1. Introduction
SARS-CoV-2 causes COVID-19, a disease characterized by fever, cough, shortness of breath, and pneumonia. The disease can be fatal in vulnerable individuals, including immunocompromised individuals and the elderly. SARS-CoV-2 can be transmitted via aerosol and through contact with contaminated surfaces (1). Search and validation of new potential antiviral chemical and biological agents is essential for the improvement of treatments and control of severe acute respiratory syndrome (SARS). This includes new variants which arise due to mutations which render other therapeutics and vaccines less effective than before.
Currently, there are many directions being tested towards the development and application of therapeutics against SARS-CoV-2. These include antiviral compounds targeting virion structures such as spike protein (2), inhibition of RNA-dependent RNA polymerase by remdesivir (3) and favipiravir (4), inhibition of proteases like SARS-coronavirus papain-like protease (PLpro) and 3C-like protease (3Clpro) (5) (6), and targeting of type I and type III interferons (7). The use of virus-neutralizing antibodies is another effective approach against SARS-CoV-2. Polyclonal antibodies in the form of convalescent serum as well as monoclonal antibodies have been shown to be effective towards controlling infections, including SARS-CoV-2 (8) (9).
Another interesting direction is utilization of clustered regularly interspaced short palindromic repeats (CRISPR) technology as a therapeutic to target viral genome and destruction of specific viral RNA with Cas13. This approach called PACMAN (Prophylactic Antiviral CRISPR in huMAN cells) has been developed, albeit with many limitations such as delivery (10).
Most of these approaches are still in various stages of development and have either not been approved or have emergency use authorization only. Others have more prominent side effects, low availability, and high cost in addition to other limitations. In addition to these issues, there has been significant issues towards the creation of novel classes of antimicrobial drugs in the past 30 years. While some advancements have been seen, challenges specifically with antivirals are apparent. Of the more than 220 viruses that infect humans, only 10 have antiviral drugs which are clinically approved (11). Therefore, further drug discovery efforts are needed to address this problem. Here, we propose the antimicrobial peptide mimic, peptoid, as another approach to curtail the infection. Peptoids are sequence-specific poly-N-substituted glycines, mimicking the function of natural antimicrobial peptides (AMPs) with the added benefit of being resistant to proteases (12). It is well known that AMPs such as defensins and cathelicidins are a part of the innate defense against infections (13), including the sole human cathelicidin, LL-37 (14) (15). To the best of our knowledge, the mechanism of antimicrobial action of AMPs is based on disruption of the cell membrane, either by pore formation or alteration of lipid layer structure and function (16) (17), or through intracellular effects such as inhibiting transcription, translation, or protein synthesis (18) (19). Peptoids, as non-natural forms of AMPs or mimetics of AMPs, have demonstrated similar antimicrobial properties. One direct advantage of antimicrobial peptoids over antimicrobial peptides is the ability to resist protease degradation due to the movement of the R group from the α-carbon to the amide (20). Additionally, peptoids have been shown to act quickly in killing bacteria in vitro, especially compared to traditionally used antibiotics (21).
Several different peptoids have been shown to be effective against various gram-positive and gram-negative bacteria (22) (23) and viruses, including herpes simplex virus type 1 and SARS-CoV-2 (24) (25). Among the peptoids that have been tested against these two viruses, TM9 demonstrated the best antiviral effect (24). The predicted mechanism of action for TM9 peptoid against enveloped viruses is disruption of the envelope membrane structure, although this has not yet been fully elucidated (25). Murine hepatitis virus and SARS-CoV-2 are both members of the genus Betacoronavirus (26) and have similar genome structure and biology. Humans are not susceptible to MHV infection making it safer and easier to do testing with MHV since it does not require a BSL-3 laboratory setting (27) (28) (29). Virulence and organotropism of MHV depends on the virus strain and varies as multiple strains exist. Depending upon the tropism of the particular strain used, this virus serves as a model for human respiratory, gastrointestinal, liver, and neurologic infections (27).
In this study we use three coronavirus strains, MHV-1, MHV-A59, and recombinant MHV-A59-GFP to examine respiratory system tropism and the impacts of virulence on therapeutic efficacy. The MHV coronaviruses have been shown to generate severe acute respiratory syndrome (SARS) in mice that demonstrates pathological similarities to human SARS (30) (31). We observed antiviral activity of a novel peptoid TM9 for all three MHV strains, suggesting that TM9 will be effective against all coronaviruses, including SARS-CoV-2. We utilize an aerosol route of infection for MHV in mice via a Madison chamber that produces a dry aerosol, making this model closely mimic the natural route of SARS-CoV-2 transmission (32) (33) and confirming that MHV represents an inexpensive, rapid, and straightforward model for COVID-19 therapeutic screening.
2. Materials and Methods
Cell culture: Murine lung carcinoma-derived cell line L2 lung carcinoma derived fibroblastic cells (34), human A549 (human lung carcinoma (ATCC CCL-185), and HULEC-5a (human microvascular endothelium (ATCC CRL-3244)) cell lines were used in this study. Cells were cultured in cell culture dishes with Dulbecco’s Modification of Eagle’s Medium (DMEM) (Corning) containing 4.5 g/L glucose, L-glutamine, and sodium pyruvate and supplemented with 10% of FBS, 100 U/ml penicillin, 10 μg/ml streptomycin (Lonza), GlutaMax (Gibco), 0.01 M HEPES buffer solution (Gibco) and MEM nonessential amino acids (Corning).
Synthesis of TM9 peptoid: Peptoid was synthesized as described previously (24) (21) (35). Briefly, peptoid synthesis was carried out using a Symphony X (Gyros Protein Technologies, Tucson, AZ) peptide synthesizer located at the Molecular Foundry in the Lawrence Berkeley National Laboratory, Berkeley, CA. Peptoids were synthesized on a Rink amide MBHA resin (EMD Biosciences, Gibbstown, NJ). All reagents were purchased from Sigma Aldrich (St. Louis, MO). Synthesis followed the submonomer protocol from Zuckermann, et al (35). Peptoids were cleaved from the resin by treating with trifluoroacetic acid (TFA):triisopropylsilane:water (95:2.5:2.5 volume ratio) for 30 minutes. A C18 column in a reversed-phase high performance liquid chromatography (HPLC) system (Waters Corporation, Milford, MA) was used for purification with a linear acetonitrile and water gradient with a compound purity greater than 98% as measured by analytical reverse-phased HPLC. Confirmation of the peptoid synthesis was determined using electrospray ionization mass spectrometry.
Test of efficacy and toxicity of TM9 peptoid: 15,000 cells/well of L2, A549 and HULEC-5a cell lines were plated in 96 well plates with 5 replicates for each concentration of peptoid and incubated at 37°C, 5% CO2 for 24 hours. After incubation 10 two-fold serial dilutions of TM9 were prepared from 0.98 μg/ml to 500 µg/ml. Each dilution of TM9 was added to respective wells with cell cultures and to separate plates with viruses. The cells and viruses were incubated for 2 hours. Then, preincubated viruses with MOI: 0.1 were added to the cells containing respective concentrations of peptoids and incubated for 3 hours and 40 hours. After incubation the supernatant of each replicate was collected for plaque assay.
Cell viability was measured using 10% alamarBlue containing growth medium. It was added to each well and incubated for 2 hours. After the incubation, the alamarBlue containing medium was transferred to a new clear 96-well plate and the absorbance was measured at 490 nm.
Virus: The following three strains of Murine hepatitis virus: MHV-1 (ATCC VR-261) and MHV-A59 (ATCC VR-764) and MHV-A59-GFP were used. MHV-A-59-GFP is a recombinant version of MHV-A59 generated by Susan Weiss’s lab where the ns4 gene has been replaced by the EGFP sequence (36). Stock viruses were stored at -80oC.
Plaque assay: L2 cells were plated in 12-well plates at 0.4 x 106 cells per well. Cells were seeded in complete DMEM with 10% FBS, 1 ml per well, and incubated at 37 oC for two days. Serial ten-fold dilutions of virus in DMEM in 5 ml snap cap tubes were prepared. DMEM was aspirated from the L2 cell monolayers and 200 ul of virus dilutions were added to each well. Plates were incubated with the virus inoculum at room temperature for 60 minutes with periodic shaking. After incubation, 1.6% agarose melted and cooled to 50oC was mixed with the same volume of 2x EMEM (Quality Biological) to make an overlay solution, 1 ml of which was added to each well. After adding overlays, cells were incubated for 2 days at 37oC, 5% CO2 and humidity. After the incubation, overlays were carefully removed and cells fixed with 10% neutral buffered formalin (Sigma) for 15 min, stained with 0.1% crystal violet in 20% ethanol for 5 min and washed.
Generation of the animal model for respiratory infection: 5-7-week-old female BALB/c (Envigo) mice were used as a model of the infection according to the protocol approved by Texas A&M Animal Care and Use Committee. Animals were randomly distributed into 4 groups: Group 1 - 5 animals - uninfected control group; group 2 - 10 mice infected with MHV-1; group 3 - 10 mice infected with MHV-A59; group 4 – 10 mice infected with MHV-A59-GFP. The Madison aerosol chamber was used to infect the animals. Viral supernatants in DMEM were used in the following titers: MHV-1 - 105 PFU/ml; MHV-A59 - 107 PFU/ml; MHV-A59-GFP - 107 PFU/ml. Each group of mice was exposed to the corresponding virus for a total of 15 min by running 3 consecutive 5-minute infection cycles. Clinical observations were made before infection and every 12 hours for 5 days after the infection.
Every 24 hours two animals from each experimental group and one animal from the control group were sacrificed. Necropsy and gross pathological examination were performed. Lungs, brain, olfactory bulbs, heart, liver, spleen, and kidneys were homogenized with 1 ml of DMEM. Homogenates were used for plaque assay to quantify the virus.
Statistical analysis and graphics: Statistics and data management were performed using Microsoft Excel and Graphpad Prism software. BioRender was used to prepare
Figure 5.
3. Results
3.1. The Effect of TM9 Peptoid Was Similar on Different Cell Lines But Dependent on the Duration of Exposure
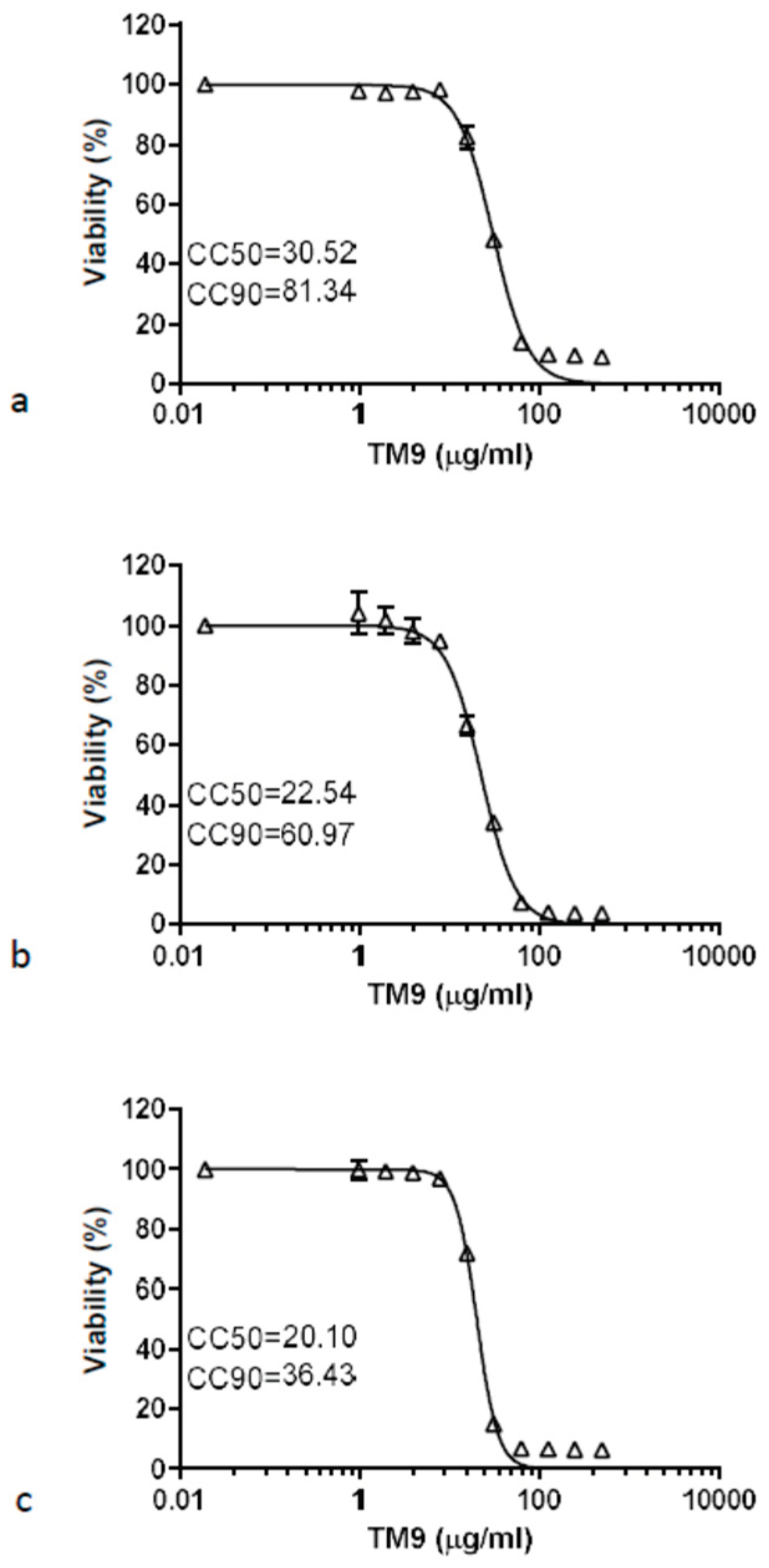
Both a murine lung carcinoma-derived cell line (L2) and two types of human cells, lung carcinoma (A549) and microvascular endothelial cells (HULEC-5a), demonstrated similar sensitivity to the peptoid TM9 3 hours post-incubation (
Figure 1). The cytotoxic concentration with 50% viability (CC50) was measured to be between 20 and 30 μg/ml for all cell lines. The cytotoxic concentration where there was 90% cell viability (CC90) was measured at 81.34 μg/ml for L2 cells, 60.97 for A549 cells, and 36.43 μg/ml for HULEC-5a cells.
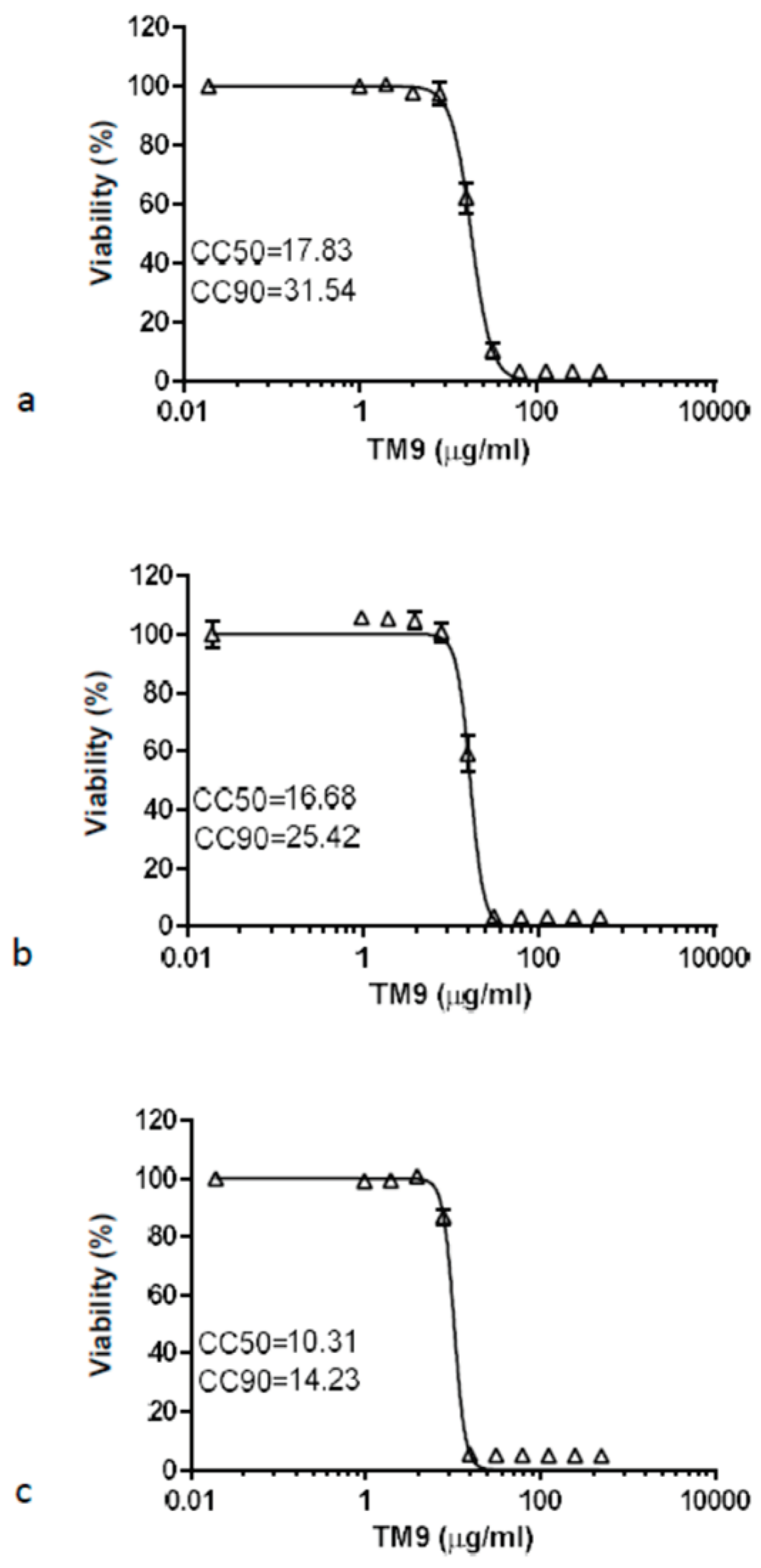
Comparing the 40-hour time point to the 3-hour time point, the CC50 reduced by 43% in L2 cells, down to 17.83 μg/ml and the CC90 reduced to 31.54 μg/ml. For A549 cells the CC50 decreased to 16.68 μg/ml while the CC90 reduced to 25.42 μg/ml. Lastly, HULEC-5a cells showed a reduction of the CC50 to 10.31 μg/ml and CC90 to 14.23 μg/ml. The ratio of CC90 to CC50 was almost three for both the murine lung carcinoma cells and human lung carcinoma cells after three hours. Human intravascular endothelial cells were more sensitive to the peptoid as these cells had the lowest ratio (1.81) of CC90 to CC50. For all three cell lines, exposure to TM9 for 40 hours resulted in greater cytotoxic effects at lower concentrations of peptoid and caused the ratio of CC90 to CC50 to decrease.
Figure 2.
Prolonged toxicity test of TM9 peptoid in vitro. Cells were incubated with twofold serial dilutions of the peptoid in cell culture medium for 40 hours. Cell viability measured using 10% alamarBlue reagent. Cell viability was calculated as a percentage of metabolic activity of corresponding untreated controls. Error bars show standard deviation of 5 experimental replicates. A – L2 cells; b – A549 cells; c – HULEC-5a cells. CC50 – cytotoxic concentration reducing viability by 50%; CC90 – cytotoxic concentration reducing viability by 90%.
Figure 2.
Prolonged toxicity test of TM9 peptoid in vitro. Cells were incubated with twofold serial dilutions of the peptoid in cell culture medium for 40 hours. Cell viability measured using 10% alamarBlue reagent. Cell viability was calculated as a percentage of metabolic activity of corresponding untreated controls. Error bars show standard deviation of 5 experimental replicates. A – L2 cells; b – A549 cells; c – HULEC-5a cells. CC50 – cytotoxic concentration reducing viability by 50%; CC90 – cytotoxic concentration reducing viability by 90%.
3.2. The Effective Antiviral Concentration of TM9 Was within a Therapeutic Window in L2 Cell Culture
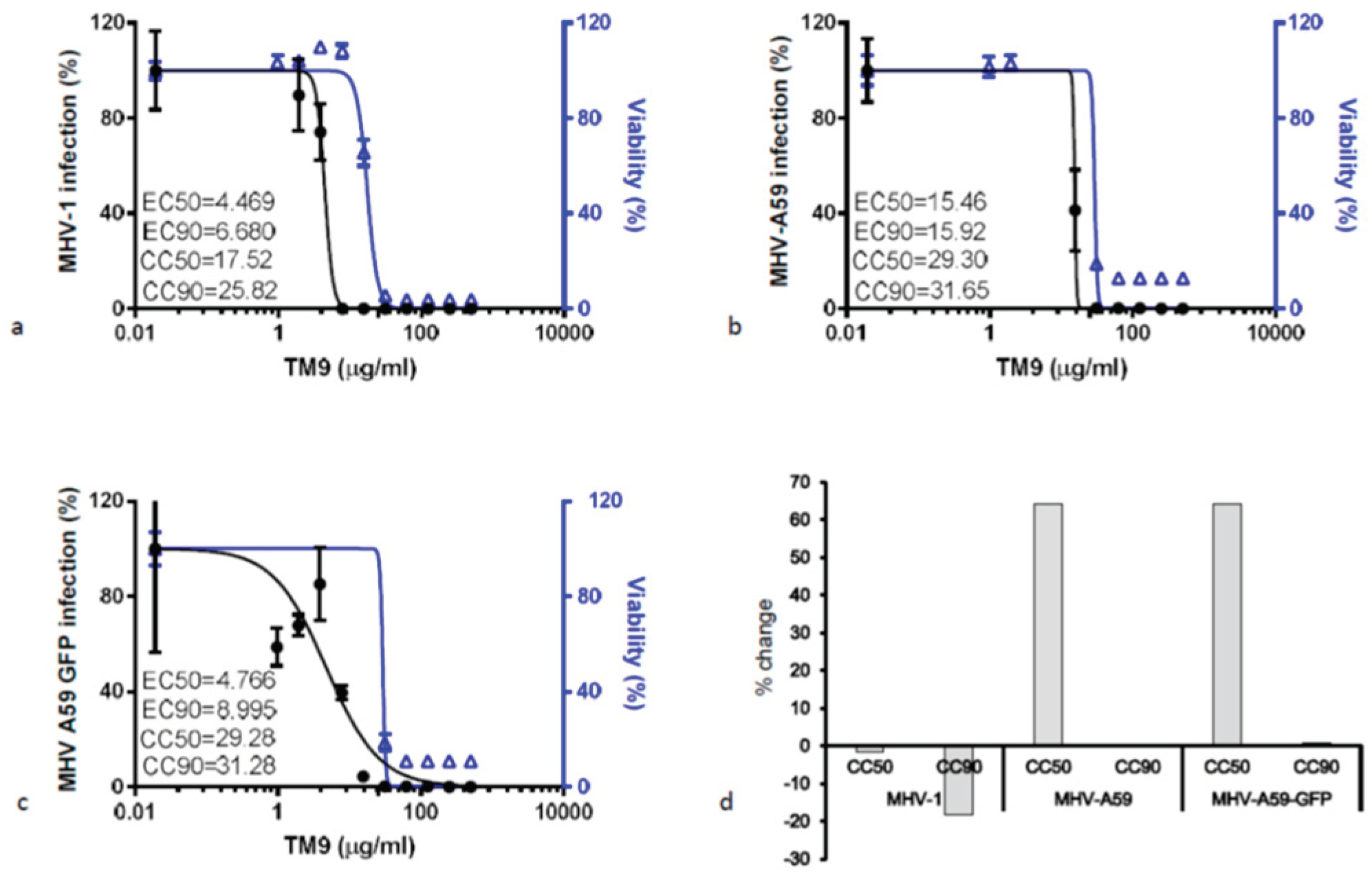
L2 cells were preincubated with serial dilutions of TM9 before infection with either MHV-1, MHV-A59, or MHV-A59-GFP (
Figure 3). The CC50 of L2 cells infected with MHV-1 was 17.52 μg/ml, while the effective concentration of the peptoid against virus (EC50) was 6.68 μg/ml, yielding a therapeutic index (CC50/EC90) of 2.62. MHV-A59-infected cells yielded a CC50 of 29.30 μg/ml and an EC-90 or 15.92 μg/ml resulting in a therapeutic index of 1.84. Lastly, L2 cells infected with MHV-A59-GFP had a CC50 of 29.28 μg/ml and an EC90 of 8.99 μg/ml, resulting in a therapeutic index of 3.25, the highest among all three viruses.
3.3. Respiratory MHV Infection Was Effectively Generated in All Experimental Animals by Aerosol Inoculation Using Madison Chamber
Mice were infected with one of the three MHV viral strains using a Madison Chamber at 105 PFU/ml for MHV-1 and 107 PFU/ml for both MHV-A59 and MHV-A59-GFP. Mice that were exposed to aerosols of one of these viruses demonstrated signs of the disease starting from the first day post-infection, lasting up to the 3rd day post-infection. Clinical signs of the infection, including lethargy, dyspnea, nasal discharge, and fever (39-40 oC) were observed throughout these three days, subsiding afterwards.
To determine the effect that virus had on these mice, 2 mice from each group were euthanized daily. Subsequently, necropsies were performed to determine pathological effect on the mice and the viral titers seen in various organs. Necropsy revealed pathological changes in the lungs including swelling in lung tissue and redness due to vascular dilation. There were no visible gross-pathological changes in other organs during the entire observation period.
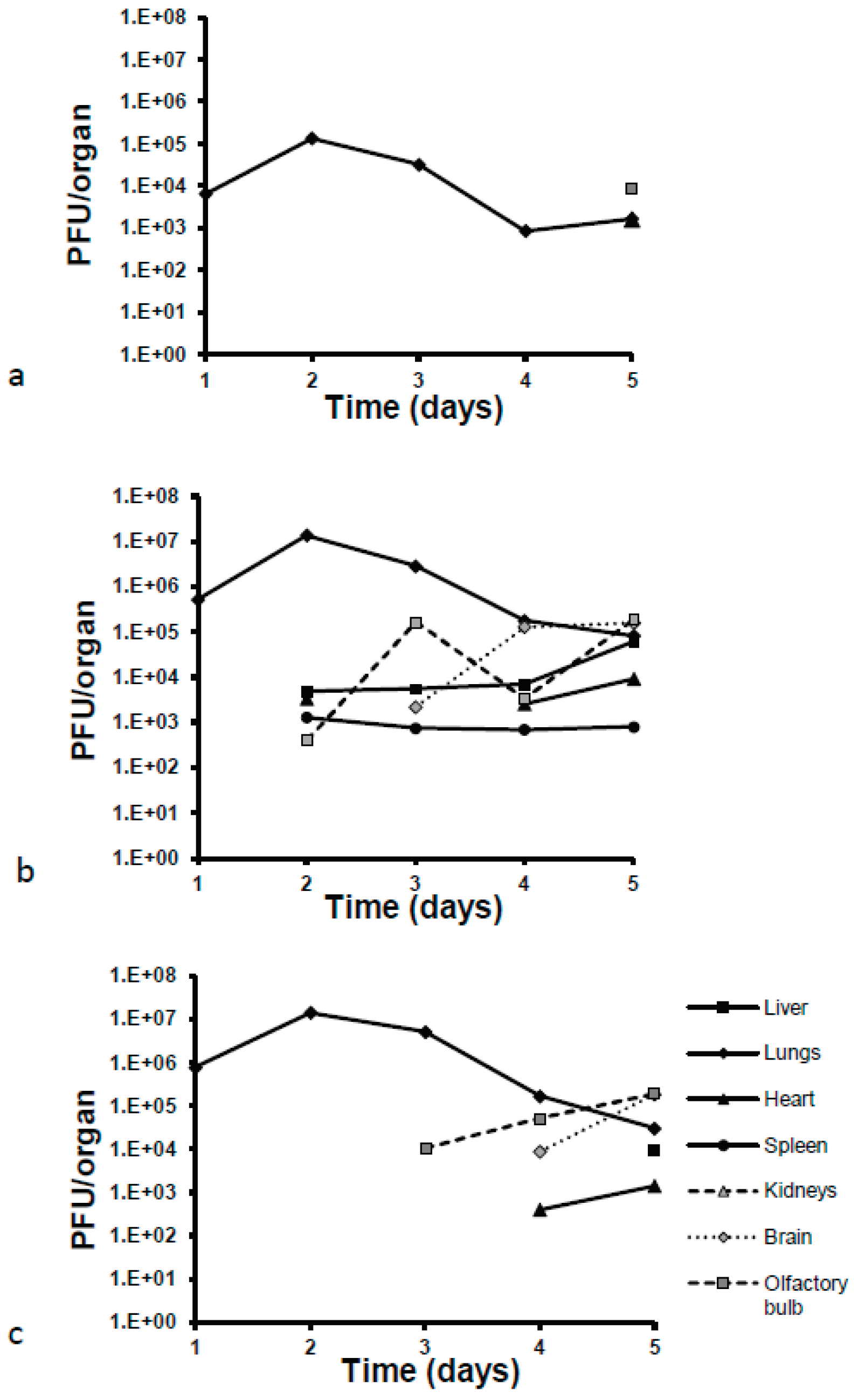
Viral loads were 100-fold and 10-fold lower in lung homogenates for MHV-1 than those observed for both MHV-A549 strains starting from day one post-infection (
Figure 4). All mice treated with virus increased viral loads by day two post-infection, with increases approximately one log higher than on day one. Viral titers decreased from this point on until the final day of observation, day 5, resulting in a final decrease of one log compared to the original titers of day 1. All MHV-1 viral loads were two logs lower than both MHV-A59 strains. Interestingly, another difference seen between these strains was the ability for systemic dissemination to other organs. In fact, the only other organ which showed MHV-1 virus as detected and quantified by plaque assay was the olfactory bulbs, seen exclusively on day 5 post-infection. Conversely, MHV-A59 virus was detected in olfactory bulbs, liver, heart, and spleen on day two post-infection. From day 3 onward, this strain was detectable in the brain, with a significant increase in viral loads (2 logs) on day 4. MHV-A59-GFP was initially detected in olfactory bulbs on day 3 post-infection. From here onward, the viral load increased until the end of the observation period. Virus was detectable in the brain and heart starting on day 4 and in the liver starting on day 5.
4. Discussion
In this work, we determined the therapeutic window and therapeutic index for TM9 peptoid
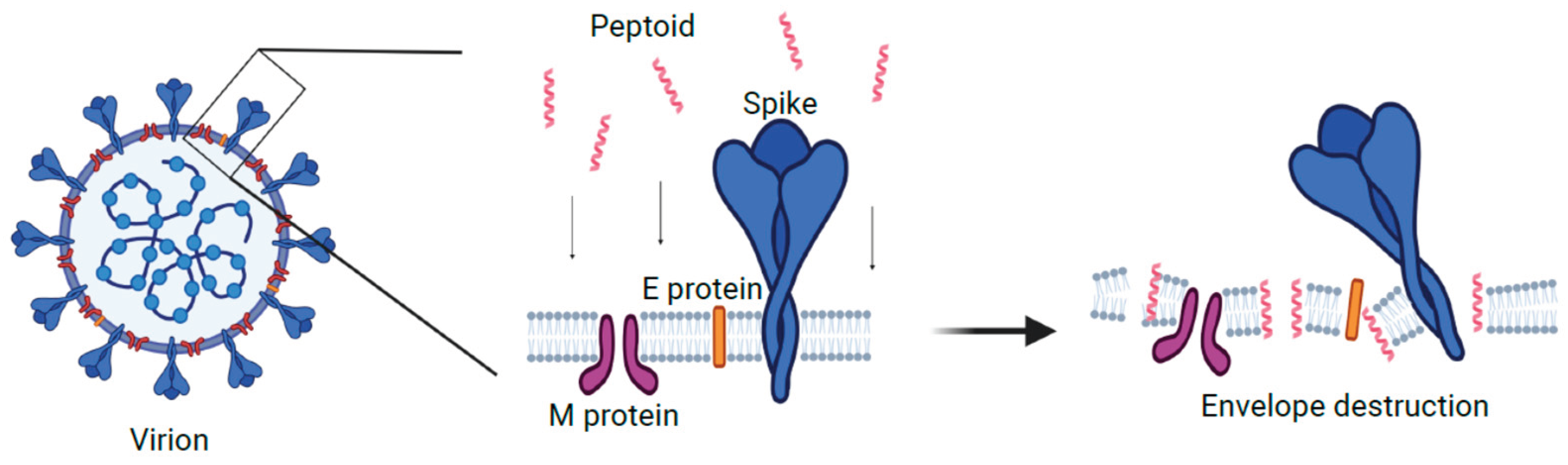
in vitro. TM9 has been shown to have a similar mechanism to the human cathelicidin LL-37. It has been demonstrated that LL-37 causes disruption in the cell membrane of bacteria, causing pore formation which leads to cytoplasmic leaking, osmotic disbalance, and multiple functional dysregulations (37). As such, we can speculate that the effect of this peptoid on the membrane of the viral envelope is similar to its effect on the bacterial membrane (
Figure 5). While this model needs further investigation, we believe that the similarities of virion envelope structure likely means that TM9 acts against virions in a similar manner to how it acts against bacteria. There are several potential ways in which TM9 may affect the ability of MHV virions from being viable. The first is that TM9 prevents virions from being able to attach to the host cell receptors by blocking or binding to receptors on the virion. If this were to be the case, virions would be unable to bind to the surface receptors of the cells they were infecting, resulting in reduced ability to infect cells. A second possibility is that TM9 binds to the membrane component of the virion and exposes the internal parts of the virion to an extracellular environment. This would result in lowered viability as virions sensitive RNA genome would be exposed to any RNAses in the media or have key components leaking into the environment, likely before they are able to infect cells. As the virion envelope is typically derived from the mammalian cell outer or inner membrane and has similar structure, this model likely is involved. It is possible that both mechanisms are occurring simultaneously. If this is the case, the most important question is how the antiviral effect of the peptoid is related to its cytotoxic effect. Recently it has been shown that phosphatidylserine in the viral envelope plays an important role in membrane susceptibility to the disruption by the peptoid, and may thus be a potential way to optimize future peptoid design towards virions over mammalian cells (25).
Figure 5.
Proposed model of mechanism of action for TM9 peptoid. Peptoid molecules destabilize structure and organization of the lipid bilayer and destroy viral envelope and/or disrupt its functions. Created with BioRender.com.
Figure 5.
Proposed model of mechanism of action for TM9 peptoid. Peptoid molecules destabilize structure and organization of the lipid bilayer and destroy viral envelope and/or disrupt its functions. Created with BioRender.com.
Here we demonstrated that the effect of the TM9 peptoid on mammalian cells showed a difference between cytotoxic concentration and maximum nontoxic concentration. Both murine and human lung carcinoma cells had similar levels of tolerance to TM9. According to our results, the CC50 of TM9 3 hours post-incubation was 20 – 30 μg/ml for both lung carcinoma (A549) cells and L2 cells, while the CC90 was approximately 61 μg/ml and 81 μg/ml respectively, while microvascular endothelial cells had a CC90 of approximately 36 μg/ml. This suggests that human intravascular endothelial cells were more sensitive to the peptoid, which should be considered upon parenteral administration of the peptoid. Other studies demonstrated that tumor cells can be more sensitive to cationic antimicrobial peptides due to higher metabolic activity of these cells (38).
Cultured cells have the distinct disadvantage of being less accurate measures of cytotoxicity as they are missing many of the tight junctions involved in tissues. As such, as the next step in this work, it may be necessary to try alternative methods of cell culture to get a better understanding of how cytotoxic TM9 is. One such method would be to do testing via air-liquid interface (ALI), which has been shown to cause changes on the surface properties of alveolar epithelial cells (39). Subsequent or concurrent experiments in vivo may show even less toxicity and will provide insight into potential adverse systemic effects. Local application in the form of aerosol inhalation should be also considered as an alternative or additional therapeutic administration procedure. Local aerosol administration has demonstrated promising results in application of antimicrobial peptides against other infections of the respiratory system (40) (41).
The therapeutic index of TM9, as a ratio of CC50 to EC50, depends on the virulence of the viral strain according to our results. MHV-A59 was the most virulent strain and the effective concentration of TM9 against this virus was the highest. MHV-1 was the most susceptible to peptoid and demonstrated the highest therapeutic index. Further investigation is required as TM9 demonstrated a relatively low therapeutic index in traditional cell culture, particularly in the case of the most virulent strain of MHV. Therefore, cytotoxicity tests using only a cell culture model are likely not sufficient. In this case, a more comprehensive and integrative approach is required, including hemolytic activity assays, cytotoxicity tests in vivo, and apoptosis tests. This will all have to be done using peripheral blood mononuclear cells as it was proposed previously to determine therapeutic index for antimicrobial peptides (42) or in vitro using 3D cell culture (22) (24).
The second part of this work was to evaluate MHV infection as a model of human SARS-Cov-2 infection for further tests of peptoids and other antivirals in vivo. An important part of this goal was to determine if utilization of the Madison chamber was an effective method to test aerosolized MHV in mice. Madison aerosol chambers produce a dry aerosol, and the animals present are not under anesthesia, allowing for the most natural transmission of the virus in a highly accurate and reproducible manner, providing a model of respiratory coronavirus infection and testing of antimicrobial peptoids and other antiviral therapeutics. Our results have shown that utilization of the Madison chamber is an effective instrument for aerosol inoculation of mice with respiratory MHV variants. All animals showed signs of infection via respiratory and/or other symptoms as is typically seen with MHV infection. Disease severity was dependent on the MHV strain; however, disease manifestations were observed in all infected mice, with the highest levels in animals infected with MHV-A59. Infection caused by MHV-A59-GFP was similar to infections with MHV-A59 wild type based on viral load titers and their dynamics of change over time, but showed slower dissemination to other organs. Previous reports have shown that MHV-A59 tropism to multiple organs occurs, including infection of the respiratory tract (31) and the brain/liver (43). MHV-1 infection was not systemic and was only detected in the respiratory tract and the olfactory bulb. This strain demonstrated lower virulence and is considered a strain with only respiratory tropism. As a result, it is often used in murine respiratory coronavirus models, but only in highly susceptible mice lineages such as A/J mice (44) (45). All of this suggests that the use of a Madison Chamber for infection with MHV strains is not only possible, but effective. As MHV and SARS-CoV-2 are both Betacoronaviruses, it stands to reason that future studies with SARS-CoV-2 may be able to take advantage of the effectiveness of using a Madison Chamber as well.
These data demonstrate that the effective concentration of TM9 is within the therapeutic range and MHV provides a rapid, simple, and effective model for coronavirus infection and evaluation of therapeutics in a systemic and organ-specific manner. Since these data are quite promising, further studies are warranted to evaluate the therapeutic efficacy of TM9 and other, more tailored peptoids in vivo and to use other coronaviruses, including SARS-CoV-2.
Author Contributions
Conceptualization, J.D.C and A.E.B.; methodology, J.D.C, M.L. and S.K; validation, J.D.C., A.E.B., N.M., J.S.L., and J.L.L.; formal analysis, ML. and S.K.; investigation, M.L, S.K. and J.D.C.; resources, KJ.K, A.E.B and J.D.C.; data curation, M.L. and S.K; writing—original draft preparation, M.L. and A.B.B.; writing—review and editing, J.D.C., A.B.B. J.L.L., N.M., J.S.L., K.J.K and A.E.B.; visualization, M.L and S.K.; supervision, J.D.C.; project administration, J.D.C.; funding acquisition, J.D.C. and A.E.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part from funds provided by the Texas A&M University System and NIH grants EB032983, AI141607 and AI149383. A.E.B. thanks the NIH for funding this work with a Director's Pioneer Award, grant # 1DP1 OD029517.
Institutional Review Board Statement
The animal study protocol was approved by Texas A&M Animal Care and Use Committee. (protocol 2021-0201, approved 09 September 2021).
Acknowledgments
AEB also acknowledges funding from Stanford University's Discovery Innovation Fund, the Cisco University Research Program Fund, and the Silicon Valley Community Foundation, Stephen Pearse, and Dr. James J. Truchard and the Truchard Foundation. We gratefully acknowledge Dr. Michael Connolly and Dr. Behzad Rad at the Molecular Foundry for assistance with peptoid synthesis and sample preparation equipment. Work at the Molecular Foundry was supported by the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Conflicts of Interest
A.E.B. is a shareholder and member of the Board of Directors of Maxwell Biosciences; N.M. is a consultant for Maxwell Biosciences; J.D.C. has stock in Maxwell Biosciences, a company that has licensed peptoid therapeutics.
References
- Atzrodt CL, Maknojia I, McCarthy RDP, Oldfield TM, Po J, Ta KTL, et al. A Guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020 Sep;287(17):3633–50. [CrossRef]
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009 Mar;7(3):226–36. [CrossRef]
- Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 Mar;30(3):269–71. [CrossRef]
- Furuta Y, Komeno T, Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B. 2017;93(7):449–63. [CrossRef]
- Báez-Santos YM, St. John SE, Mesecar AD. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015 Mar;115:21–38. [CrossRef]
- Jo S, Kim S, Shin DH, Kim MS. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem. 2020 Jan 1;35(1):145–51. [CrossRef]
- Felgenhauer U, Schoen A, Gad HH, Hartmann R, Schaubmar AR, Failing K, et al. Inhibition of SARS-CoV-2 by type I and type III interferons. J Biol Chem. 2020 Oct 9;295(41):13958–64. [CrossRef]
- Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020 Aug;584(7819):115–9. [CrossRef]
- Wang C, Li W, Drabek D, Okba NMA, van Haperen R, Osterhaus ADME, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020 May 4;11(1):2251. [CrossRef]
- Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell. 2020 May;181(4):865-876.e12.
- Adamson CS, Chibale K, Goss RJM, Jaspars M, Newman DJ, Dorrington RA. Antiviral drug discovery: preparing for the next pandemic. Chem. Soc. Rev. 2021; (50):3647. [CrossRef]
- Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, et al. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc Natl Acad Sci. 2008 Feb 26;105(8):2794–9. [CrossRef]
- Bals R, Weiner DJ, Moscioni AD, Meegalla RL, Wilson JM. Augmentation of Innate Host Defense by Expression of a Cathelicidin Antimicrobial Peptide. McGhee JR, editor. Infect Immun. 1999 Nov;67(11):6084–9. [CrossRef]
- Gudmundsson GH, Agerberth B, Odeberg J, Bergman T, Olsson B, Salcedo R. The Human Gene FALL39 and Processing of the Cathelin Precursor to the Antibacterial Peptide LL-37 in Granulocytes. Eur J Biochem. 1996;238(2):325–32. [CrossRef]
- Johansson J, Gudmundsson GH, Rottenberg ME, Berndt KD, Agerberth B. Conformation-dependent Antibacterial Activity of the Naturally Occurring Human Peptide LL-37. J Biol Chem. 1998 Feb;273(6):3718–24. [CrossRef]
- Yang L, Weiss TM, Lehrer RI, Huang HW. Crystallization of Antimicrobial Pores in Membranes: Magainin and Protegrin. Biophys J. 2000 Oct;79(4):2002–9. [CrossRef]
- Beyond structural models for the mode of action: How natural antimicrobial peptides affect lipid transport | Elsevier Enhanced Reader [Internet]. [cited 2022 Dec 30]. Available from: https://reader.elsevier.com/reader/sd/pii/S0021979720311413?token=88D9A9CD0BF4811511B3484D660C84A4F3DDB01A89765BB4AD1CFE9DF7E97A2C14D99A233184B79EF110BE250B1685A1&originRegion=us-east-1&originCreation=20221230222938.
- Nyembe PL, Ntombela T, Makatini MM. Review: Structure-Activity Relationship of Antimicrobial Peptoids. Pharmaceutics. 2023 May;15(5):1506. [CrossRef]
- Chongsiriwatana NP, Lin JS, Kapoor R, Wetzler M, Rea JAC, Didwania MK, et al. Intracellular biomass flocculation as a key mechanism of rapid bacterial killing by cationic, amphipathic antimicrobial peptides and peptoids. Sci Rep. 2017 Dec 1;7(1):16718. [CrossRef]
- Miller SM, Simon RJ, Zuckermann RN, Kerr JM, Moos WH. Comparison of the proteolytic susceptibilities of homologous L-amino acid, D-amino acid, and N-substituted glycine peptide and peptoid oligomers. Drug Dev. Res. 1995 May; 35(1):20-32.
- Benjamin AB, Moule MG, Didwania MK, Hardy J, Saenkham-Huntsinger P, Sule P, Nielsen JE, Lin JS, Contag CH, Barron AE, Cirillo JD. Efficacy of cathelicidin-mimetic antimicrobial peptoids against Staphylococcus aureus. Microbiol. Spectr. 2022 June;10(3):e00534-22. [CrossRef]
- Nielsen JE, Alford MA, Yung DBY, Molchanova N, Fortkort JA, Lin JS, et al. Self-Assembly of Antimicrobial Peptoids Impacts Their Biological Effects on ESKAPE Bacterial Pathogens. ACS Infect Dis. 2022 Mar 11;8(3):533–45. [CrossRef]
- Efficacy of Cathelicidin-Mimetic Antimicrobial Peptoids against Staphylococcus aureus [Internet]. [cited 2022 Dec 29]. Available from: https://journals.asm.org/doi/epub/10.1128/spectrum.00534-22. [CrossRef]
- Diamond G, Molchanova N, Herlan C, Fortkort JA, Lin JS, Figgins E, et al. Potent Antiviral Activity against HSV-1 and SARS-CoV-2 by Antimicrobial Peptoids. Pharmaceuticals. 2021 Apr;14(4):304. [CrossRef]
- Tate PM, Mastrodomenico V, Cunha C, McClure J, Barron AE, Diamond G, et al. Peptidomimetic Oligomers Targeting Membrane Phosphatidylserine Exhibit Broad Antiviral Activity. ACS Infect Dis. 2023 Aug 11;9(8):1508–22. [CrossRef]
- Current ICTV Taxonomy Release | ICTV [Internet]. [cited 2023 Jan 18]. Available from: https://ictv.global/taxonomy.
- Körner R, Majjouti M, Alcazar M, Mahabir E. Of Mice and Men: The Coronavirus MHV and Mouse Models as a Translational Approach to Understand SARS-CoV-2. Viruses. 2020 Aug 12;12(8):880. [CrossRef]
- De Albuquerque N, Baig E, Ma X, Zhang J, He W, Rowe A, et al. MurineHepatitis Virus Strain 1 Produces a Clinically Relevant Model of Severe Acute Respiratory Syndrome in A/J Mice. J Virol. 2006 Nov;80(21):10382–94. [CrossRef]
- Featherstone AB, Brown AC, Chitlapilly Dass S. Murine Hepatitis Virus, a Biosafety Level 2 Model for SARS-CoV-2, Can Remain Viable on Meat and Meat Packaging Materials for at Least 48 Hours. Tamber S, editor. Microbiol Spectr. 2022 Oct 26;10(5):e01862-22. [CrossRef]
- Ma XZ, Bartczak A, Zhang J, Khattar R, Chen L, Liu MF, et al. Proteasome Inhibition In Vivo Promotes Survival in a Lethal Murine Model of Severe Acute Respiratory Syndrome. J Virol. 2010 Dec;84(23):12419–28. [CrossRef]
- Yang Z, Du J, Chen G, Zhao J, Yang X, Su L, et al. Coronavirus MHV-A59 infects the lung and causes severe pneumonia in C57BL/6 mice. Virol Sin. 2014 Dec;29(6):393–402. [CrossRef]
- Kutter JS, de Meulder D, Bestebroer TM, Lexmond P, Mulders A, Richard M, et al. SARS-CoV and SARS-CoV-2 are transmitted through the air between ferrets over more than one meter distance. Nat Commun. 2021 Mar 12;12(1):1653. [CrossRef]
- Huang YC, Tu HC, Kuo HY, Shao PL, Gong YN, Weng HY, et al. Outbreak investigation in a COVID-19 designated hospital: The combination of phylogenetic analysis and field epidemiology study suggesting airborne transmission. J Microbiol Immunol Infect Wei Mian Yu Gan Ran Za Zhi. 2023 Jan 16;S1684-1182(23)00002-6. [CrossRef]
- Rothfels KH, Axelrad AA, Siminovich L, Mclulloch EA, Parker RC (1959) The origin of altered cell lines from mouse, monkey and man as indicated by chromosome and transplantation studies. Can Cancer Conf 3:189- 214.
- Connolly MD, Xuan S, Molchanova N, Zuckermann RN. Chapter 8 – Submonomer synthesis of sequence defined peptoids with diverse side-chains. In Methods in Enzymology, 1st Edition; Pyle AM, Christianson DW, Colowick SP, Kaplan NO, Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 656, pp. 241-270.
- Xhindoli D, Pacor S, Benincasa M, Scocchi M, Gennaro R, Tossi A. The human cathelicidin LL-37 — A pore-forming antibacterial peptide and host-cell modulator. Biochim Biophys Acta BBA - Biomembr. 2016 Mar 1;1858(3):546–66. [CrossRef]
- Wang K rong, Zhang B zhi, Zhang W, Yan J xi, Li J, Wang R. Antitumor effects, cell selectivity and structure–activity relationship of a novel antimicrobial peptide polybia-MPI. Peptides. 2008 Jun;29(6):963–8. [CrossRef]
- Öhlinger K, Kolesnik T, Meindl C, Gallé B, Absenger-Novak M, Kolb-Lenz D, Frhlich E. Air-liquid interface culture changes surface properties of A549 cells. Toxicol In Vitro. 2019; (60):369-382. [CrossRef]
- Falciani C, Zevolini F, Brunetti J, Riolo G, Gracia R, Marradi M, et al. Antimicrobial Peptide-Loaded Nanoparticles as Inhalation Therapy for Pseudomonas aeruginosa Infections. Int J Nanomedicine. 2020 Feb 17;15:1117–28. [CrossRef]
- Wang Y, Chang RYK, Britton WJ, Chan HK. Advances in the development of antimicrobial peptides and proteins for inhaled therapy. Adv Drug Deliv Rev. 2022 Jan 1;180:114066. [CrossRef]
- Bacalum M, Radu M. Cationic Antimicrobial Peptides Cytotoxicity on Mammalian Cells: An Analysis Using Therapeutic Index Integrative Concept. Int J Pept Res Ther. 2015 Mar 1;21(1):47–55. [CrossRef]
- Savarin C, Bergmann CC. Viral-induced suppression of self-reactive T cells: Lessons from neurotropic coronavirus-induced demyelination. J Neuroimmunol. 2017 Jul 15;308:12–6. [CrossRef]
- Paidas MJ, Mohamed AB, Norenberg MD, Saad A, Barry AF, Colon C, et al. Multi-Organ Histopathological Changes in a Mouse Hepatitis Virus Model of COVID-19. Viruses. 2021 Sep;13(9):1703. [CrossRef]
- Gong HH, Worley MJ, Carver KA, Goldstein DR, Deng JC. Neutrophils drive pulmonary vascular leakage in MHV-1 infection of susceptible A/J mice. Front Immunol [Internet]. 2023 [cited 2023 Jul 26];13. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1089064. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).