-S Cluster Biosynthesis. J Am Chem Soc 2022, 144, 17496–17515. [CrossRef]
- Herrera, M.G.; Noguera, M.E.; Sewell, K.E.; Agudelo Suarez, W.A.; Capece, L.; Klinke, S.; Santos, J. Structure of the Human ACP-ISD11 Heterodimer. Biochemistry 2019, 58, 4596–4609. [CrossRef]
- Friemel, M.; Marelja, Z.; Li, K.; Leimkuhler, S. The N-Terminus of Iron-Sulfur Cluster Assembly Factor ISD11 Is Crucial for Subcellular Targeting and Interaction with l-Cysteine Desulfurase NFS1. Biochemistry 2017, 56, 1797–1808. [CrossRef]
- Hershkovitz, T.; Kurolap, A.; Tal, G.; Paperna, T.; Mory, A.; Staples, J.; Brigatti, K.W.; Regeneron Genetics, C.; Gonzaga-Jauregui, C.; Dumin, E.; et al. A recurring NFS1 pathogenic variant causes a mitochondrial disorder with variable intra-familial patient outcomes. Mol Genet Metab Rep 2021, 26, 100699. [CrossRef]
- Patra, S.; Barondeau, D.P. Mechanism of activation of the human cysteine desulfurase complex by frataxin. Proc Natl Acad Sci U S A 2019, 116, 19421–19430. [CrossRef]
- Maio, N.; Rouault, T.A. Mammalian iron sulfur cluster biogenesis: From assembly to delivery to recipient proteins with a focus on novel targets of the chaperone and co-chaperone proteins. IUBMB Life 2022, 74, 684–704. [CrossRef]
- Kakhlon, O.; Cabantchik, Z.I. The labile iron pool: Characterization, measurement, and participation in cellular processes. Free Radical Biology and Medicine 2002, 33, 1037–1046. [CrossRef]
- Philpott, C.C.; Protchenko, O.; Wang, Y.; Novoa-Aponte, L.; Leon-Torres, A.; Grounds, S.; Tietgens, A.J. Iron-tracking strategies: Chaperones capture iron in the cytosolic labile iron pool. Front Mol Biosci 2023, 10, 1127690. [CrossRef]
- Hider, R.C.; Kong, X.L. Glutathione: a key component of the cytoplasmic labile iron pool. BioMetals 2011, 24. [CrossRef]
- Patel, S.J.; Frey, A.G.; Palenchar, D.J.; Achar, S.; Bullough, K.Z.; Vashisht, A.; Wohlschlegel, J.A.; Philpott, C.C. A PCBP1-BolA2 chaperone complex delivers iron for cytosolic [2Fe-2S] cluster assembly. Nat Chem Biol 2019, 15, 872–881. [CrossRef]
- Li, H.; Mapolelo, D.T.; Randeniya, S.; Johnson, M.K.; Outten, C.E. Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2. Biochemistry 2012, 51, 1687–1696. [CrossRef]
- Li, H.; Outten, C.E. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry 2012, 51, 4377–4389. [CrossRef]
- Sheftel, A.D.; Wilbrecht, C.; Stehling, O.; Niggemeyer, B.; Elsasser, H.P.; Muhlenhoff, U.; Lill, R. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol Biol Cell 2012, 23, 1157–1166. [CrossRef]
- Maio, N.; Singh, A.; Uhrigshardt, H.; Saxena, N.; Tong, W.H.; Rouault, T.A. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metabolism 2014, 19, 445–457. [CrossRef]
- Zheng, L.; Cash, V.L.; Flint, D.H.; Dean, D.R. Assembly of Iron-Sulfur Clusters. Journal of Biological Chemistry 1998, 273, 13264–13272. [CrossRef]
- Frazzon, J.; Dean, D.R. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr Opin Chem Biol 2003, 7, 166–173. [CrossRef]
- Saito, H.; Sargent, T.; Parker, H.G.; Lawrence, J.H. Whole-body Iron Loss In Normal Man Measured with a Gamma Spectrometer'. Journal of Nuclear Medicine 1964, 5, 571–580.
- Andrews, N.C. The iron transporter DMT1. The international journal of biochemistry & cell biology 1999, 31, 991–994. [CrossRef]
- Zoller, H.; Theurl, I.; Koch, R.; Kaser, A.; Weiss, G. Mechanisms of iron mediated regulation of the duodenal iron transporters divalent metal transporter 1 and ferroportin 1. Blood cells, molecules & diseases 2002, 29, 488–497. [CrossRef]
- Ma, Y.; Yeh, M.; Yeh, K.-y.; Glass, J. Iron Imports. V. Transport of iron through the intestinal epithelium. American Journal of Physiology-Gastrointestinal and Liver Physiology 2006, 290, G417-G422. [CrossRef]
- Andrews, N.C. Iron homeostasis: Insights from genetics and animal models. Nature Reviews Genetics 2000, 1, 208–217. [CrossRef]
- Canonne-Hergaux, F.; Zhang, A.S.; Ponka, P.; Gros, P. Characterization of the iron transporter DMT1 (NRAMP2/DCT1) in red blood cells of normal and anemic mk/mk mice. Blood 2001, 98, 3823–3830. [CrossRef]
- Shaw, G.C.; Cope, J.J.; Li, L.; Corson, K.; Hersey, C.; Ackermann, G.E.; Gwynn, B.; Lambert, A.J.; Wingert, R.A.; Traver, D.; et al. Mitoferrin is essential for erythroid iron assimilation. Nature 2006, 440, 96–100. [CrossRef]
- Chung, J.; Anderson, S.A.; Gwynn, B.; Deck, K.M.; Chen, M.J.; Langer, N.B.; Shaw, G.C.; Huston, N.C.; Boyer, L.F.; Datta, S.; et al. Iron regulatory protein-1 protects against mitoferrin-1-deficient porphyria. Journal of Biological Chemistry 2014, 289, 7835–7843. [CrossRef]
- Friend, C.; Scher, W.; Holland, J.G.; Sato, T. Hemoglobin synthesis in murine virus-induced leukemic cells in vitro: stimulation of erythroid differentiation by dimethyl sulfoxide. Proceedings of the National Academy of Sciences of the United States of America 1971, 68, 378–382. [CrossRef]
- Levi, S.; Rovida, E. The role of iron in mitochondrial function. Biochimica et Biophysica Acta - General Subjects 2009, 1790, 629–636. [CrossRef]
- Hentze, M.W.; Muckenthaler, M.U.; Andrews, N.C. Balancing Acts: Molecular Control of Mammalian Iron Metabolism. Cell 2004, 117, 285–297. [CrossRef]
- Andrews, N.C.; Andrews, N.C. Forging a field : the golden age of iron biology ASH 50th anniversary review Forging a field : the golden age of iron biology. Blood 2009, 112, 219–230. [CrossRef]
- Knutson, M.D.; Oukka, M.; Koss, L.M.; Aydemir, F.; Wessling-Resnick, M. Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin. Proceedings of the National Academy of Sciences 2005, 102, 1324–1328. [CrossRef]
- Donovan, A.; Brownlie, A.; Zhou, Y.; Shepard, J.; Pratt, S.J.; Moynihan, J.; Paw, B.H.; Drejer, A.; Barut, B.; Zapata, A.; et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 2000, 403, 776–781. [CrossRef]
- Sabelli, M.; Montosi, G.; Garuti, C.; Caleffi, A.; Oliveto, S.; Biffo, S.; Pietrangelo, A. Human macrophage ferroportin biology and the basis for the ferroportin disease. Hepatology 2017, 65, 1512–1525. [CrossRef]
- Garrick, M.D.; Garrick, L.M. Cellular iron transport. Biochimica et Biophysica Acta - General Subjects 2009, 1790, 309–325. [CrossRef]
- Korolnek, T.; Hamza, I. Macrophages and iron trafficking at the birth and death of red cells. Blood 2015, 125, 2893–2897. [CrossRef]
- Garrick, M.D. Human iron transporters. Genes and Nutrition 2011, 6, 45–54. [CrossRef]
- Lane, D.J.R.; Merlot, A.M.; Huang, M.L.H.; Bae, D.H.; Jansson, P.J.; Sahni, S.; Kalinowski, D.S.; Richardson, D.R. Cellular iron uptake, trafficking and metabolism: Key molecules and mechanisms and their roles in disease. Biochimica et Biophysica Acta - Molecular Cell Research 2015, 1853, 1130–1144. [CrossRef]
- Kim, K.S.; Zhang, D.L.; Kovtunovych, G.; Ghosh, M.C.; Ollivierre, H.; Eckhaus, M.A.; Rouault, T.A. Infused wild-type macrophages reside and self-renew in the liver to rescue the hemolysis and anemia of Hmox1-deficient mice. Blood Adv 2018, 2, 2732–2743. [CrossRef]
- Kovtunovych, G.; Eckhaus, M.A.; Ghosh, M.C.; Ollivierre-Wilson, H.; Rouault, T.A. Dysfunction of the heme recycling system in heme oxygenase 1-deficient mice: effects on macrophage viability and tissue iron distribution. Blood 2010, 116, 6054–6062. [CrossRef]
- Kovtunovych, G.; Ghosh, M.C.; Ollivierre, W.; Weitzel, R.P.; Eckhaus, M.A.; Tisdale, J.F.; Yachie, A.; Rouault, T.A. Wild-type macrophages reverse disease in heme oxygenase 1-deficient mice. Blood 2014, 124, 1522–1530. [CrossRef]
- Papanikolaou, G.; Pantopoulos, K. Iron metabolism and toxicity. Toxicology and Applied Pharmacology 2005, 202, 199–211. [CrossRef]
- Cherayil, B.J. Iron and immunity: Immunological consequences of iron deficiency and overload. Archivum Immunologiae et Therapiae Experimentalis 2010, 58, 407–415. [CrossRef]
- Gattermann, N.; Muckenthaler, M.U.; Kulozik, A.E.; Metzgeroth, G.; Hastka, J. The Evaluation of Iron Deficiency and Iron Overload. Dtsch Arztebl Int 2021, 118, 847–856. [CrossRef]
- Pasricha, S.R.; Tye-Din, J.; Muckenthaler, M.U.; Swinkels, D.W. Iron deficiency. Lancet 2021, 397, 233–248. [CrossRef]
- Bresgen, N.; Eckl, P.M. Oxidative stress and the homeodynamics of iron metabolism. Biomolecules 2015, 5, 808–847. [CrossRef]
- Steinbicker, A.U.; Muckenthaler, M.U. Out of balance-systemic iron homeostasis in iron-related disorders. Nutrients 2013, 5, 3034–3061. [CrossRef]
- Mackenzie, E.L.; Iwasaki, K.; Tsuji, Y. Intracellular Iron Transport and Storage: From Molecular Mechanisms to Health Implications. Antioxidants & Redox Signaling 2008, 10, 997–1030. [CrossRef]
- Aisen, P.; Enns, C.; Wessling-Resnick, M. Chemistry and biology of eukaryotic iron metabolism. International Journal of Biochemistry and Cell Biology 2001, 33, 940–959. [CrossRef]
- Pietrangelo, A. Hereditary Hemochromatosis — A New Look at an Old Disease. New England Journal of Medicine 2004, 350, 2383–2397. [CrossRef]
- Haley, H.M.S.S.; Hill, A.G.; Greenwood, A.I.; Woerly, E.M.; Rienstra, C.M.; Burke, M.D. Peridinin Is an Exceptionally Potent and Membrane-Embedded Inhibitor of Bilayer Lipid Peroxidation. Journal of the American Chemical Society 2018, 140, 15227–15240. [CrossRef]
- Muckenthaler, M.U.; Galy, B.; Hentze, M.W. Systemic Iron Homeostasis and the Iron-Responsive Element/Iron-Regulatory Protein (IRE/IRP) Regulatory Network. Annual Review of Nutrition 2008, 28, 197–213. [CrossRef]
- Hentze, M.W.; Muckenthaler, M.U.; Galy, B.; Camaschella, C. Two to Tango: Regulation of Mammalian Iron Metabolism. Cell 2010, 142, 24–38. [CrossRef]
- Rouault, T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nature Chemical Biology 2006, 2, 406–414. [CrossRef]
- Nairz, M.; Haschka, D.; Demetz, E.; Weiss, G. Iron at the interface of immunity and infection. Frontiers in Pharmacology 2014, 5 JUL, 1-11. [CrossRef]
- Nairz, M.; Weiss, G. Iron in infection and immunity. Mol Aspects Med 2020, 75, 100864. [CrossRef]
- Marchetti, M.; De Bei, O.; Bettati, S.; Campanini, B.; Kovachka, S.; Gianquinto, E.; Spyrakis, F.; Ronda, L. Iron Metabolism at the Interface between Host and Pathogen: From Nutritional Immunity to Antibacterial Development. Int J Mol Sci 2020, 21. [CrossRef]
- Gerner, R.R.; Nuccio, S.P.; Raffatellu, M. Iron at the host-microbe interface. Mol Aspects Med 2020, 75, 100895. [CrossRef]
- Iatsenko, I.; Marra, A.; Boquete, J.P.; Pena, J.; Lemaitre, B. Iron sequestration by transferrin 1 mediates nutritional immunity in Drosophila melanogaster. Proc Natl Acad Sci USA 2020, 117, 7317–7325. [CrossRef]
- Caza, M.; Kronstad, J.W. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Frontiers in Cellular and Infection Microbiology 2013, 3, 1–23. [CrossRef]
- Lesuisse, E.; Blaiseau, P.L.; Dancis, A.; Camadro, J.M. Siderophore uptake and use by the yeast Saccharomyces cerevisiae. Microbiology (Reading) 2001, 147 (Pt 12), 289–298. [CrossRef]
- Wilson, B.R.; Bogdan, A.R.; Miyazawa, M.; Hashimoto, K.; Tsuji, Y. Siderophores in Iron Metabolism: From Mechanism to Therapy Potential. Trends Mol Med 2016, 22, 1077–1090. [CrossRef]
- Martinelli, M.; Strisciuglio, C.; Alessandrella, A.; Rossi, F.; Auricchio, R.; Campostrini, N.; Girelli, D.; Nobili, B.; Staiano, A.; Perrotta, S.; et al. Serum hepcidin and iron absorption in paediatric inflammatory bowel disease. Journal of Crohn's and Colitis 2016, 10, 566–574. [CrossRef]
- Nakashige, T.G.; Zhang, B.; Krebs, C.; Nolan, E.M. Human calprotectin is an iron-sequestering host-defense protein. Nature Chemical Biology 2015, 11, 765–771. [CrossRef]
- Arosio, P.; Elia, L.; Poli, M. Ferritin, cellular iron storage and regulation. IUBMB Life 2017, 69, 414–422. [CrossRef]
- Andrews, N. Disorders of Iron Metabolism. The New England journal of medicine 1999, 341, 1986–1995. [CrossRef]
- Jabara, H.H.; Boyden, S.E.; Chou, J.; Ramesh, N.; Massaad, M.J.; Benson, H.; Bainter, W.; Fraulino, D.; Rahimov, F.; Sieff, C.; et al. A missense mutation in TFRC, encoding transferrin receptor 1, causes combined immunodeficiency. Nat Genet 2016, 48, 74–78. [CrossRef]
- Wessling-Resnick, M. Nramp1 and other transporters involved in metal withholding during infection. Journal of Biological Chemistry 2015, 290, 18984–18990. [CrossRef]
- Brittenham, G.M.; Andersson, M.; Egli, I.; Foman, J.T.; Zeder, C.; Westerman, M.E.; Hurrell, R.F. Circulating non-transferrin-bound iron after oral administration of supplemental and fortification doses of iron to healthy women: a randomized study. Am J Clin Nutr 2014, 100, 813–820. [CrossRef]
- Weinberg, E.D. Nutritional immunity. Host's attempt to withold iron from microbial invaders. JAMA 1975, 231, 39–41. [CrossRef]
- Murdoch, C.C.; Skaar, E.P. Nutritional immunity: the battle for nutrient metals at the host-pathogen interface. Nat Rev Microbiol 2022, 20, 657–670. [CrossRef]
- Anderson, G.J. Ironing Out Disease: Inherited Disorders of Iron Homeostasis. IUBMB Life (International Union of Biochemistry and Molecular Biology: Life) 2001, 51, 11–17. [CrossRef]
- Brissot, P.; Bardou-Jacquet, E.; Jouanolle, A.M.; Loréal, O. Iron disorders of genetic origin: A changing world. Trends in Molecular Medicine 2011, 17, 707–713. [CrossRef]
- Grillo, A.S.; SantaMaria, A.M.; Kafina, M.D.; Cioffi, A.G.; Huston, N.C.; Han, M.; Seo, Y.A.; Yien, Y.Y.; Nardone, C.; Menon, A.V.; et al. Restored iron transport by a small molecule promotes absorption and hemoglobinization in animals. Science 2017, 356, 608–616. [CrossRef]
- Gangat, N.; Wolanskyj, A.P. Anemia of chronic disease. Seminars in Hematology 2013, 50, 232–238. [CrossRef]
- Rivera, S.; Ganz, T. Animal Models of Anemia of Inflammation. Seminars in Hematology 2009, 46, 351–357. [CrossRef]
- Anderson, Sheila A.; Nizzi, Christopher P.; Chang, Y.-I.; Deck, Kathryn M.; Schmidt, Paul J.; Galy, B.; Damnernsawad, A.; Broman, Aimee T.; Kendziorski, C.; Hentze, Matthias W.; et al. The IRP1-HIF-2α Axis Coordinates Iron and Oxygen Sensing with Erythropoiesis and Iron Absorption. Cell Metabolism 2013, 17. [CrossRef]
- Crack, J.C.; Green, J.; Thomson, A.J.; Le Brun, N.E. Iron-sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide. Acc Chem Res 2014, 47, 3196–3205. [CrossRef]
- Ghosh, M.C.; Zhang, D.; Ollivierre, H.; Noguchi, A.; Springer, D.; Linehan, W.M.; Rouault, T.A. Therapeutic inhibition of HIF2α reverses polycythemia and pulmonary hypertension in murine models of two human diseases. Blood 2020, 10.1182/blood.2020009138. [CrossRef]
- Wilkinson, N.; Pantopoulos, K. IRP1 regulates erythropoiesis and systemic iron homeostasis by controlling HIF2α mRNA translation. Blood 2013, 122. [CrossRef]
- Nandal, A.; Ruiz, J.C.; Subramanian, P.; Ghimire-Rijal, S.; Sinnamon, R.A.; Stemmler, T.L.; Bruick, R.K.; Philpott, C.C. Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metab 2011, 14, 647–657. [CrossRef]
- Wang, D.; Wang, L.H.; Zhao, Y.; Lu, Y.P.; Zhu, L. Hypoxia regulates the ferrous iron uptake and reactive oxygen species level via divalent metal transporter 1 (DMT1) exon1B by hypoxia-inducible factor-1. IUBMB Life 2010, 62, 629–636. [CrossRef]
- Mastrogiannaki, M.; Matak, P.; Keith, B.; Simon, M.C.; Vaulont, S.; Peyssonnaux, C. HIF-2α, but not HIF-1α, promotes iron absorption in mice. Journal of Clinical Investigation 2009, 119. [CrossRef]
- Sanchez, M.; Galy, B.; Muckenthaler, M.U.; Hentze, M.W. Iron-regulatory proteins limit hypoxia-inducible factor-2α expression in iron deficiency. Nature Structural and Molecular Biology 2007, 14, 420–426. [CrossRef]
- Ghosh, M.C.; Zhang, D.-L.; Ollivierre, H.; Eckhaus, M.A.; Rouault, T.A. Translational repression of HIF2α expression in mice with Chuvash polycythemia reverses polycythemia. Journal of Clinical Investigation 2018, 128. [CrossRef]
- Holmes-Hampton, G.P.; Ghosh, M.C.; Rouault, T.A. Methods for Studying Iron Regulatory Protein 1: An Important Protein in Human Iron Metabolism. Methods Enzymol 2018, 599, 139–155. [CrossRef]
- Johnson, N.B.; Deck, K.M.; Nizzi, C.P.; Eisenstein, R.S. A synergistic role of IRP1 and FBXL5 proteins in coordinating iron metabolism during cell proliferation. Journal of Biological Chemistry 2017, 292. [CrossRef]
- Sanchez, M.; Galy, B.; Schwanhaeusser, B.; Blake, J.; Bähr-Ivacevic, T.; Benes, V.; Selbach, M.; Muckenthaler, M.U.; Hentze, M.W. Iron regulatory protein-1 and -2: transcriptome-wide definition of binding mRNAs and shaping of the cellular proteome by iron regulatory proteins. Blood 2011, 118. [CrossRef]
- Pantopoulos, K. Iron Metabolism and the IRE/IRP Regulatory System: An Update. Annals of the New York Academy of Sciences 2004, 1012, 1–13. [CrossRef]
- Pellicer Martinez, M.T.; Martinez, A.B.; Crack, J.C.; Holmes, J.D.; Svistunenko, D.A.; Johnston, A.W.B.; Cheesman, M.R.; Todd, J.D.; Le Brun, N.E. Sensing iron availability via the fragile [4Fe-4S] cluster of the bacterial transcriptional repressor RirA. Chem Sci 2017, 8, 8451–8463. [CrossRef]
- Hibbing, M.E.; Fuqua, C. Antiparallel and interlinked control of cellular iron levels by the Irr and RirA regulators of Agrobacterium tumefaciens. J Bacteriol 2011, 193, 3461–3472. [CrossRef]
- Rudolph, G.; Hennecke, H.; Fischer, H.M. Beyond the Fur paradigm: iron-controlled gene expression in rhizobia. FEMS Microbiol Rev 2006, 30, 631–648. [CrossRef]
- Vergnes, A.; Viala, J.P.; Ouadah-Tsabet, R.; Pocachard, B.; Loiseau, L.; Meresse, S.; Barras, F.; Aussel, L. The iron-sulfur cluster sensor IscR is a negative regulator of Spi1 type III secretion system in Salmonella enterica. Cell Microbiol 2017, 19. [CrossRef]
- Saninjuk, K.; Romsang, A.; Duang-Nkern, J.; Vattanaviboon, P.; Mongkolsuk, S. Transcriptional regulation of the Pseudomonas aeruginosa iron-sulfur cluster assembly pathway by binding of IscR to multiple sites. PLoS One 2019, 14, e0218385. [CrossRef]
- Outten, C.E.; Albetel, A.N. Iron sensing and regulation in Saccharomyces cerevisiae: Ironing out the mechanistic details. Curr Opin Microbiol 2013, 16, 662–668. [CrossRef]
- Yamaguchi-Iwai, Y.; Dancis, A.; Klausner, R.D. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J 1995, 14, 1231–1239. [CrossRef]
- Gupta, M.; Outten, C.E. Iron-sulfur cluster signaling: The common thread in fungal iron regulation. Curr Opin Chem Biol 2020, 55, 189–201. [CrossRef]
- Poor, C.B.; Wegner, S.V.; Li, H.; Dlouhy, A.C.; Schuermann, J.P.; Sanishvili, R.; Hinshaw, J.R.; Riggs-Gelasco, P.J.; Outten, C.E.; He, C. Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc Natl Acad Sci U S A 2014, 111, 4043–4048. [CrossRef]
- Shen, M.; Goforth, J.B.; Eisenstein, R.S. Iron-dependent post transcriptional control of mitochondrial aconitase expression. Metallomics 2023, 15. [CrossRef]
- Zimmer, M.; Ebert, B.L.; Neil, C.; Brenner, K.; Papaioannou, I.; Melas, A.; Tolliday, N.; Lamb, J.; Pantopoulos, K.; Golub, T.; et al. Small-molecule inhibitors of HIF-2a translation link its 5'UTR iron-responsive element to oxygen sensing. Mol Cell 2008, 32, 838–848. [CrossRef]
- Zhang, D.L.; Hughes, R.M.; Ollivierre-Wilson, H.; Ghosh, M.C.; Rouault, T.A. A Ferroportin Transcript that Lacks an Iron-Responsive Element Enables Duodenal and Erythroid Precursor Cells to Evade Translational Repression. Cell Metabolism 2009, 9, 461–473. [CrossRef]
- Garza, K.R.; Clarke, S.L.; Ho, Y.H.; Bruss, M.D.; Vasanthakumar, A.; Anderson, S.A.; Eisenstein, R.S. Differential translational control of 5' IRE-containing mRNA in response to dietary iron deficiency and acute iron overload. Metallomics 2020, 12, 2186–2198. [CrossRef]
- Cmejla, R.; Petrak, J.; Cmejlova, J. A novel iron responsive element in the 3′UTR of human MRCKα. Biochemical and Biophysical Research Communications 2006, 341. [CrossRef]
- Sanchez, M.; Galy, B.; Dandekar, T.; Bengert, P.; Vainshtein, Y.; Stolte, J.; Muckenthaler, M.U.; Hentze, M.W. Iron Regulation and the Cell Cycle. Journal of Biological Chemistry 2006, 281. [CrossRef]
- Gunshin, H.; Mackenzie, B.; Berger, U.V.; Gunshin, Y.; Romero, M.F.; Boron, W.F.; Nussberger, S.; Gollan, J.L.; Hediger, M.A. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 1997, 388. [CrossRef]
- Tybl, E.; Gunshin, H.; Gupta, S.; Barrientos, T.; Bonadonna, M.; Celma Nos, F.; Palais, G.; Karim, Z.; Sanchez, M.; Andrews, N.C.; et al. Control of Systemic Iron Homeostasis by the 3' Iron-Responsive Element of Divalent Metal Transporter 1 in Mice. Hemasphere 2020, 4, e459. [CrossRef]
- Qatato, M.; Bonadonna, M.; Palais, G.; Ertl, A.; Schmidt, G.; Polycarpou-Schwarz, M.; Karim, Z.; Galy, B. IRE-dependent Regulation of Intestinal Dmt1 Prevails During Chronic Dietary Iron Deficiency but is Dispensable in Conditions of Acute Erythropoietic Stress. Hemasphere 2022, 6, e693. [CrossRef]
- Garrick, M.D.; Dolan, K.G.; Horbinski, C.; Ghio, A.J.; Higgins, D.; Porubcin, M.; Moore, E.G.; Hainsworth, L.N.; Umbreit, J.N.; Conrad, M.E.; et al. DMT1: A mammalian transporter for multiple metals. BioMetals 2003, 16, 41–54. [CrossRef]
- Ducamp, S.; Luscieti, S.; Ferrer-Cortès, X.; Nicolas, G.; Manceau, H.; Peoc’h, K.; Yien, Y.Y.; Kannengiesser, C.; Gouya, L.; Puy, H.; et al. A mutation in the iron-responsive element of ALAS2 is a modifier of disease severity in a patient suffering from CLPX associated erythropoietic protoporphyria. Haematologica 2021, 10.3324/haematol.2020.272450. [CrossRef]
- Wilkinson, N.; Pantopoulos, K. The IRP/IRE system in vivo: insights from mouse models. Front Pharmacol 2014, 5, 176. [CrossRef]
- Kaptain, S.; Downey, W.E.; Tang, C.; Philpott, C.; Haile, D.; Orloff, D.G.; Harford, J.B.; Rouault, T.A.; Klausner, R.D. A regulated RNA binding protein also possesses aconitase activity. Proc Natl Acad Sci U S A 1991, 88, 10109–10113. [CrossRef]
- Rouault, T.A.; Maio, N. How Oxidation of a Unique Iron-Sulfur Cluster in FBXL5 Regulates IRP2 Levels and Promotes Regulation of Iron Metabolism Proteins. Mol Cell 2020, 78, 1–3. [CrossRef]
- Zumbrennen-Bullough, K.B.; Becker, L.; Garrett, L.; Hölter, S.M.; Calzada-Wack, J.; Mossbrugger, I.; Quintanilla-Fend, L.; Racz, I.; Rathkolb, B.; Klopstock, T.; et al. Abnormal Brain Iron Metabolism in Irp2 Deficient Mice Is Associated with Mild Neurological and Behavioral Impairments. PLoS ONE 2014, 9. [CrossRef]
- Henderson, B.R. Iron regulatory proteins 1 and 2. Bioessays 1996, 18, 739–746. [CrossRef]
- Meyron-Holtz, E.G.; Ghosh, M.C.; Iwai, K.; LaVaute, T.; Brazzolotto, X.; Berger, U.V.; Land, W.; Ollivierre-Wilson, H.; Grinberg, A.; Love, P.; et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. The EMBO Journal 2004, 23. [CrossRef]
- Cluster, O.-r.; Wang, H.; Shi, H.; Rajan, M.; Stoll, S.; Leibold, E.A. Article FBXL5 Regulates IRP2 Stability in Iron Homeostasis. Molecular Cell 2020, 10.1016/j.molcel.2020.02.011, 1-11. [CrossRef]
- Holland, L.Z.; Ocampo Daza, D. A new look at an old question: when did the second whole genome duplication occur in vertebrate evolution? Genome Biol 2018, 19, 209. [CrossRef]
- Hsiao, T.L.; Vitkup, D. Role of duplicate genes in robustness against deleterious human mutations. PLoS Genetics 2008, 4. [CrossRef]
- Wang, H.; Shi, H.; Rajan, M.; Canarie, E.R.; Hong, S.; Simoneschi, D.; Pagano, M.; Bush, M.F.; Stoll, S.; Leibold, E.A.; et al. FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster. Mol Cell 2020, 78, 31–41 e35. [CrossRef]
- Jiao, Q.; Du, X.; Wei, J.; Li, Y.; Jiang, H. Oxidative Stress Regulated Iron Regulatory Protein IRP2 Through FBXL5-Mediated Ubiquitination-Proteasome Way in SH-SY5Y Cells. Front Neurosci 2019, 13, 20. [CrossRef]
- Chollangi, S.; Thompson, J.W.; Ruiz, J.C.; Gardner, K.H.; Bruick, R.K. Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5) Communicates Cellular Iron and Oxygen Availability by Distinct Mechanisms. Journal of Biological Chemistry 2012, 287. [CrossRef]
- Moroishi, T.; Nishiyama, M.; Takeda, Y.; Iwai, K.; Nakayama, Keiichi I. The FBXL5-IRP2 Axis Is Integral to Control of Iron Metabolism In Vivo. Cell Metabolism 2011, 14. [CrossRef]
- Leon-Sicairos, C.R.; Figueroa-Angulo, E.E.; Calla-Choque, J.S.; Arroyo, R. The Non-Canonical Iron-Responsive Element of IRE-tvcp12 Hairpin Structure at the 3'-UTR of Trichomonas vaginalis TvCP12 mRNA That Binds TvHSP70 and TvACTN-3 Can Regulate mRNA Stability and Amount of Protein. Pathogens 2023, 12. [CrossRef]
- Del Rey, M.Q.; Mancias, J.D. NCOA4-mediated ferritinophagy: A potential link to neurodegeneration. In Frontiers in Neuroscience, Frontiers Media S.A.: 2019; Vol. 13.
- Mancias, J.D.; Wang, X.; Gygi, S.P.; Harper, J.W.; Kimmelman, A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 2014, 508, 105–109. [CrossRef]
- Santana-Codina, N.; Mancias, J.D. The role of NCOA4-mediated ferritinophagy in health and disease; 2018; Vol. 11.
- Santana-Codina, N.; Del Rey, M.Q.; Kapner, K.S.; Zhang, H.; Gikandi, A.; Malcolm, C.; Poupault, C.; Kuljanin, M.; John, K.M.; Biancur, D.E.; et al. NCOA4-Mediated Ferritinophagy Is a Pancreatic Cancer Dependency via Maintenance of Iron Bioavailability for Iron-Sulfur Cluster Proteins. Cancer Discov 2022, 12, 2180–2197. [CrossRef]
- Kuno, S.; Iwai, K. Oxygen modulates iron homeostasis by switching iron sensing of NCOA4. J Biol Chem 2023, 299, 104701. [CrossRef]
- Zhao, H.; Lu, Y.; Zhang, J.; Sun, Z.; Cheng, C.; Liu, Y.; Wu, L.; Zhang, M.; He, W.; Hao, S.; et al. NCOA4 requires a [3Fe-4S] to sense and maintain the iron homeostasis. J Biol Chem 2023, 300, 105612. [CrossRef]
- Andrews, S.C.; Robinson, A.K.; Rodriguez-Quinones, F. Bacterial iron homeostasis. FEMS Microbiol Rev 2003, 27, 215–237. [CrossRef]
- Bradley, J.M.; Svistunenko, D.A.; Wilson, M.T.; Hemmings, A.M.; Moore, G.R.; Le Brun, N.E. Bacterial iron detoxification at the molecular level. J Biol Chem 2020, 295, 17602–17623. [CrossRef]
- Small, S.K.; Puri, S.; Sangwan, I.; O'Brian, M.R. Positive control of ferric siderophore receptor gene expression by the Irr protein in Bradyrhizobium japonicum. J Bacteriol 2009, 191, 1361–1368. [CrossRef]
- Yeoman, K.H.; Curson, A.R.; Todd, J.D.; Sawers, G.; Johnston, A.W. Evidence that the Rhizobium regulatory protein RirA binds to cis-acting iron-responsive operators (IROs) at promoters of some Fe-regulated genes. Microbiology (Reading) 2004, 150 (Pt 12), 4065–4074. [CrossRef]
- Pankievicz, V.C.S.; Irving, T.B.; Maia, L.G.S.; Ane, J.M. Are we there yet? The long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops. BMC Biol 2019, 17, 99. [CrossRef]
- Sevilla, E.; Bes, M.T.; Gonzalez, A.; Peleato, M.L.; Fillat, M.F. Redox-Based Transcriptional Regulation in Prokaryotes: Revisiting Model Mechanisms. Antioxid Redox Signal 2019, 30, 1651–1696. [CrossRef]
- Shimberg, G.D.; Pritts, J.D.; Michel, S.L.J. Iron-Sulfur Clusters in Zinc Finger Proteins. Methods Enzymol 2018, 599, 101–137. [CrossRef]
- Kaplan, C.D.; Kaplan, J. Iron acquisition and transcriptional regulation. Chem Rev 2009, 109, 4536–4552. [CrossRef]
- Pijuan, J.; Moreno, D.F.; Yahya, G.; Moisa, M.; Ul Haq, I.; Krukiewicz, K.; Mosbah, R.; Metwally, K.; Cavalu, S. Regulatory and pathogenic mechanisms in response to iron deficiency and excess in fungi. Microb Biotechnol 2023, 16, 2053–2071. [CrossRef]
- Ramos-Alonso, L.; Romero, A.M.; Martinez-Pastor, M.T.; Puig, S. Iron Regulatory Mechanisms in Saccharomyces cerevisiae. Front Microbiol 2020, 11, 582830. [CrossRef]
- Askwith, C.; Eide, D.; Van Ho, A.; Bernard, P.S.; Li, L.; Davis-Kaplan, S.; Sipe, D.M.; Kaplan, J. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 1994, 76, 403–410. [CrossRef]
- Stearman, R.; Yuan, D.S.; Yamaguchi-Iwai, Y.; Klausner, R.D.; Dancis, A. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 1996, 271, 1552–1557. [CrossRef]
- Heymann, P.; Ernst, J.F.; Winkelmann, G. Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol Lett 2000, 186, 221–227. [CrossRef]
- Yun, C.W.; Tiedeman, J.S.; Moore, R.E.; Philpott, C.C. Siderophore-iron uptake in saccharomyces cerevisiae. Identification of ferrichrome and fusarinine transporters. J Biol Chem 2000, 275, 16354–16359. [CrossRef]
- Martinez-Pastor, M.T.; Perea-Garcia, A.; Puig, S. Mechanisms of iron sensing and regulation in the yeast Saccharomyces cerevisiae. World J Microbiol Biotechnol 2017, 33, 75. [CrossRef]
- Rodrigues-Pousada, C.; Devaux, F.; Caetano, S.M.; Pimentel, C.; da Silva, S.; Cordeiro, A.C.; Amaral, C. Yeast AP-1 like transcription factors (Yap) and stress response: a current overview. Microb Cell 2019, 6, 267–285. [CrossRef]
- Rietzschel, N.; Pierik, A.J.; Bill, E.; Lill, R.; Muhlenhoff, U. The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe-2S] clusters. Mol Cell Biol 2015, 35, 370–378. [CrossRef]
- Sorribes-Dauden, R.; Peris, D.; Martinez-Pastor, M.T.; Puig, S. Structure and function of the vacuolar Ccc1/VIT1 family of iron transporters and its regulation in fungi. Comput Struct Biotechnol J 2020, 18, 3712–3722. [CrossRef]
- Velez-Bermudez, I.C.; Schmidt, W. Iron sensing in plants. Front Plant Sci 2023, 14, 1145510. [CrossRef]
- Senoura, T.; Kobayashi, T.; An, G.; Nakanishi, H.; Nishizawa, N.K. Defects in the rice aconitase-encoding OsACO1 gene alter iron homeostasis. Plant Mol Biol 2020, 104, 629–645. [CrossRef]
- Inoue, H.; Higuchi, K.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J 2003, 36, 366–381. [CrossRef]
- Inoue, H.; Kobayashi, T.; Nozoye, T.; Takahashi, M.; Kakei, Y.; Suzuki, K.; Nakazono, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J Biol Chem 2009, 284, 3470–3479. [CrossRef]
- Ogo, Y.; Itai, R.N.; Nakanishi, H.; Kobayashi, T.; Takahashi, M.; Mori, S.; Nishizawa, N.K. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J 2007, 51, 366–377. [CrossRef]
- Battistuzzi, F.U.; Feijao, A.; Hedges, S.B. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol 2004, 4, 44. [CrossRef]
- Brochier-Armanet, C.; Forterre, P.; Gribaldo, S. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr Opin Microbiol 2011, 14, 274–281. [CrossRef]
- Hoshino, Y.; Villanueva, L. Four billion years of microbial terpenome evolution. FEMS Microbiol Rev 2023, 47. [CrossRef]
- Johnson, C.; England, A.; Munro-Ehrlich, M.; Colman, D.R.; DuBois, J.L.; Boyd, E.S. Pathways of Iron and Sulfur Acquisition, Cofactor Assembly, Destination, and Storage in Diverse Archaeal Methanogens and Alkanotrophs. J Bacteriol 2021, 203, e0011721. [CrossRef]
- Roche, B.; Aussel, L.; Ezraty, B.; Mandin, P.; Py, B.; Barras, F. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta 2013, 1827, 455–469. [CrossRef]
- Baker, B.J.; De Anda, V.; Seitz, K.W.; Dombrowski, N.; Santoro, A.E.; Lloyd, K.G. Diversity, ecology and evolution of Archaea. Nat Microbiol 2020, 5, 887–900. [CrossRef]
- Iwasaki, T. Iron-sulfur world in aerobic and hyperthermoacidophilic archaea Sulfolobus. Archaea 2010, 2010. [CrossRef]
- Saini, J.; Deere, T.M.; Lessner, D.J. The minimal SUF system is not required for Fe-S cluster biogenesis in the methanogenic archaeon Methanosarcina acetivorans. Sci Rep 2023, 13, 15120. [CrossRef]
- Uhrigshardt, H.; Zoske, S.; Anemuller, S. Sulfolobus aconitase, a regulator of iron metabolism? Biochem Soc Trans 2002, 30, 685–687. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




