Submitted:
22 February 2024
Posted:
23 February 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Tn5 transposon tagmentation of accessible chromatin (ATAC-seq)
Nicking enzyme assisted accessible chromatin sequencing (NicE-seq)
Competing Interests
Acknowledgements
Abbreviations
References
- Berson, A., Nativio, R., Berger, S.L., and Bonini, N.M. (2018). Epigenetic Regulation in Neurodegenerative Diseases. Trends Neurosci 41, 587-598. [CrossRef]
- Pal, S., and Tyler, J.K. (2016). Epigenetics and aging. Sci Adv 2, e1600584. [CrossRef]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403, 41-45. [CrossRef]
- Bhat, K.P., Umit Kaniskan, H., Jin, J., and Gozani, O. (2021). Epigenetics and beyond: targeting writers of protein lysine methylation to treat disease. Nat Rev Drug Discov 20, 265-286. [CrossRef]
- Isbel, L., Grand, R.S., and Schubeler, D. (2022). Generating specificity in genome regulation through transcription factor sensitivity to chromatin. Nat Rev Genet 23, 728-740. [CrossRef]
- Klemm, S.L., Shipony, Z., and Greenleaf, W.J. (2019). Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 20, 207-220. [CrossRef]
- Shirvaliloo, M. (2022). The landscape of histone modifications in epigenomics since 2020. Epigenomics 14, 1465-1477. [CrossRef]
- Luger, K., Rechsteiner, T.J., Flaus, A.J., Waye, M.M., and Richmond, T.J. (1997). Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol 272, 301-311. [CrossRef]
- Ioshikhes, I.P., Albert, I., Zanton, S.J., and Pugh, B.F. (2006). Nucleosome positions predicted through comparative genomics. Nat Genet 38, 1210-1215. [CrossRef]
- Mavrich, T.N., Jiang, C., Ioshikhes, I.P., Li, X., Venters, B.J., Zanton, S.J., Tomsho, L.P., Qi, J., Glaser, R.L., Schuster, S.C., et al. (2008). Nucleosome organization in the Drosophila genome. Nature 453, 358-362. [CrossRef]
- Schones, D.E., Cui, K., Cuddapah, S., Roh, T.Y., Barski, A., Wang, Z., Wei, G., and Zhao, K. (2008). Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887-898. [CrossRef]
- Kraushaar, D.C., Jin, W., Maunakea, A., Abraham, B., Ha, M., and Zhao, K. (2013). Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol 14, R121. [CrossRef]
- Li, S., Wei, T., and Panchenko, A.R. (2023). Histone variant H2A.Z modulates nucleosome dynamics to promote DNA accessibility. Nat Commun 14, 769. [CrossRef]
- Weiner, A., Hsieh, T.H., Appleboim, A., Chen, H.V., Rahat, A., Amit, I., Rando, O.J., and Friedman, N. (2015). High-resolution chromatin dynamics during a yeast stress response. Mol Cell 58, 371-386. [CrossRef]
- Consortium, E.P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57-74. [CrossRef]
- Consortium, E.P., Moore, J.E., Purcaro, M.J., Pratt, H.E., Epstein, C.B., Shoresh, N., Adrian, J., Kawli, T., Davis, C.A., Dobin, A., et al. (2020). Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699-710. [CrossRef]
- Shimizu, M., Roth, S.Y., Szent-Gyorgyi, C., and Simpson, R.T. (1991). Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J 10, 3033-3041. [CrossRef]
- Weintraub, H., and Groudine, M. (1976). Chromosomal subunits in active genes have an altered conformation. Science 193, 848-856. [CrossRef]
- Boyle, A.P., Davis, S., Shulha, H.P., Meltzer, P., Margulies, E.H., Weng, Z., Furey, T.S., and Crawford, G.E. (2008). High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311-322. [CrossRef]
- Simpson, R.T. (1998). Chromatin structure and analysis of mechanisms of activators and repressors. Methods 15, 283-294. [CrossRef]
- Vierstra, J., and Stamatoyannopoulos, J.A. (2016). Genomic footprinting. Nat Methods 13, 213-221. [CrossRef]
- Crawford, G.E., Holt, I.E., Whittle, J., Webb, B.D., Tai, D., Davis, S., Margulies, E.H., Chen, Y., Bernat, J.A., Ginsburg, D., et al. (2006). Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res 16, 123-131. [CrossRef]
- Albert, I., Mavrich, T.N., Tomsho, L.P., Qi, J., Zanton, S.J., Schuster, S.C., and Pugh, B.F. (2007). Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572-576. [CrossRef]
- Tsompana, M., and Buck, M.J. (2014). Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7, 33. [CrossRef]
- Klein, D.C., and Hainer, S.J. (2020). Genomic methods in profiling DNA accessibility and factor localization. Chromosome Res 28, 69-85. [CrossRef]
- Mansisidor, A.R., and Risca, V.I. (2022). Chromatin accessibility: methods, mechanisms, and biological insights. Nucleus 13, 236-276. [CrossRef]
- Waldron, L., Simpson, P., Parmigiani, G., and Huttenhower, C. (2012). Report on emerging technologies for translational bioinformatics: a symposium on gene expression profiling for archival tissues. BMC Cancer 12, 124. [CrossRef]
- Kokkat, T.J., Patel, M.S., McGarvey, D., LiVolsi, V.A., and Baloch, Z.W. (2013). Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank 11, 101-106. [CrossRef]
- Blum, F. (1894). Notiz über die Anwendung des Formaldehyds (Formol) als Härtungs-und Konservierungsmittel. Anat. Anz 9, 229-231.
- Donczo, B., and Guttman, A. (2018). Biomedical analysis of formalin-fixed, paraffin-embedded tissue samples: The Holy Grail for molecular diagnostics. J Pharm Biomed Anal 155, 125-134. [CrossRef]
- Steiert, T.A., Parra, G., Gut, M., Arnold, N., Trotta, J.R., Tonda, R., Moussy, A., Gerber, Z., Abuja, P.M., Zatloukal, K., et al. (2023). A critical spotlight on the paradigms of FFPE-DNA sequencing. Nucleic Acids Res 51, 7143-7162. [CrossRef]
- Guyard, A., Boyez, A., Pujals, A., Robe, C., Tran Van Nhieu, J., Allory, Y., Moroch, J., Georges, O., Fournet, J.C., Zafrani, E.S., and Leroy, K. (2017). DNA degrades during storage in formalin-fixed and paraffin-embedded tissue blocks. Virchows Arch 471, 491-500. [CrossRef]
- Nicieza, R.G., Huergo, J., Connolly, B.A., and Sanchez, J. (1999). Purification, characterization, and role of nucleases and serine proteases in Streptomyces differentiation. Analogies with the biochemical processes described in late steps of eukaryotic apoptosis. J Biol Chem 274, 20366-20375. [CrossRef]
- Koohy, H., Down, T.A., and Hubbard, T.J. (2013). Chromatin accessibility data sets show bias due to sequence specificity of the DNase I enzyme. PLoS One 8, e69853. [CrossRef]
- Nordstrom, K.J.V., Schmidt, F., Gasparoni, N., Salhab, A., Gasparoni, G., Kattler, K., Muller, F., Ebert, P., Costa, I.G., consortium, D., et al. (2019). Unique and assay specific features of NOMe-, ATAC- and DNase I-seq data. Nucleic Acids Res 47, 10580-10596. [CrossRef]
- Yardimci, G.G., Frank, C.L., Crawford, G.E., and Ohler, U. (2014). Explicit DNase sequence bias modeling enables high-resolution transcription factor footprint detection. Nucleic Acids Res 42, 11865-11878. [CrossRef]
- Gusmao, E.G., Allhoff, M., Zenke, M., and Costa, I.G. (2016). Analysis of computational footprinting methods for DNase sequencing experiments. Nat Methods 13, 303-309. [CrossRef]
- Martins, A.L., Walavalkar, N.M., Anderson, W.D., Zang, C., and Guertin, M.J. (2018). Universal correction of enzymatic sequence bias reveals molecular signatures of protein/DNA interactions. Nucleic Acids Res 46, e9. [CrossRef]
- Schmidt, F., Gasparoni, N., Gasparoni, G., Gianmoena, K., Cadenas, C., Polansky, J.K., Ebert, P., Nordstrom, K., Barann, M., Sinha, A., et al. (2017). Combining transcription factor binding affinities with open-chromatin data for accurate gene expression prediction. Nucleic Acids Res 45, 54-66. [CrossRef]
- Jin, W., Tang, Q., Wan, M., Cui, K., Zhang, Y., Ren, G., Ni, B., Sklar, J., Przytycka, T.M., Childs, R., et al. (2015). Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528, 142-146. [CrossRef]
- Cooper, J., Ding, Y., Song, J., and Zhao, K. (2017). Genome-wide mapping of DNase I hypersensitive sites in rare cell populations using single-cell DNase sequencing. Nat Protoc 12, 2342-2354. [CrossRef]
- Axel, R. (1975). Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry 14, 2921-2925. [CrossRef]
- Valouev, A., Ichikawa, J., Tonthat, T., Stuart, J., Ranade, S., Peckham, H., Zeng, K., Malek, J.A., Costa, G., McKernan, K., et al. (2008). A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res 18, 1051-1063. [CrossRef]
- Li, Z., Schug, J., Tuteja, G., White, P., and Kaestner, K.H. (2011). The nucleosome map of the mammalian liver. Nat Struct Mol Biol 18, 742-746. [CrossRef]
- Gutierrez, G., Millan-Zambrano, G., Medina, D.A., Jordan-Pla, A., Perez-Ortin, J.E., Penate, X., and Chavez, S. (2017). Subtracting the sequence bias from partially digested MNase-seq data reveals a general contribution of TFIIS to nucleosome positioning. Epigenetics Chromatin 10, 58. [CrossRef]
- Mieczkowski, J., Cook, A., Bowman, S.K., Mueller, B., Alver, B.H., Kundu, S., Deaton, A.M., Urban, J.A., Larschan, E., Park, P.J., et al. (2016). MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat Commun 7, 11485. [CrossRef]
- Henikoff, J.G., Belsky, J.A., Krassovsky, K., MacAlpine, D.M., and Henikoff, S. (2011). Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci U S A 108, 18318-18323. [CrossRef]
- Chereji, R.V., Bryson, T.D., and Henikoff, S. (2019). Quantitative MNase-seq accurately maps nucleosome occupancy levels. Genome Biol 20, 198. [CrossRef]
- Nagy, P.L., Cleary, M.L., Brown, P.O., and Lieb, J.D. (2003). Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci U S A 100, 6364-6369. [CrossRef]
- Giresi, P.G., Kim, J., McDaniell, R.M., Iyer, V.R., and Lieb, J.D. (2007). FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17, 877-885. [CrossRef]
- Simon, J.M., Giresi, P.G., Davis, I.J., and Lieb, J.D. (2012). Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc 7, 256-267. [CrossRef]
- Nagy, P.L., and Price, D.H. (2009). Formaldehyde-assisted isolation of regulatory elements. Wiley Interdiscip Rev Syst Biol Med 1, 400-406. [CrossRef]
- Zhou, W., Ji, Z., Fang, W., and Ji, H. (2019). Global prediction of chromatin accessibility using small-cell-number and single-cell RNA-seq. Nucleic Acids Res 47, e121. [CrossRef]
- Giresi, P.G., and Lieb, J.D. (2009). Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements). Methods 48, 233-239. [CrossRef]
- Gaulton, K.J., Nammo, T., Pasquali, L., Simon, J.M., Giresi, P.G., Fogarty, M.P., Panhuis, T.M., Mieczkowski, P., Secchi, A., Bosco, D., et al. (2010). A map of open chromatin in human pancreatic islets. Nat Genet 42, 255-259. [CrossRef]
- Buchert, E.M., Fogarty, E.A., Uyehara, C.M., McKay, D.J., and Buttitta, L.A. (2023). A tissue dissociation method for ATAC-seq and CUT&RUN in Drosophila pupal tissues. Fly (Austin) 17, 2209481. [CrossRef]
- Berg, D.E. (2017). Julian Davies and the discovery of kanamycin resistance transposon Tn5. J Antibiot (Tokyo) 70, 339-346. [CrossRef]
- Reznikoff, W.S. (2003). Tn5 as a model for understanding DNA transposition. Mol Microbiol 47, 1199-1206. [CrossRef]
- Li, N., Jin, K., Bai, Y., Fu, H., Liu, L., and Liu, B. (2020). Tn5 Transposase Applied in Genomics Research. Int J Mol Sci 21. [CrossRef]
- Buenrostro, J.D., Giresi, P.G., Zaba, L.C., Chang, H.Y., and Greenleaf, W.J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213-1218. [CrossRef]
- Kaya-Okur, H.S., Wu, S.J., Codomo, C.A., Pledger, E.S., Bryson, T.D., Henikoff, J.G., Ahmad, K., and Henikoff, S. (2019). CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10, 1930. [CrossRef]
- Buenrostro, J.D., Wu, B., Chang, H.Y., and Greenleaf, W.J. (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21-21 29 29. [CrossRef]
- Wolpe, J.B., Martins, A.L., and Guertin, M.J. (2023). Correction of transposase sequence bias in ATAC-seq data with rule ensemble modeling. NAR Genom Bioinform 5, lqad054. [CrossRef]
- Li, Z., Schulz, M.H., Look, T., Begemann, M., Zenke, M., and Costa, I.G. (2019). Identification of transcription factor binding sites using ATAC-seq. Genome Biol 20, 45. [CrossRef]
- Corces, M.R., Trevino, A.E., Hamilton, E.G., Greenside, P.G., Sinnott-Armstrong, N.A., Vesuna, S., Satpathy, A.T., Rubin, A.J., Montine, K.S., Wu, B., et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959-962. [CrossRef]
- Grandi, F.C., Modi, H., Kampman, L., and Corces, M.R. (2022). Chromatin accessibility profiling by ATAC-seq. Nat Protoc 17, 1518-1552. [CrossRef]
- Henikoff, S., Henikoff, J.G., Ahmad, K., Paranal, R.M., Janssens, D.H., Russell, Z.R., Szulzewsky, F., Kugel, S., and Holland, E.C. (2023). Epigenomic analysis of formalin-fixed paraffin-embedded samples by CUT&Tag. Nat Commun 14, 5930. [CrossRef]
- Yadav, R.P., Polavarapu, V.K., Xing, P., and Chen, X. (2022). FFPE-ATAC: A Highly Sensitive Method for Profiling Chromatin Accessibility in Formalin-Fixed Paraffin-Embedded Samples. Curr Protoc 2, e535. [CrossRef]
- Zhang, H., Polavarapu, V.K., Xing, P., Zhao, M., Mathot, L., Zhao, L., Rosen, G., Swartling, F.J., Sjoblom, T., and Chen, X. (2022). Profiling chromatin accessibility in formalin-fixed paraffin-embedded samples. Genome Res 32, 150-161. [CrossRef]
- Zhao, L., Polavarapu, V.K., Yadav, R.P., Xing, P., and Chen, X. (2022). A Highly Sensitive Method to Efficiently Profile the Histone Modifications of FFPE Samples. Bio Protoc 12. [CrossRef]
- Zhao, L., Xing, P., Polavarapu, V.K., Zhao, M., Valero-Martinez, B., Dang, Y., Maturi, N., Mathot, L., Neves, I., Yildirim, I., et al. (2021). FACT-seq: profiling histone modifications in formalin-fixed paraffin-embedded samples with low cell numbers. Nucleic Acids Res 49, e125. [CrossRef]
- Amatori, S., and Fanelli, M. (2022). The Current State of Chromatin Immunoprecipitation (ChIP) from FFPE Tissues. Int J Mol Sci 23. [CrossRef]
- Oba, U., Kohashi, K., Sangatsuda, Y., Oda, Y., Sonoda, K.H., Ohga, S., Yoshimoto, K., Arai, Y., Yachida, S., Shibata, T., et al. (2022). An efficient procedure for the recovery of DNA from formalin-fixed paraffin-embedded tissue sections. Biol Methods Protoc 7, bpac014. [CrossRef]
- Ponnaluri, V.K.C., Zhang, G., Esteve, P.O., Spracklin, G., Sian, S., Xu, S.Y., Benoukraf, T., and Pradhan, S. (2017). NicE-seq: high resolution open chromatin profiling. Genome Biol 18, 122. [CrossRef]
- Chin, H.G., Sun, Z., Vishnu, U.S., Hao, P., Cejas, P., Spracklin, G., Esteve, P.O., Xu, S.Y., Long, H.W., and Pradhan, S. (2020). Universal NicE-seq for high-resolution accessible chromatin profiling for formaldehyde-fixed and FFPE tissues. Clin Epigenetics 12, 143. [CrossRef]
- Esteve, P.O., Vishnu, U.S., Chin, H.G., and Pradhan, S. (2020). Visualization and Sequencing of Accessible Chromatin Reveals Cell Cycle and Post-HDAC inhibitor Treatment Dynamics. J Mol Biol 432, 5304-5321. [CrossRef]
- Vishnu, U.S., Esteve, P.O., Chin, H.G., and Pradhan, S. (2021). One-pot universal NicE-seq: all enzymatic downstream processing of 4% formaldehyde crosslinked cells for chromatin accessibility genomics. Epigenetics Chromatin 14, 53. [CrossRef]
- Chan, S.H., Zhu, Z., Dunigan, D.D., Van Etten, J.L., and Xu, S.Y. (2006). Cloning of Nt.CviQII nicking endonuclease and its cognate methyltransferase: M.CviQII methylates AG sequences. Protein Expr Purif 49, 138-150. [CrossRef]
- Pranzatelli, T.J.F., Michael, D.G., and Chiorini, J.A. (2018). ATAC2GRN: optimized ATAC-seq and DNase1-seq pipelines for rapid and accurate genome regulatory network inference. BMC Genomics 19, 563. [CrossRef]
- Smith, J.P., and Sheffield, N.C. (2020). Analytical Approaches for ATAC-seq Data Analysis. Curr Protoc Hum Genet 106, e101. [CrossRef]
- Yan, F., Powell, D.R., Curtis, D.J., and Wong, N.C. (2020). From reads to insight: a hitchhiker's guide to ATAC-seq data analysis. Genome Biol 21, 22. [CrossRef]
- Bolger, A.M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. [CrossRef]
- Langmead, B., and Salzberg, S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357-359. [CrossRef]
- Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., Durbin, R., and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078-2079. [CrossRef]
- Quinlan, A.R. (2014). BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics 47, 11 12 11-34. [CrossRef]
- Quinlan, A.R., and Hall, I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. [CrossRef]
- Amemiya, H.M., Kundaje, A., and Boyle, A.P. (2019). The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci Rep 9, 9354. [CrossRef]
- Liu, T. (2014). Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol 1150, 81-95. [CrossRef]
- Ramirez, F., Ryan, D.P., Gruning, B., Bhardwaj, V., Kilpert, F., Richter, A.S., Heyne, S., Dundar, F., and Manke, T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160-165. [CrossRef]
- Robinson, J.T., Thorvaldsdottir, H., Turner, D., and Mesirov, J.P. (2023). igv.js: an embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). Bioinformatics 39. [CrossRef]
- Robinson, M.D., McCarthy, D.J., and Smyth, G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. [CrossRef]
- Talebi, A., Thiery, J.P., and Kerachian, M.A. (2021). Fusion transcript discovery using RNA sequencing in formalin-fixed paraffin-embedded specimen. Crit Rev Oncol Hematol 160, 103303. [CrossRef]
- Siegel, E.M., Berglund, A.E., Riggs, B.M., Eschrich, S.A., Putney, R.M., Ajidahun, A.O., Coppola, D., and Shibata, D. (2014). Expanding epigenomics to archived FFPE tissues: an evaluation of DNA repair methodologies. Cancer Epidemiol Biomarkers Prev 23, 2622-2631. [CrossRef]
- Cejas, P., Li, L., O'Neill, N.K., Duarte, M., Rao, P., Bowden, M., Zhou, C.W., Mendiola, M., Burgos, E., Feliu, J., et al. (2016). Chromatin immunoprecipitation from fixed clinical tissues reveals tumor-specific enhancer profiles. Nat Med 22, 685-691. [CrossRef]
- DeTure, M.A., and Dickson, D.W. (2019). The neuropathological diagnosis of Alzheimer's disease. Mol Neurodegener 14, 32. [CrossRef]
- Willroider, M., Roeber, S., Horn, A.K.E., Arzberger, T., Scheifele, M., Respondek, G., Sabri, O., Barthel, H., Patt, M., Mishchenko, O., et al. (2021). Superiority of Formalin-Fixed Paraffin-Embedded Brain Tissue for in vitro Assessment of Progressive Supranuclear Palsy Tau Pathology With [(18)F]PI-2620. Front Neurol 12, 684523. [CrossRef]
- Clark, S.J., Statham, A., Stirzaker, C., Molloy, P.L., and Frommer, M. (2006). DNA methylation: bisulphite modification and analysis. Nat Protoc 1, 2353-2364. [CrossRef]
- Vaisvila, R., Ponnaluri, V.K.C., Sun, Z., Langhorst, B.W., Saleh, L., Guan, S., Dai, N., Campbell, M.A., Sexton, B.S., Marks, K., et al. (2021). Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA. Genome Res 31, 1280-1289. [CrossRef]
- Skene, P.J., and Henikoff, S. (2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6. [CrossRef]
- Jiang, S., and Mortazavi, A. (2018). Integrating ChIP-seq with other functional genomics data. Brief Funct Genomics 17, 104-115. [CrossRef]
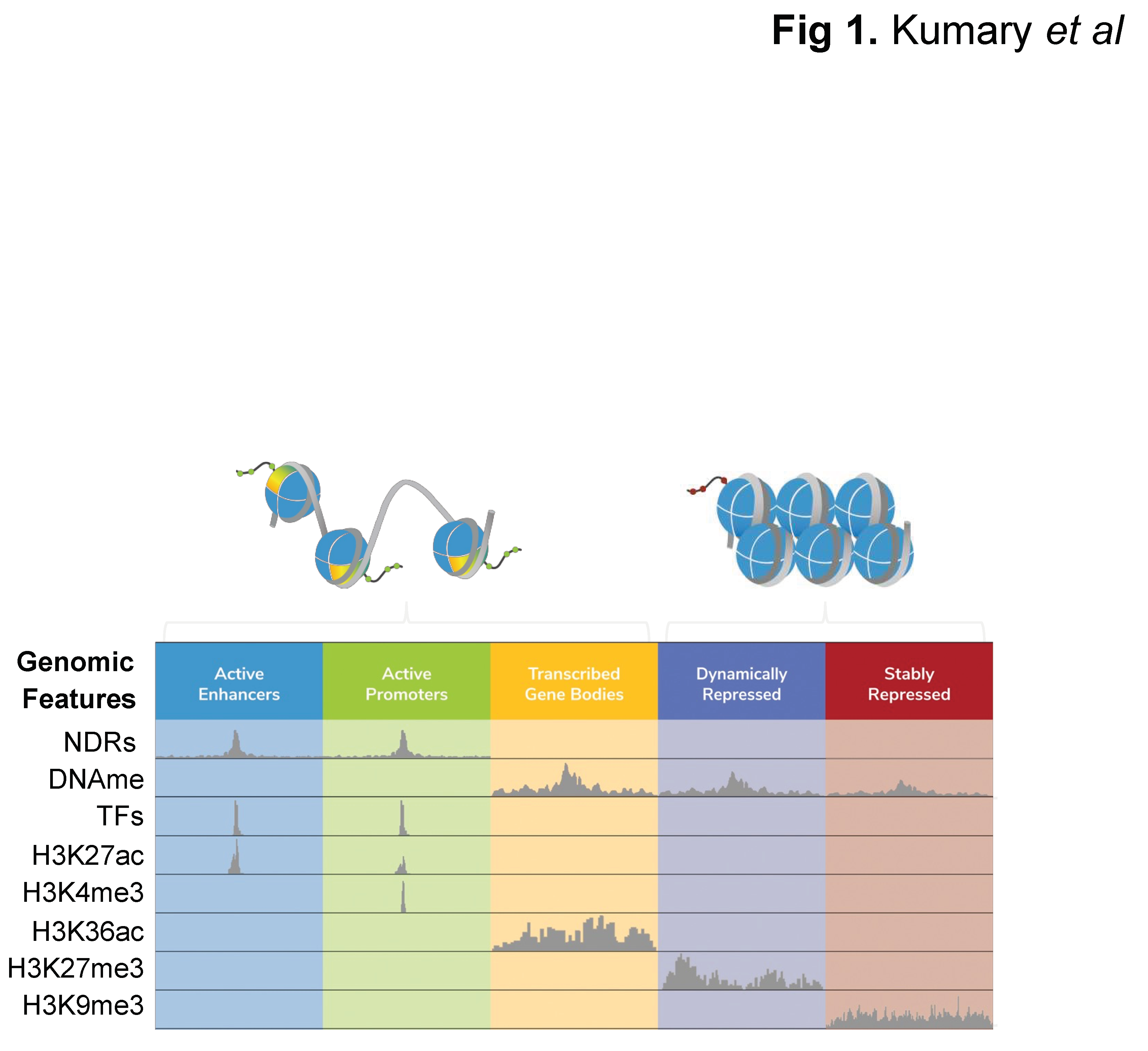
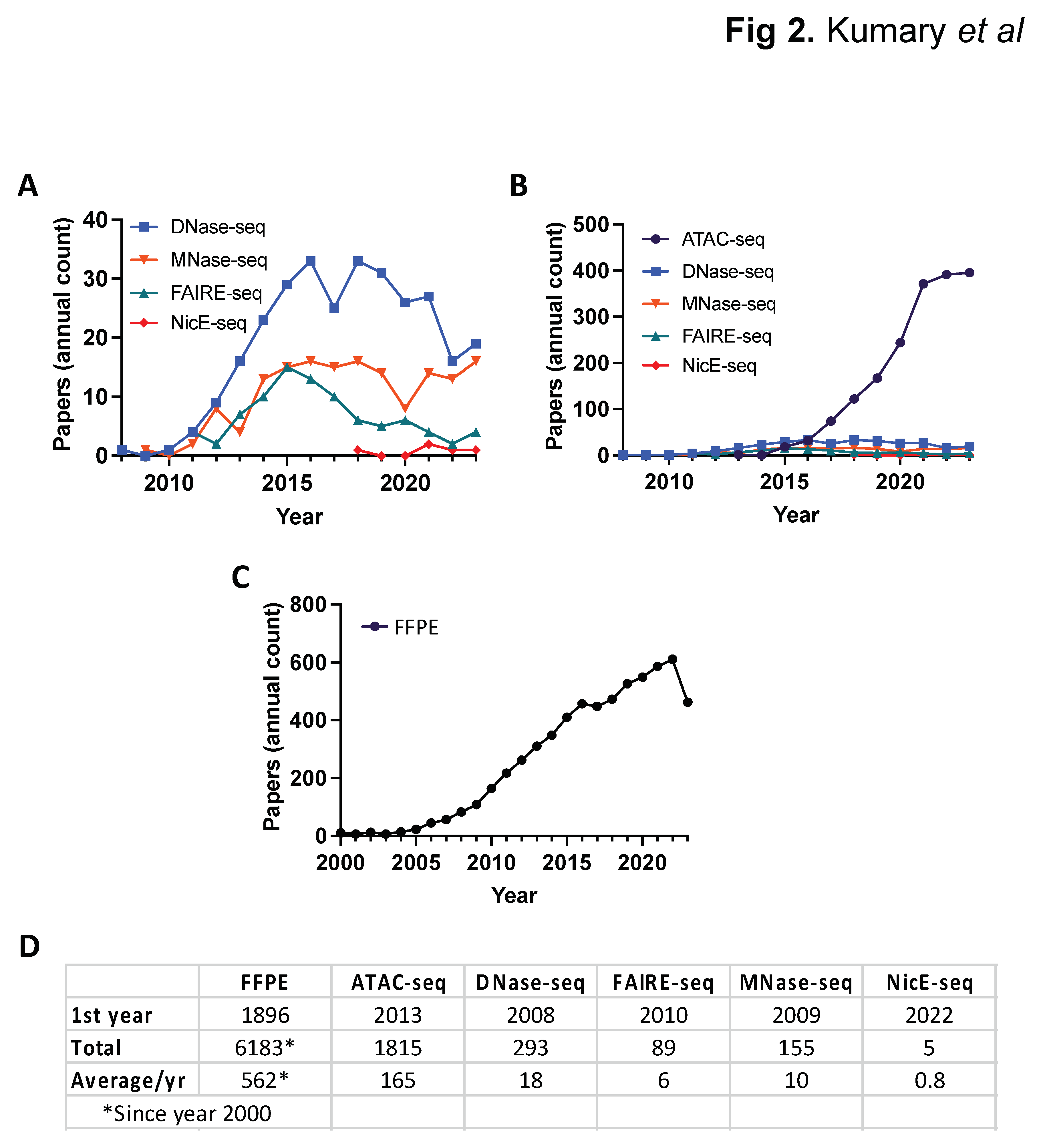
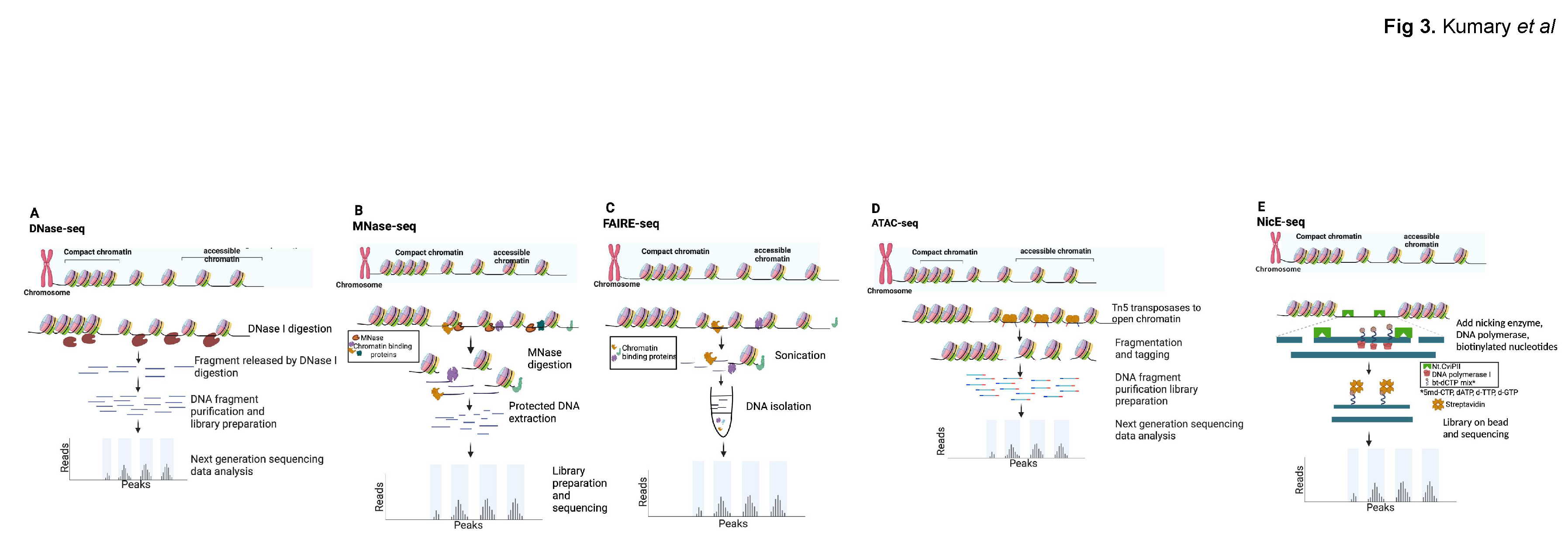
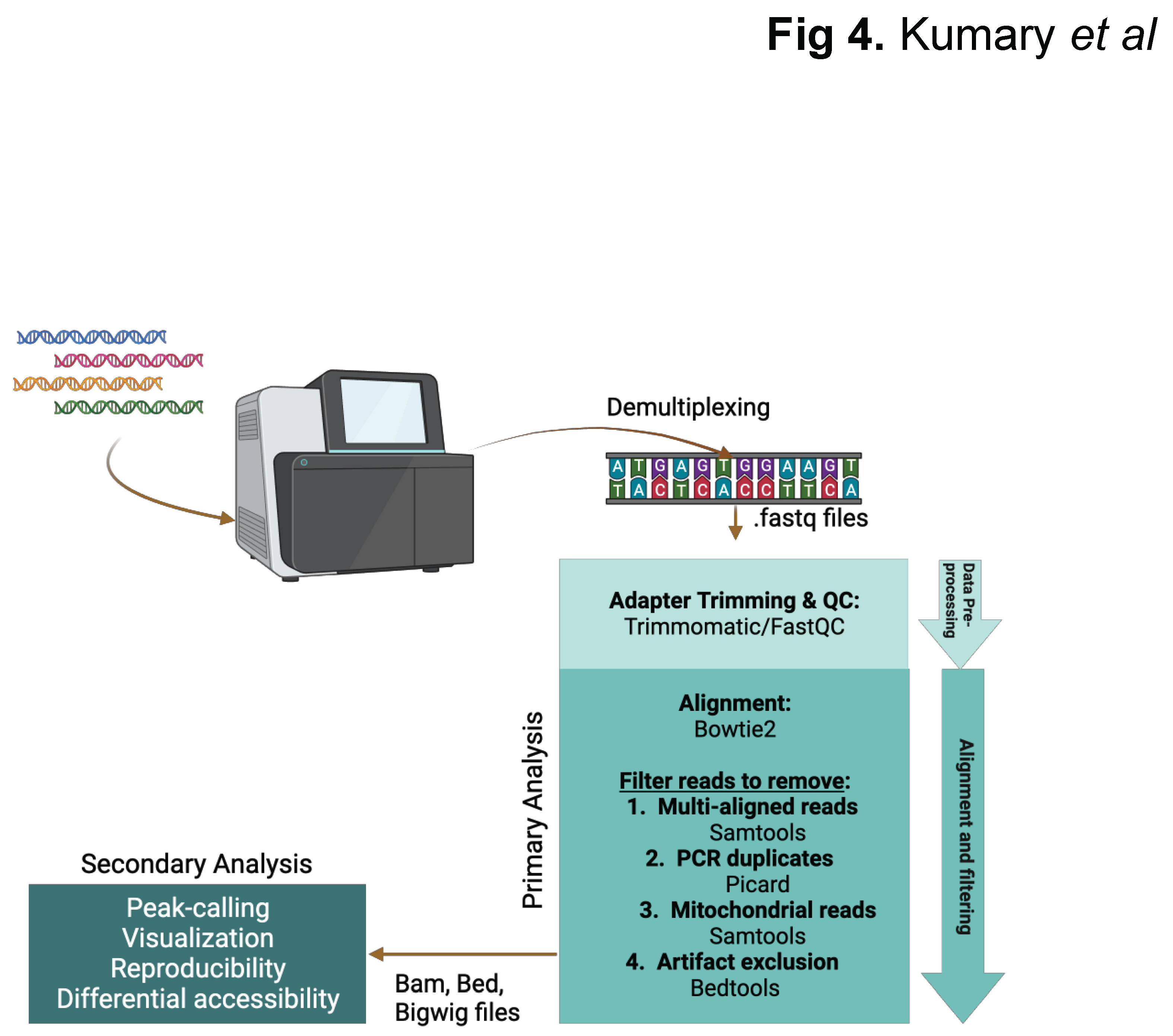
| DNase-seq | MNase-seq | FAIRE-seq | ATAC-seq | NicE-seq | |
|---|---|---|---|---|---|
| Type of input cells/tissue | Fresh/Formaldehyde cross-linked/FFPE (Formalin Fixed Paraffin Embedded) | Fresh/formaldehyde cross-linked | Formaldehyde cross-linked | Fresh/formaldehyde cross-linked (less efficient in fixed) | Formaldehyde cross-linked/FFPE |
| Application to FFPE (PubMed) | 1 | 0 | 1 | 2 | 2 |
| Number of input cells | 1M-10M | 10K-10M | 10K-10M | 1 cell – 50k | 25 cells – 100k |
| Approach | DNase I (endonuclease) cuts unprotected DNA | MNase (endo-exonuclease) digests unprotected DNA | Sonicate unprotected DNA in crosslinked material | Tn5 transposase tagments open region with DNA adapters | Nt-CviPII nickase cuts/labels CCD sites in unprotected DNA |
| Sequencing type | Single/Paired End | Single/Paired End | Single/Paired End | Single/Paired End | Single/Paired End |
| Target region | NDR | Linker DNA between Nucleosomes | NDR | NDR | NDR |
| Sequencing depth (human genome; ~3B bp) | 20-50 M mapped reads | 150-200 M mapped reads | 20-50 M mapped reads | 25-30 million (M) mapped non-mitochondrial (mito) reads | 20-30 M mapped reads |
| Cleavage bias | Yes | Yes | No | Yes | Yes |
|
Advantages / Disadvantages |
No prior knowledge of the sequence or binding protein is required / Time consuming. Requires laborious enzyme titrations and calibrations. Requires high sequencing depth. |
Nucleosome positioning can be inferred / Requires laborious enzyme titrations and calibrations. Requires high sequencing depth. Indirect profiling of open regions. |
No enzymes optimization or titration required / Low signal-to-noise. Relatively complex computational data analysis and interpretation. Results are highly fixation dependent. |
Simple, fast and sensitive approach. High signal-to-noise. / High mito DNA counts (unless nuclei isolated). Requires two independent tagmentation events in opposite orientation. Tn5 sequence bias and promoter-enrichment bias. |
Simple enzymatic approach. 5% mito DNA counts. Optimal in fixed or FFPE samples. Can be used in clinical settings. Efficiently profiles promoters and enhancers. / AT-rich sequences may be underrepresented. |
| References | [19] | [9–11,43,44] | [50] | [60,62] | [74–77] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





