Introduction
Banana and plantain (
Musa spp.) are crops of vital importance to hundreds of millions of people around the world. Most edible cultivars of banana are derived from two wild species, namely
M. acuminata (A genome) and
M. balbisiana (B genome). According to Simmonds [
1], during their long-term evolution, these two wild species hybridized and interbred, evolving into modern banana through continuous natural selection and artificial selection. Simmonds and Shepherd developed a banana hybrid identification system using 15 prominent traits, among which 13 traits are related to the reproductive organs and are highly polymorphic between
M. acuminata and
M. balbisiana [
2]. Based on these phenotypic scoring systems, the
M. acuminata ×
M. balbisiana hybrids could be characterized into different ploidy levels (diploid, triploid, and tetraploid) and several genome groups (e.g., AA, AB, AAA, AAB, and ABB). Generally, one genome group can be divided into several subgroups; for example, the AAA genome group contains Cavendish, Gros Michel, Red, Lakatan, and Ibota Bota. According to morphological markers, highly similar cultivated varieties of banana can be classified into a subgroup, such as Cavendish banana‘Brazil’,‘Williams’, and‘Pei Chiao’. The identification of banana germplasm is the premise for the evaluation and utilization of banana germplasm resources. However, the management of banana germplasm resources has relied immensely on identification using local names and morphological characters, and the extent of the genetic diversity of banana has not been established with molecular markers. Most of the morphological markers are polygenic and highly influenced by the environment. Moreover, morphological observation is time-consuming and laborious. Despite this, morphological markers are still used in banana breeding due to the unavailability of other markers.
Molecular marker technology has been widely used in germplasm resource identification over the past decades. As described by Kaemmer, DNA oligonucleotide and amplification fingerprinting has been successfully used to detect genetic polymorphisms in 15 representative species and cultivars of the genus
Musa comprising the AA, AAA, AAAA, AAB, ABB, and BB genotypes [
3]. Risterucci [
4] demonstrated the usefulness of diversity arrays technology (DArT)—a DNA hybridization-based molecular marker technique that can simultaneously detect variation at numerous genomic loci without sequence information—for genetic diversity analyses of
Musa genotypes. Numerous studies have utilized few simple sequence repeat (SSR) markers, which have relatively limited genome coverage, in the classification of banana [
5,
6,
7,
8]. Using SSR and amplified fragment length polymorphism (AFLP) markers, several studies analyzed
Musa genome groups [
9,
10,
11,
12]. However, due to objective and subjective reasons, the repeatability of SSRs cannot be guaranteed.
Single nucleotide polymorphisms (SNPs), another class of markers, can accurately distinguish highly similar crop germplasm resources [
13]. Moreover, SNPs have higher genetic stability and are the most promising molecular marker at present for differentiating crop germplasm. They are referred to as the third-generation molecular marker. Due to their abundance and genome-wide coverage, and particularly with the advent of high-throughput genotyping methods such as genotyping-by-sequencing, SNP markers have been employed in population genetics studies in banana [
14,
15]. Alberto [
14] used SNP markers in DNA sequencing data related to restriction enzyme sites to study and compare the chromosome structures of 36 banana varieties belonging to the ABB genotype (including different subspecies). Gardoce [
16] developed a 1 K SNP genotyping panel, effectively distinguishing between genomic groups, based on the filtering of high-quality genome-wide SNPs from the Musa Germplasm Information System, and used it to assess the genetic diversity and population structure of 183
Musa spp. accessions. However, the further application of SNPs in banana germplasm identification needs to be explored.
Fragment variation analysis of the internal transcribed spacer (ITS) in the ribosomal RNA (rRNA) coding gene is widely used to evaluate phylogenetic relationships at a lower taxonomic level, since this region has experienced limited natural selection pressure and exhibits great variation, even among closely related species. Nwakanma [
17] analyzed the ITS fragments of nine banana genotypes (AA, BB, AB, AAA, AAB, ABB, AAAA, AAAB, and AABB) by restriction fragment length polymorphism (RFLP) and found that the A and B genomes could be distinguished by Rsa I digestion. In addition, Dita [
18] found that the banana
ACTIN2 gene could also be used as a molecular marker to identify the A and B genomes. The above two markers can be used to identify whether banana resources contain A and B genomes, but they are unable to determine the copy number of A and B genomes in polyploids (for example, AAB and ABB cannot be distinguished). Teo [
19] used inter-retrotransposon amplified polymorphism (IRAP) to identify the A and B genomes. Subsequently, Nair [
20] used copia-IRAP primers to amplify banana germplasm resources which, together with Alu I digestion, could effectively identify AAB and ABB. However, the above methods cannot be used for classification below the genome type. The ITS sequences of 36 banana species (42 accessions from the ingroup representing three genera) together with 10 ingroup accessions retrieved from the GenBank database and four outgroup accessions were used to construct the phylogeny of the banana family [
21]. However, it remains unclear whether ITS can be used for the classification of cultivated banana varieties.
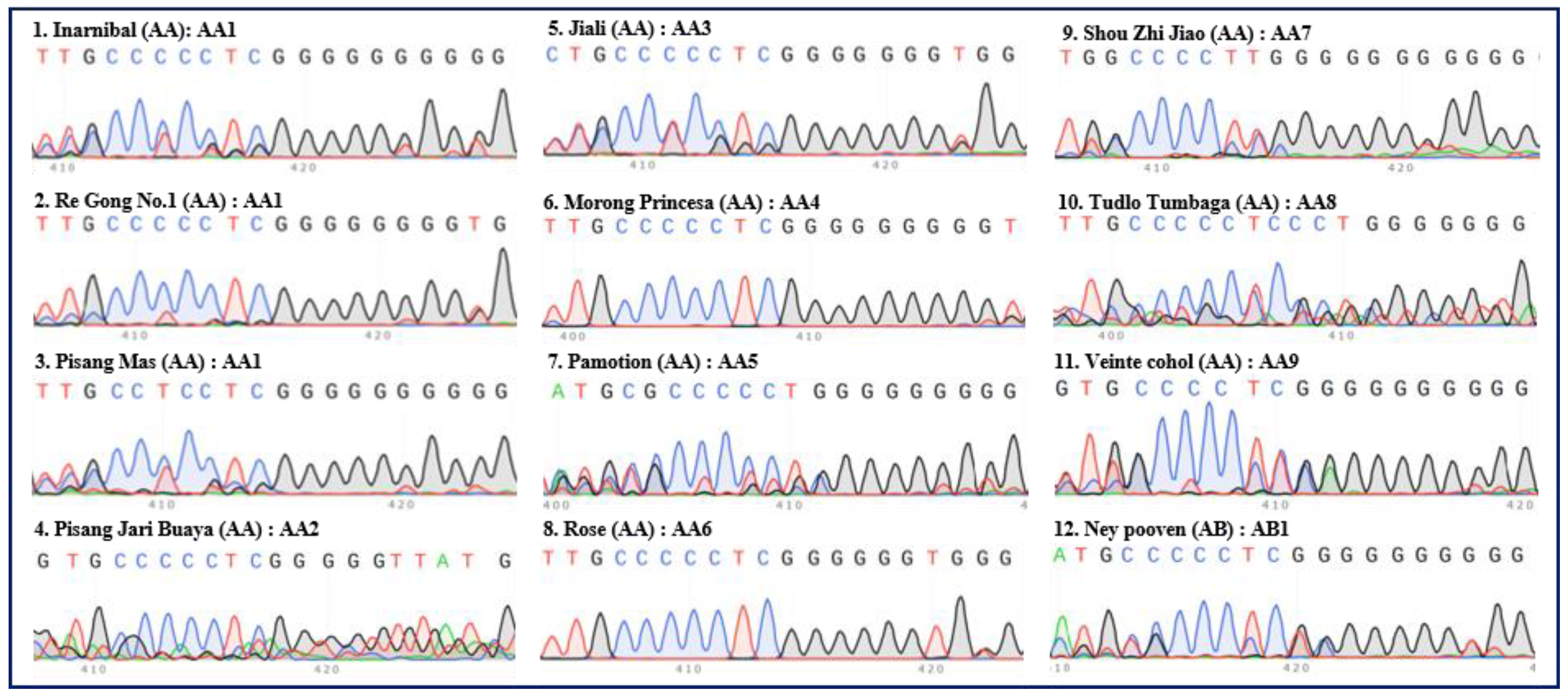
The National Litchi and Banana Germplasm Resources Garden in China manages and conserves more than 400 accessions of local and introduced cultivars and wild species of Musa in the field and through in vitro conservation. The genetic characterization of the germplasm collection has not been explored extensively at the molecular level. In this paper, we discovered that the ITS sequencing peaks (especially the “420 bp region”) of banana exhibited recognizable and repeatable polymorphism characteristics. Based on this finding, we developed a new method for identifying banana germplasm resources and successfully divided 62 accessions of banana into 44 “ITS types”.
Discussion
The genetic background of modern cultivated banana is complex, which is primarily because (1) during evolution, the ancestors of cultivated banana were formed by different wild species and interspecific hybridization, which endowed them with a rich genetic diversity; and (2) similarly, the long-term vegetative reproduction of cultivated banana varieties resulted in the accumulation of significant genetic diversity. Therefore, the differences in ITS sequences in cultivated banana can reflect the differences in cultivation types to some extent. In the present analysis, the superposition results of the SNP sets from the genomes of banana in the first-generation sequencing peak map of the ITS fragment were used as a fingerprint of an ITS SNP polymorphism. It was confirmed by our experiments that the ITS sequencing peaks, and particularly the “420 bp region”, of different banana cultivars could accurately reflect their genetic background to some extent. Using the “420 bp region” as a marker, 62 accessions of banana were clustered into 44 types. The improvement of banana cultivars through crossbreeding is promising [
22]. Our method appears to be effective for identifying the offspring and therefore will be useful for the early detection of hybrid banana.
A simple and reliable genotyping method for banana clones, hybrids, species, and relatives will facilitate germplasm management and support breeding initiatives toward a marker-based approach. Various molecular marker technologies have been applied in banana: RFLP, variable number of tandem repeats (VNTR), random amplified polymorphic DNA (RAPD), inter simple sequence repeats (ISSR), ITS, IRAP, AFLP, SSR, DArT, and EcoTILLING [
23]. However, these methods are generally complicated. Even though a variety of tools can be used, the classification of banana remains a challenge. For example, the classification of cultivated ABB is necessary, since the more popular names (Saba, Pisang Awak, Peyan, Bluggoe, Monthan) represent a cluster of closely related cultivars generated by somatic variation [
14]. The nomenclature of the entire ABB group is difficult to resolve given that the only source of information is the local name of each variant in Asia [
14]. This difficulty was confirmed by Saraswathi [
24], who combined morpho-taxonomic descriptors and SSR markers and attempted to discriminate the Indian subgroups. Using DArT and SSR markers on a wider sample, researchers confirmed that the classification was consistent for accessions belonging to the subgroups Pelipita, Klue Teparod, and Pisang Awak [
15,
25]. However, Sardos [
15] found that accessions classified as belonging to the subgroups Saba, Monthan, Bluggoe, Ney Mannan, or Peyan were often misclassified. Herein, using ITS sequencing, we revealed that the classification of most accessions was in accordance with traditional classification. For example, 11 of 12 Cavendish banana used in this study were clustered into one ITS type (
Figure 2), and all four Pisang Awak bananas were clustered into one ITS type (
Figure 4). Though there were some exceptions regarding consistency with the traditional classification, this could be clarified using further analysis.
The method outlined in this paper can effectively improve the efficiency of banana germplasm identification as an auxiliary means of character identification and genome type identification. However, it needs to be improved in the future. Sequencing peaks with weak signals occurred frequently and needed to be repeatedly verified. ITS sequencing might also be useful in the identification of other crops. Using this method, further sequencing peak maps of other sequences can be mined and developed as molecular markers for germplasm identification.
Figure 1.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of 12 accessions of diploid banana. Eleven accessions of AA group banana (‘Inarnibal’, ‘Re Gong No.1’, ‘Pisang Mas’, ‘Pisang Jari Buaya’, ‘Jia Li’, ‘Morong Princesa’, ‘Pamotion’, ‘Rose’, ‘Shou Zhi Jiao’, ‘Tudlo Tumbaga’, and ‘Veinte Cohol’) and one accession (‘Ney Pooven’) of AB group banana were tested. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
Figure 1.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of 12 accessions of diploid banana. Eleven accessions of AA group banana (‘Inarnibal’, ‘Re Gong No.1’, ‘Pisang Mas’, ‘Pisang Jari Buaya’, ‘Jia Li’, ‘Morong Princesa’, ‘Pamotion’, ‘Rose’, ‘Shou Zhi Jiao’, ‘Tudlo Tumbaga’, and ‘Veinte Cohol’) and one accession (‘Ney Pooven’) of AB group banana were tested. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
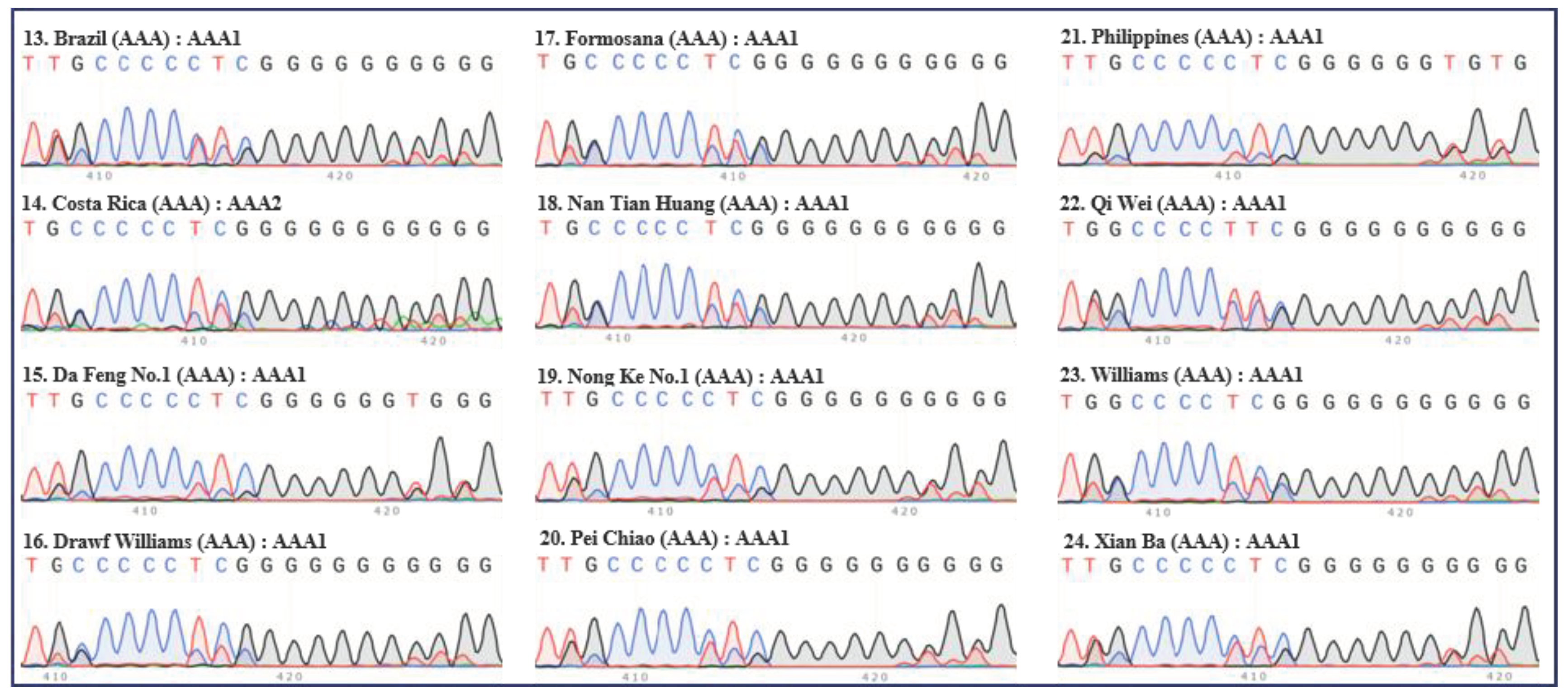
Figure 2.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of 12 accessions of Cavendish banana. Twelve accessions of Cavendish (‘Brazil’, ‘Costa Rica’, ‘Da Feng No.1’, ‘Drawf Williams’, ‘Formosana’, ‘Nan Tian Huang’, ‘Nong Ke No.1’, ‘Pei Chiao’, ‘Philippines’, ‘Qi Wei’, ‘Williams’, and ‘Xian Ba’) were tested. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
Figure 2.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of 12 accessions of Cavendish banana. Twelve accessions of Cavendish (‘Brazil’, ‘Costa Rica’, ‘Da Feng No.1’, ‘Drawf Williams’, ‘Formosana’, ‘Nan Tian Huang’, ‘Nong Ke No.1’, ‘Pei Chiao’, ‘Philippines’, ‘Qi Wei’, ‘Williams’, and ‘Xian Ba’) were tested. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
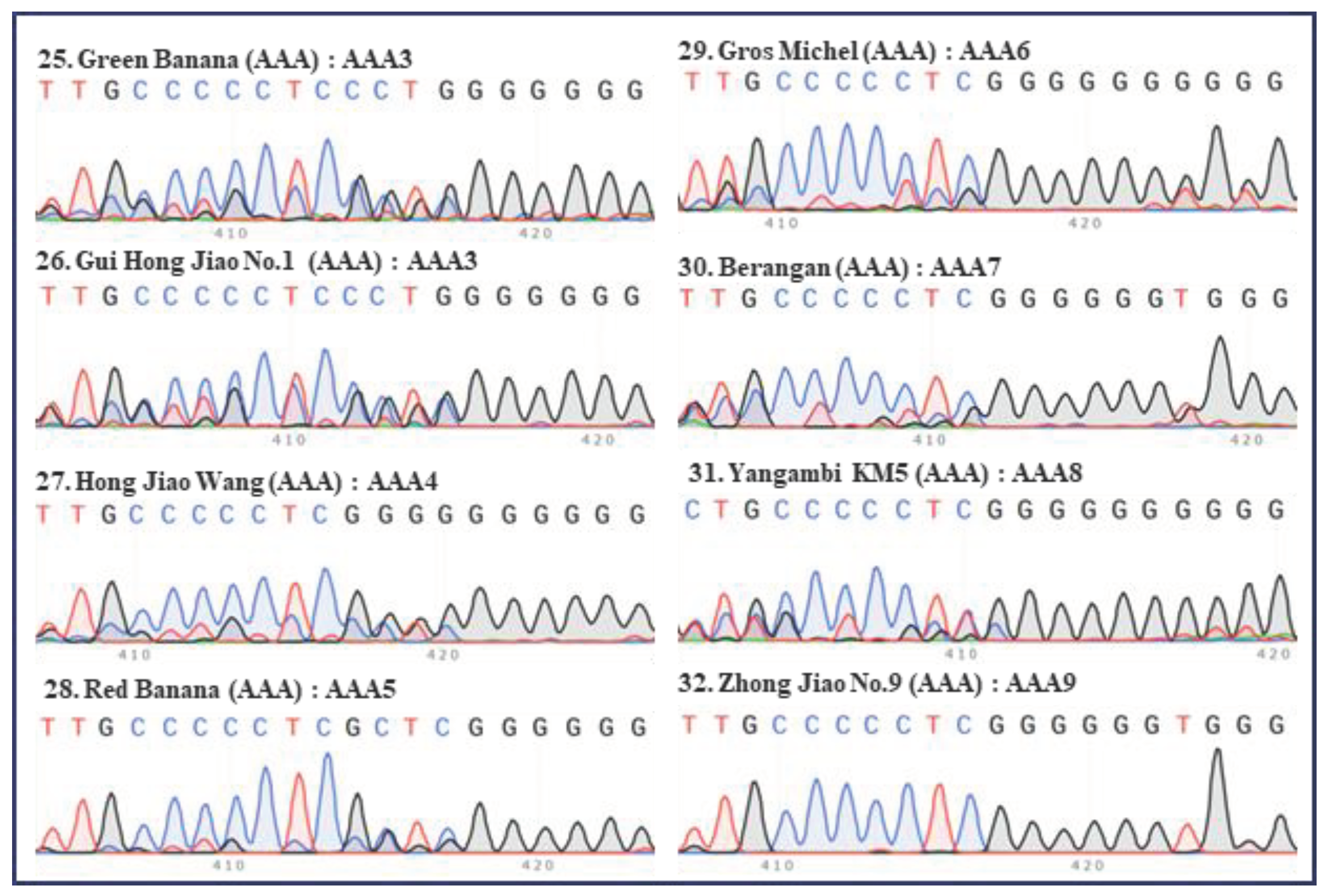
Figure 3.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of eight accessions of AAA group banana, excluding Cavendish. Four accessions of Red (‘Green Banana’, ‘Gui Hong Jiao No.1’, ‘Hong Jiao Wang’, and ‘Red Banana’), one accession of Gros Michel (‘Gros Michel’), one accession of Lakatan (‘Berangan’), one accession of Ibota Bota (‘Yangambi KM5’), and one accessions of the hybrid banana (‘Zhong Jiao No.9’) were tested. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
Figure 3.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of eight accessions of AAA group banana, excluding Cavendish. Four accessions of Red (‘Green Banana’, ‘Gui Hong Jiao No.1’, ‘Hong Jiao Wang’, and ‘Red Banana’), one accession of Gros Michel (‘Gros Michel’), one accession of Lakatan (‘Berangan’), one accession of Ibota Bota (‘Yangambi KM5’), and one accessions of the hybrid banana (‘Zhong Jiao No.9’) were tested. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
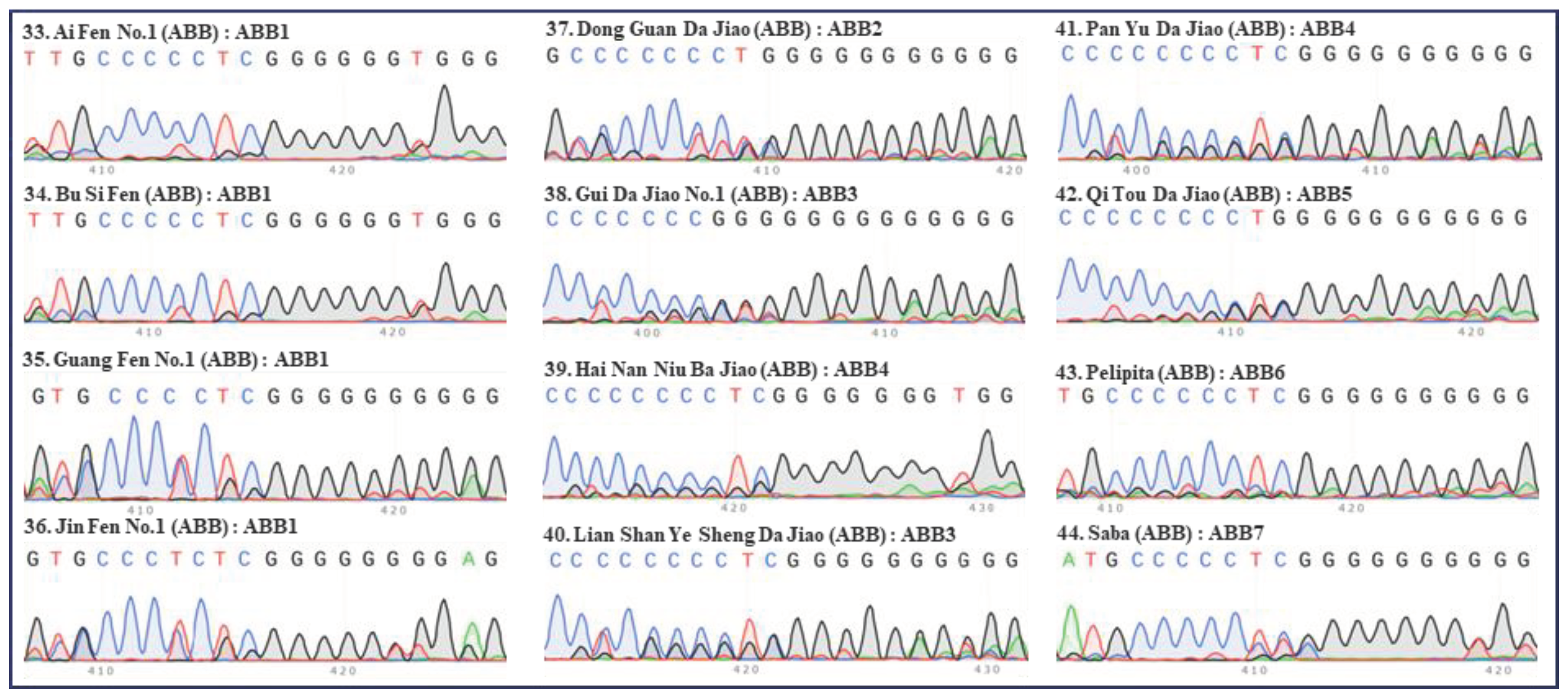
Figure 4.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of 12 accessions of ABB group banana. Four accessions of Pisang Awak (‘Ai Fen No.1’, ‘Bu Si Fen’, ‘Guang Fen No.1’ and ‘Jin Fen No.1’), six accessions of Da Jiao (‘Dong Guan Da Jiao’, ‘Gui Da Jiao No.1’, ‘Hai Nan Niu Ba Jiao’, ‘Lian Shan Ye Sheng Da Jiao’, ‘Pan Yu Da Jiao’ and ‘Qi Tou Da Jiao’), one accessions of Pelipita (‘Pelipita’) and one accessions of Saba (‘Saba’) were tested. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
Figure 4.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of 12 accessions of ABB group banana. Four accessions of Pisang Awak (‘Ai Fen No.1’, ‘Bu Si Fen’, ‘Guang Fen No.1’ and ‘Jin Fen No.1’), six accessions of Da Jiao (‘Dong Guan Da Jiao’, ‘Gui Da Jiao No.1’, ‘Hai Nan Niu Ba Jiao’, ‘Lian Shan Ye Sheng Da Jiao’, ‘Pan Yu Da Jiao’ and ‘Qi Tou Da Jiao’), one accessions of Pelipita (‘Pelipita’) and one accessions of Saba (‘Saba’) were tested. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
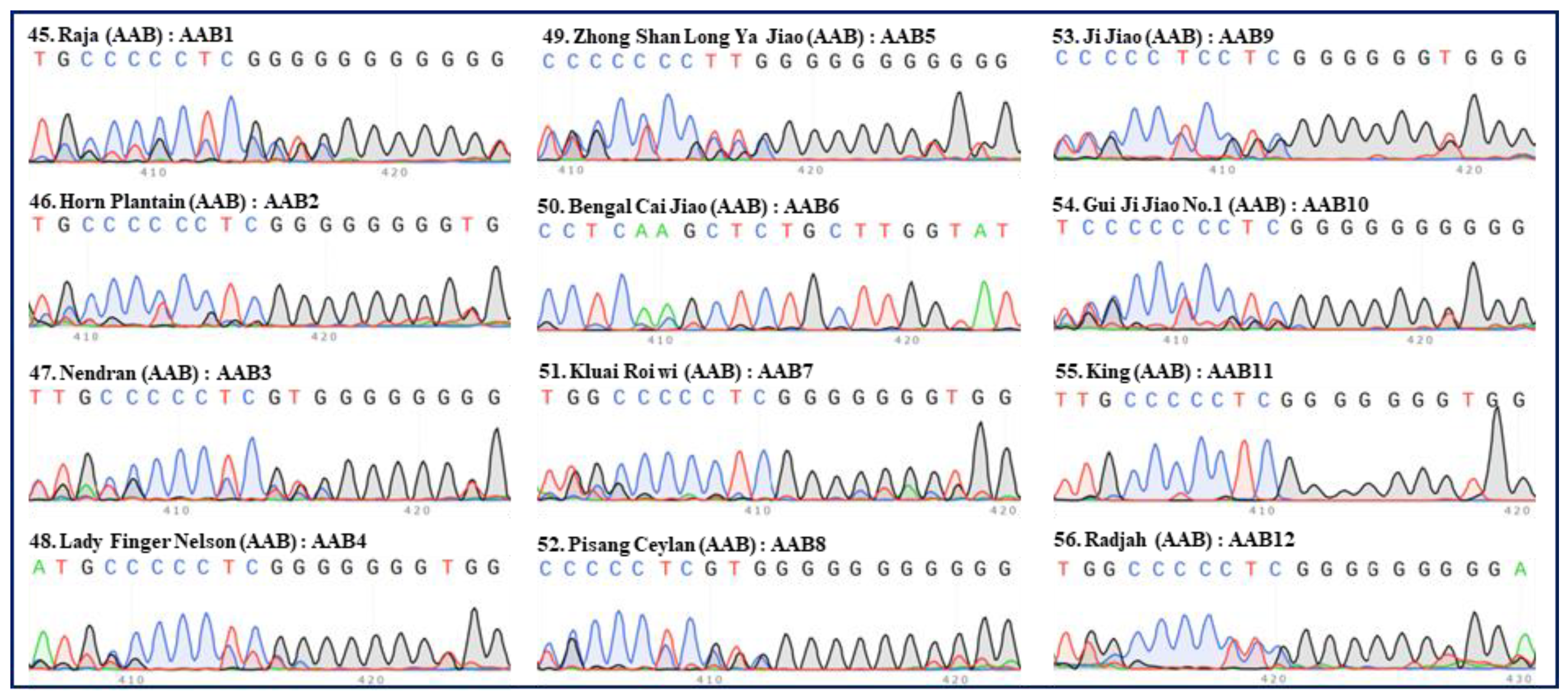
Figure 5.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of 12 accessions of AAB group banana. One accessions of Pisang Raja (‘Raja’), six accessions of Plantain (‘Horn Plantain’, and ‘Qi Tou Da Jiao’), one accessions of Pome (‘Lady Finger Nelson’) and one accessions of Silk (‘Zhong Shan Long Ya Jiao’) and six accessions of unknown subgroups (‘Bengal Cai Jiao’, ‘Kluai Roi wi’, ‘Pisang Ceylan’, ‘Ji Jiao’, ‘Gui Ji Jiao No.1’, ‘King’ and ‘Radjah’) were tested. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
Figure 5.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of 12 accessions of AAB group banana. One accessions of Pisang Raja (‘Raja’), six accessions of Plantain (‘Horn Plantain’, and ‘Qi Tou Da Jiao’), one accessions of Pome (‘Lady Finger Nelson’) and one accessions of Silk (‘Zhong Shan Long Ya Jiao’) and six accessions of unknown subgroups (‘Bengal Cai Jiao’, ‘Kluai Roi wi’, ‘Pisang Ceylan’, ‘Ji Jiao’, ‘Gui Ji Jiao No.1’, ‘King’ and ‘Radjah’) were tested. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
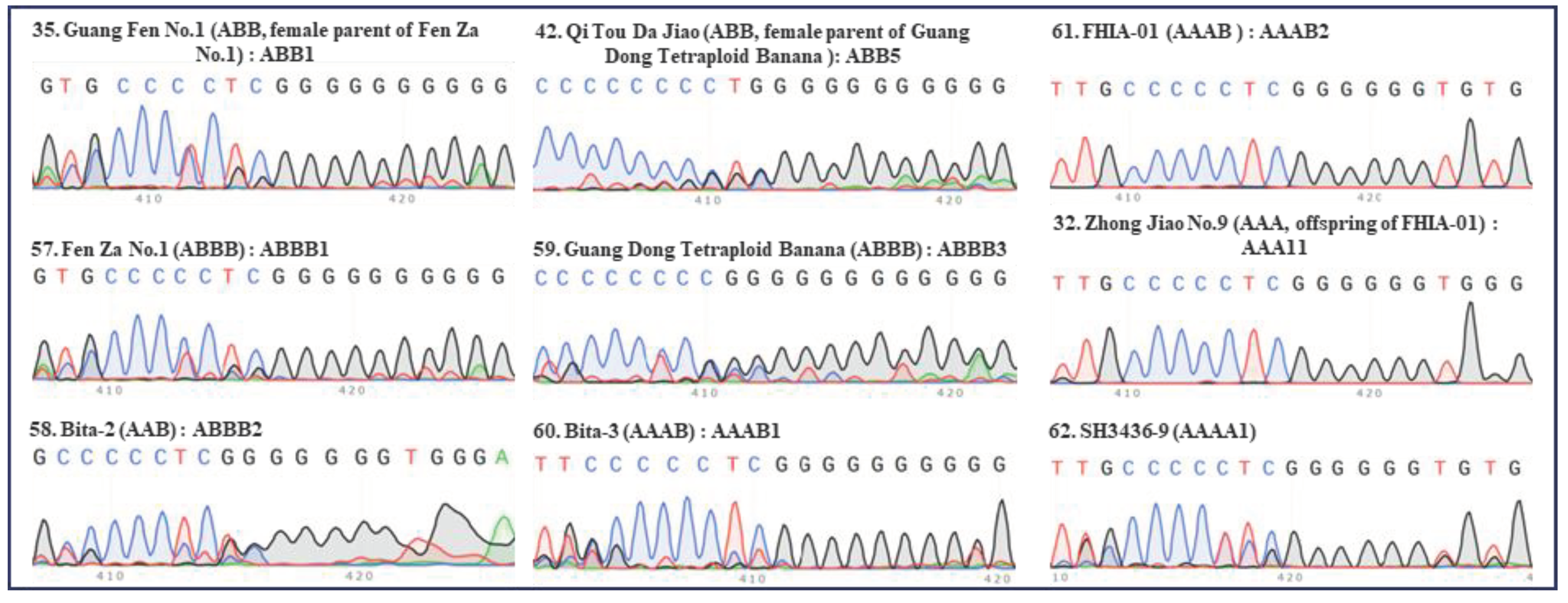
Figure 6.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of several accessions of tetraploid banana and their related cultivars. Seven accessions of tetraploid banana (‘Fen Za No.1’, ‘Bita-2’, ‘Guang Dong Tetraploid Banana’, ‘Bita-3’, ‘FHIA-01’, ‘Zhong Jiao No.9’, ‘SH3436-9’) were tested. Results of ‘Guang Fen No.1’ (female parent of ‘Fen Za No.1’), ‘Qi Tou Da Jiao’ (female parent of ‘Guang Dong Tetraploid Banana’), ‘Zhong Jiao No.9’ (offspring of ‘FHIA-01’) were shown for comparisons. Serial number, name, group, and ITS type of each accession was indicated in each panel. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
Figure 6.
Internal transcribed spacers (ITS) sequence peaks (420 bp region) of several accessions of tetraploid banana and their related cultivars. Seven accessions of tetraploid banana (‘Fen Za No.1’, ‘Bita-2’, ‘Guang Dong Tetraploid Banana’, ‘Bita-3’, ‘FHIA-01’, ‘Zhong Jiao No.9’, ‘SH3436-9’) were tested. Results of ‘Guang Fen No.1’ (female parent of ‘Fen Za No.1’), ‘Qi Tou Da Jiao’ (female parent of ‘Guang Dong Tetraploid Banana’), ‘Zhong Jiao No.9’ (offspring of ‘FHIA-01’) were shown for comparisons. Serial number, name, group, and ITS type of each accession was indicated in each panel. The serial number, name, group, and ITS type of each accession are indicated in each panel. Green, red, blue, and black peaks represent “A”, “T”, “C”, and “G”, respectively.
Table 1.
Information on the 62 accessions of banana.
Table 1.
Information on the 62 accessions of banana.
| Serial no. |
Names |
Groups |
Subgroups |
ITS types |
| 1 |
Inarnibal |
AA |
Inarnibal |
AA1 |
| 2 |
Re Gong No.1 |
AA |
Inarnibal |
AA1 |
| 3 |
Pisang Mas |
AA |
Sucrier |
AA1 |
| 4 |
Pisang Jari Buaya |
AA |
Pisang Jari Buaya |
AA2 |
| 5 |
Jia Li (mutant of Kluai lep mu nang) |
AA |
Unknown |
AA3 |
| 6 |
Morong Princesa |
AA |
Unknown |
AA4 |
| 7 |
Pamotion |
AA |
Unknown |
AA5 |
| 8 |
Rose |
AA |
Unknown |
AA6 |
| 9 |
Shou Zhi Jiao |
AA |
Unknown |
AA7 |
| 10 |
Tudlo Tumbaga |
AA |
Unknown |
AA8 |
| 11 |
Veinte cohol |
AA |
Unknown |
AA9 |
| 12 |
Ney Pooven |
AB |
Ney Pooven |
AB1 |
| 13 |
Brazil |
AAA |
Cavendish |
AAA1 |
| 14 |
Costa Rica |
AAA |
Cavendish |
AAA2 |
| 15 |
Da Feng No.1 |
AAA |
Cavendish |
AAA1 |
| 16 |
Drawf Williams |
AAA |
Cavendish |
AAA1 |
| 17 |
Formosana |
AAA |
Cavendish |
AAA1 |
| 18 |
Nan Tian Huang |
AAA |
Cavendish |
AAA1 |
| 19 |
Nong Ke No.1 |
AAA |
Cavendish |
AAA1 |
| 20 |
Pei Chiao |
AAA |
Cavendish |
AAA1 |
| 21 |
Philippines |
AAA |
Cavendish |
AAA1 |
| 22 |
Qi Wei |
AAA |
Cavendish |
AAA1 |
| 23 |
Williams |
AAA |
Cavendish |
AAA1 |
| 24 |
Xian Ba |
AAA |
Cavendish |
AAA1 |
| 25 |
Green Banana |
AAA |
Red |
AAA3 |
| 26 |
Gui Hong Jiao No.1 |
AAA |
Red |
AAA3 |
| 27 |
Hong Jiao Wang |
AAA |
Red |
AAA4 |
| 28 |
Red Banana |
AAA |
Red |
AAA5 |
| 29 |
Gros Michel |
AAA |
Gros Michel |
AAA6 |
| 30 |
Berangan |
AAA |
Lakatan |
AAA7 |
| 31 |
Yangambi KM5 |
AAA |
Ibota Bota |
AAA8 |
| 32 |
Zhong Jiao No.9 |
AAA |
FHIA-01×SH-3142 |
AAA9 |
| 33 |
Ai Fen No.1 |
ABB |
Pisang Awak |
ABB1 |
| 34 |
Bu Si Fen |
ABB |
Pisang Awak |
ABB1 |
| 35 |
Guang Fen No.1 |
ABB |
Pisang Awak |
ABB1 |
| 36 |
Jin Fen No.1 |
ABB |
Pisang Awak |
ABB1 |
| 37 |
Dong Guan Da Jiao |
ABB |
Da Jiao |
ABB2 |
| 38 |
Gui Da Jiao No.1 |
ABB |
Da Jiao |
ABB3 |
| 39 |
Hai Nan Niu Ba Jiao |
ABB |
Da Jiao |
ABB4 |
| 40 |
Lian Shan Ye Sheng Da Jiao |
ABB |
Da Jiao |
ABB3 |
| 41 |
Pan Yu Da Jiao |
ABB |
Da Jiao |
ABB4 |
| 42 |
Qi Tou Da Jiao |
ABB |
Da Jiao |
ABB5 |
| 43 |
Pelipita |
ABB |
Pelipita |
ABB6 |
| 44 |
Saba |
ABB |
Saba |
ABB7 |
| 45 |
Raja |
AAB |
Pisang Raja |
AAB1 |
| 46 |
Horn Plantain |
AAB |
Plantain |
AAB2 |
| 47 |
Nendran |
AAB |
Plantain |
AAB3 |
| 48 |
Lady Finger Nelson |
AAB |
Pome |
AAB4 |
| 49 |
Zhong Shan Long Ya Jiao |
AAB |
Silk |
AAB5 |
| 50 |
Bengal Cai Jiao |
AAB |
Unknown |
AAB6 |
| 51 |
Kluai Roi wi |
AAB |
Unknown |
AAB7 |
| 52 |
Pisang Ceylan |
AAB |
Unknown |
AAB8 |
| 53 |
Ji Jiao |
AAB |
Unknown |
AAB9 |
| 54 |
Gui Ji Jiao No.1 |
AAB |
Unknown |
AAB10 |
| 55 |
King |
AAB |
Unknown |
AAB11 |
| 56 |
Radjah |
AAB |
Unknown |
AAB12 |
| 57 |
Fen Za No.1 |
ABBB |
Guang Fen No.1×Musa Balbisiana |
ABBB1 |
| 58 |
Bita-2 |
ABBB |
Fougamou(AAB, Pisang Awak) x Musa Balbisiana 1-63 |
ABBB2 |
| 59 |
Guang Dong Tetraploid Banana |
ABBB |
Qi Tou Da Jiao×BB |
ABBB3 |
| 60 |
Bita-3 |
AAAB |
(AAAB, Laknau (AAB, Laknau) x Tjau Lagada(AA)) |
AAAB1 |
| 61 |
FHIA-01 |
AAAB |
Prata Ana×SH-3142 |
AAAB2 |
| 62 |
SH3436-9 |
AAAA |
Unknown |
AAAA1 |