1. Introduction
Infections with antibiotic-resistant (ABR) bacteria increase mortality, morbidity, social and economic consequences[
1]. Besides from the discovery of novel antimicrobial medications, existing antimicrobials must be utilized correctly because they promote the evolution of resistance strains through positive selection pressure[
2]. This is raising antimicrobial resistance (AMR) not only in humans but animals and the environment also, which makes it a global rather than a local issue, as AMR can spread between countries or continents. The improper use of antibiotics by people, firms, and farms, inadequate hygiene and sanitation, and ineffective infection prevention and control in healthcare settings are all considered major contributors to ABR bacteria formation and spread[
3]. Understanding the exact burden of resistance is a critical difficulty in combating AMR, especially in areas with limited surveillance and data. The primary causes of drug-resistant infections are assumed to include misuse, self-medication, and illogical antimicrobial use.
Growing evidence suggests that the widespread use of antibiotics in agriculture and aquaculture may be a factor in the emergence of resistance to antibiotics frequently used in human treatment; this is primarily a major issue considering the overlap between the antibiotics used for these many uses[
1]. ABR bacteria, which may have resistance genes, can, for instance, emerge in bacteria treated with antibiotics in animals. These bacteria can subsequently be passed from animals to humans. Food, direct contact between humans and livestock, or common environmental sources like sewage water can all be sources of this interspecies transmission[
4].
Antimicrobial resistance is predicted to be responsible for 10 million deaths annually by 2050, with a total monetary cost of US
$100 trillion.[
5] Although others have criticized these assumptions[
6]. AMR bacteria caused more than 670,000 illnesses in European Union (EU) and European Economic Area (EEA) nations in 2015, resulting in an estimated 33,000 mortality[
7]. World health organization (WHO) and numerous other organizations and researchers concur that the development of AMR is an important issue that requires a global, coordinated action plan to address.[
8,
9,
10] A lack of knowledge and inadequately trained professionals may all be linked to the prevalent resistance in these areas. Despite this knowledge gap, many nations, especially those in the EU, have made significant efforts to reduce the overall use of antibiotics in animals raised for food.
To address this issue, policymakers are focused on minimizing antibiotic use by expanding infection control information, adopting and implementing hospital infection control policies, and developing active hospital infection control teams in each hospital to monitor and contain illness transmission[
11]. The WHO plays an important role in monitoring antibiotic use and providing the necessary data to combat AMR. The WHO, the Food and Agriculture Organization (FAO), and the World Organization for Animal Health (OIE) have formed a tripartite partnership to coordinate global and national action plans (NAPs) to combat AMR. All the countries, including India and Germany, are now implementing AMR NAPs through multisectoral collaboration to enable comprehensive surveillance, monitoring, and policy implementation across human, animal, and environmental domains[
12].
It is essential to understand that AMR is a complex issue that necessitates a multifaceted strategy for control. The One Health concept emphasizes the connections between human health, animal health, food safety, and the environment and encourages cooperation between the health agencies responsible for these areas[
13]. In this systematic meta-analysis, we represented the Antimicrobial resistance burden in India and Germany in 2022 in three compartments: human, animal and environment. The findings of this analysis suggest research gaps for additional exploration as well as potential initiatives to minimize AMR in India and Germany along with One Health perspective.
2. Materials and Methods
2.1. Study methodology
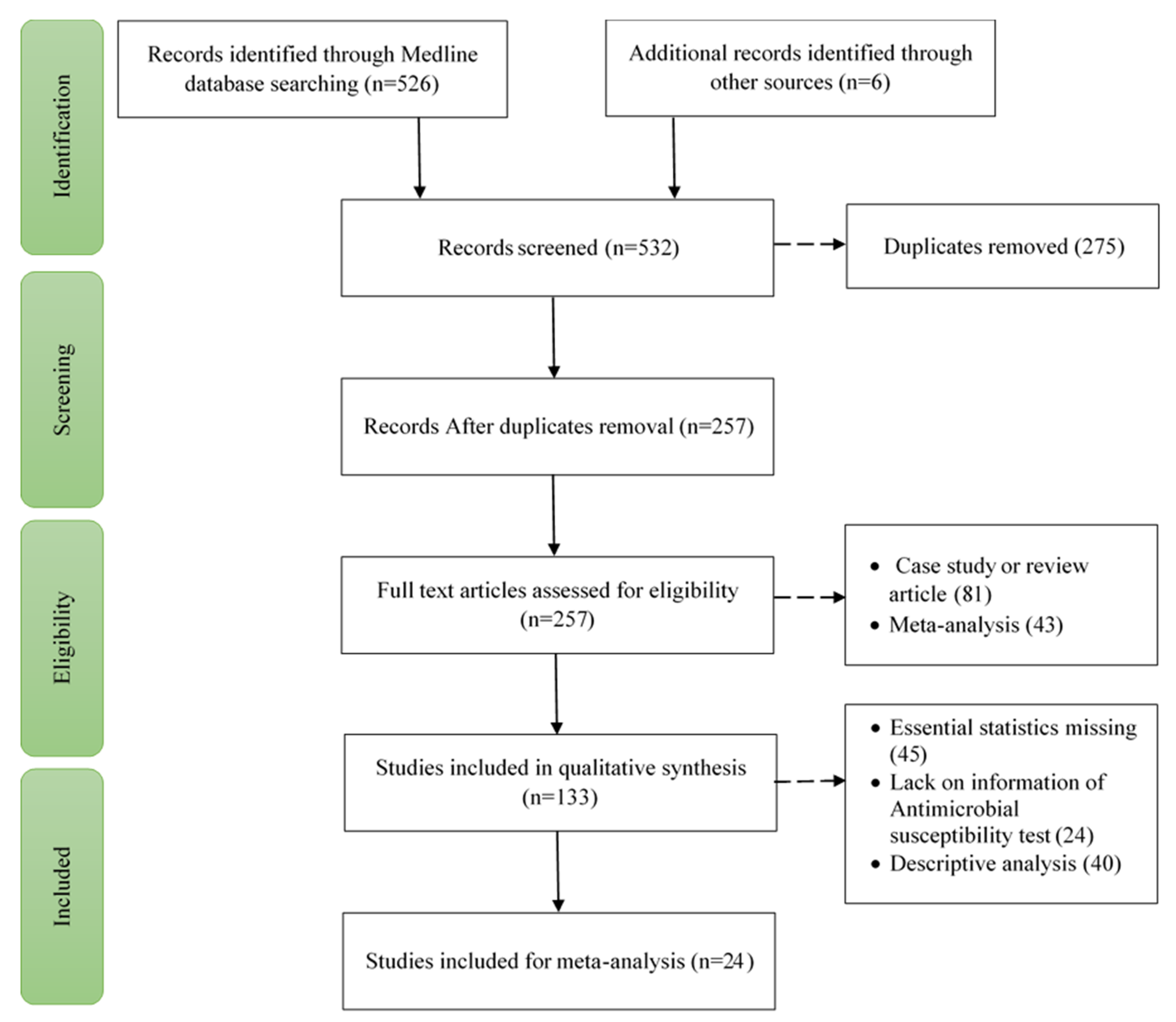
A systematic review was carried out in accordance with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analysis) guidelines[
14]. All 24 checklist literature were addressed in the study, is depicted in figure 1.
2.2. Search strategy
We conducted a comprehensive, systematic search of web databases for relevant literature, including PubMed (
https://pubmed.ncbi.nlm.nih.gov/), Google Scholar (
https://scholar.google.com/), and Science Direct (
https://www.sciencedirect.com/), published in the year of To search the articles, MeSH terms and Boolean Logic tools with the connectors ’AND’ and ’OR’ were used, including (antimicrobial resistance OR antibiotic resistance) AND (human OR animal OR environment), AND
(E. coli OR
Escherichia coli OR
Salmonella OR
Mycobacterium avium subsp. Paratuberculosis OR
Citrobacter OR
Klebsiella OR
Clostridium difficile OR
Proteus OR
Staphylococcus OR
Bacilli OR
Morganella OR
Chlamydia spp. OR
Trichinella spp. OR
Listeria spp. OR
Vibrio spp. OR
Aeromonas spp.) AND (dairy OR meat OR beef OR pig OR pork OR chicken OR fish OR shrimp OR octopus OR lobster OR marine mammals OR vulture OR ostrich OR camel) AND (environment OR soil OR wastewater OR water OR drainage) AND (India OR Germany). For the study to be considered in our analysis, the available abstract must be written in English.
2.3. Selection Criteria
Title, abstract, and full text were the three criteria for evaluating the articles. The prevalence of AMR in various microorganisms and sample collection must be discussed in the articles was from (1) humans (healthy/diseased people in hospitals); (2) animals) animals (both terrestrial and aquatic); animal food items (meat, seafood and dairy products); and (3) the environment (water, drainage, soil). Furthermore, all investigations must follow the Clinical and Laboratory Standards Institute (CLSI) criteria for antimicrobial susceptibility testing (AST).
2.4. Data extraction and Quality assessment
The following information was extracted and recorded in a spreadsheet (Microsoft Excel® 2013) after reviewing the data from each retrieved publication: (a) locality of India and Germany (East/West/North/South/Central) (b) year of publication (2022) (c) host (humans/ animals/ environment/ integrated studies between animals, humans and environment); (d) sample type (fecal sludge/ water/ retail/ veterinary clinics/ ice/ nasopharyngeal swab/ stool/faeces/ rectal swab/ urine/ vaginal swab/ other: abscess, appendix, small intestines/ colon/ stomach, gall bladder/ liver, peritoneal fluid/ placenta/ tissue) (e) microbial species (
E. coli,
Escherichia coli,
Salmonella,
Mycobacterium avium subsp. Paratuberculosis,
Citrobacter,
Klebsiella,
Clostridium difficile,
Proteus,
Staphylococcus,
Bacilli,
Morganella,
Chlamydia spp.,
Trichinella spp.,
Listeria spp.,
Vibrio spp.,
Aeromonas spp.) (f) the number of isolates (g) animal species (dairy, meat, beef, pig, pork, chicken, fish, shrimp, octopus, lobster, marine mammals, vulture, ostrich and camel) (h) laboratory methods for AST (MIC or disk diffusion); (i) WHO categorized the prevalence of ABR bacteria, antimicrobial drugs, and antimicrobial classes (highest priority critically important/high priority critically important/highly important/important)[
14]. (j) The proportion of multi-drug resistance (MDR) isolates resistant to at least one antimicrobial agent from more than three antimicrobial classes. The results were evaluated, and a pie chart was created using Mendeley to display the AMR data and the mentioned literature (version 1.19.8).
3. Result
3.1. Study selection
We categorized 526 papers published in 2022 using three internet databases. After a preliminary evaluation of the title and abstract, 275 articles from these were excluded due to their relevancy and redundancy; however, the complete texts of the remaining 257 articles were examined. The following factors led to the exclusion of 124 of the 257 articles: 51 due to case study or review articles and 43 due to meta-analysis articles. 109 of these 133 full-text items were again excluded. 45 due to the absence of essential statistics, 24 due to lack of information on AST and 40 due to descriptive analysis. Finally, 24 studies total were incorporated into this meta-analysis and systematic review (Supplementary Data 1). The method of study selection follows the PRISMA flow diagram and is illustrated in
Figure 1.
3.2. Qualitative synthesis of selected studies
24 articles were chosen for quantitative study published in the year of Out of this 14 [
15,
16,
17,
18,
19,
20,
21,
22,
23,
24,
25,
26,
27] studies were from India and 10 [
2,
28,
29,
30,
31,
32,
33,
34,
35] study were from Germany.
3.2.1. AMR studies in different region of countries
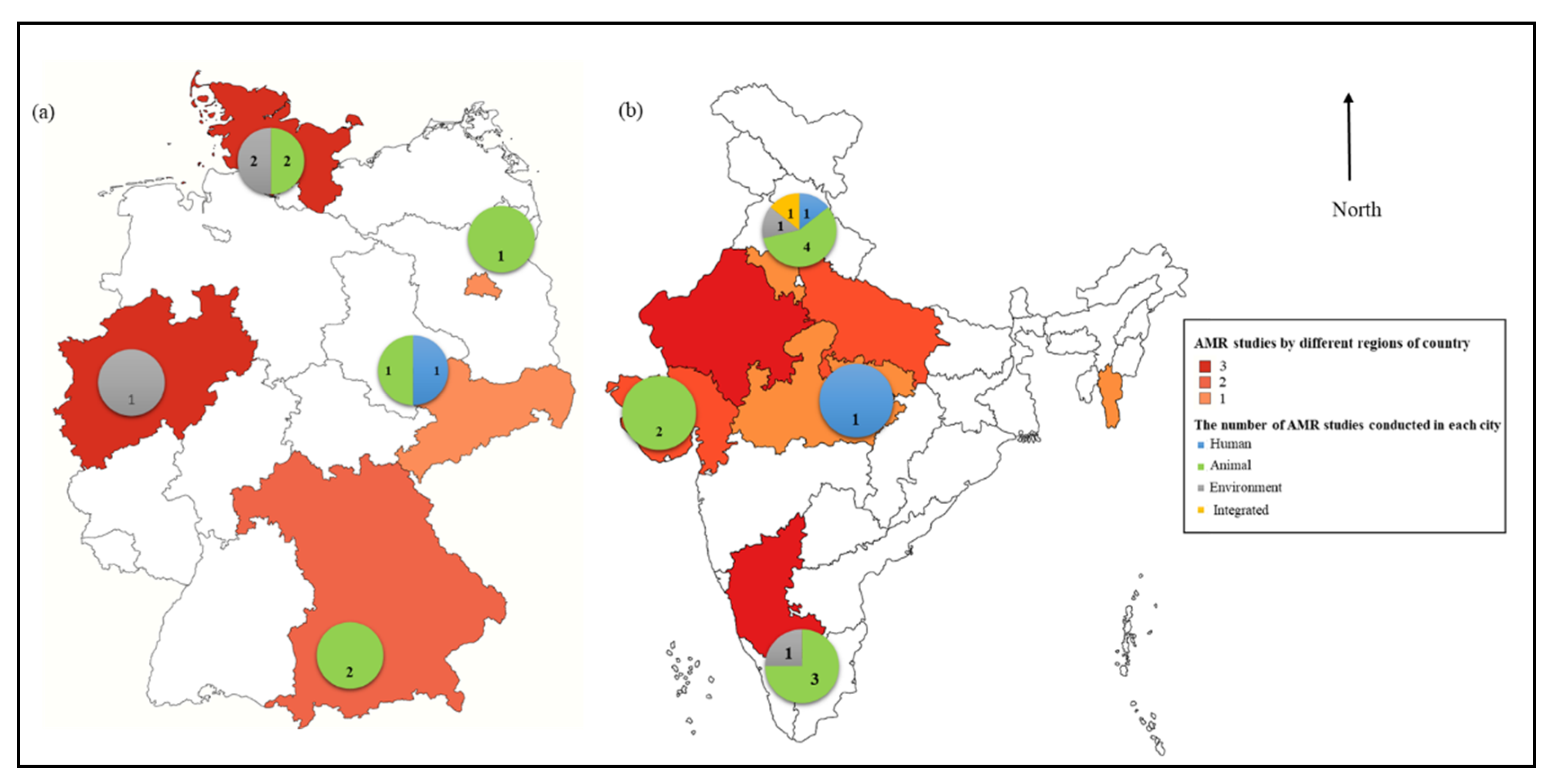
A summary of the studies chosen for qualitative synthesis is provided in Table All 24 selected studies were published in
Figure 2 depicts the study sites and the number of papers published in three regions. In India, most of the studies were published in Northern (7/14, 77.78%), followed by Southern (3/14, 33.33), Western (2, 14.29%) and Central (2, 14.29%) part. However, in Germany, most of the studies were published in Northern (4/10, 40), followed by Southern (2/10, 20%), Central (2/10, 20%), Eastern (1/10, 10%) and Western (1/10, 10%) region.
Figure.
The number of articles and study sites in each compartment i.e. Human, Animal and Environment. (a) Germany (b) India.
Figure.
The number of articles and study sites in each compartment i.e. Human, Animal and Environment. (a) Germany (b) India.
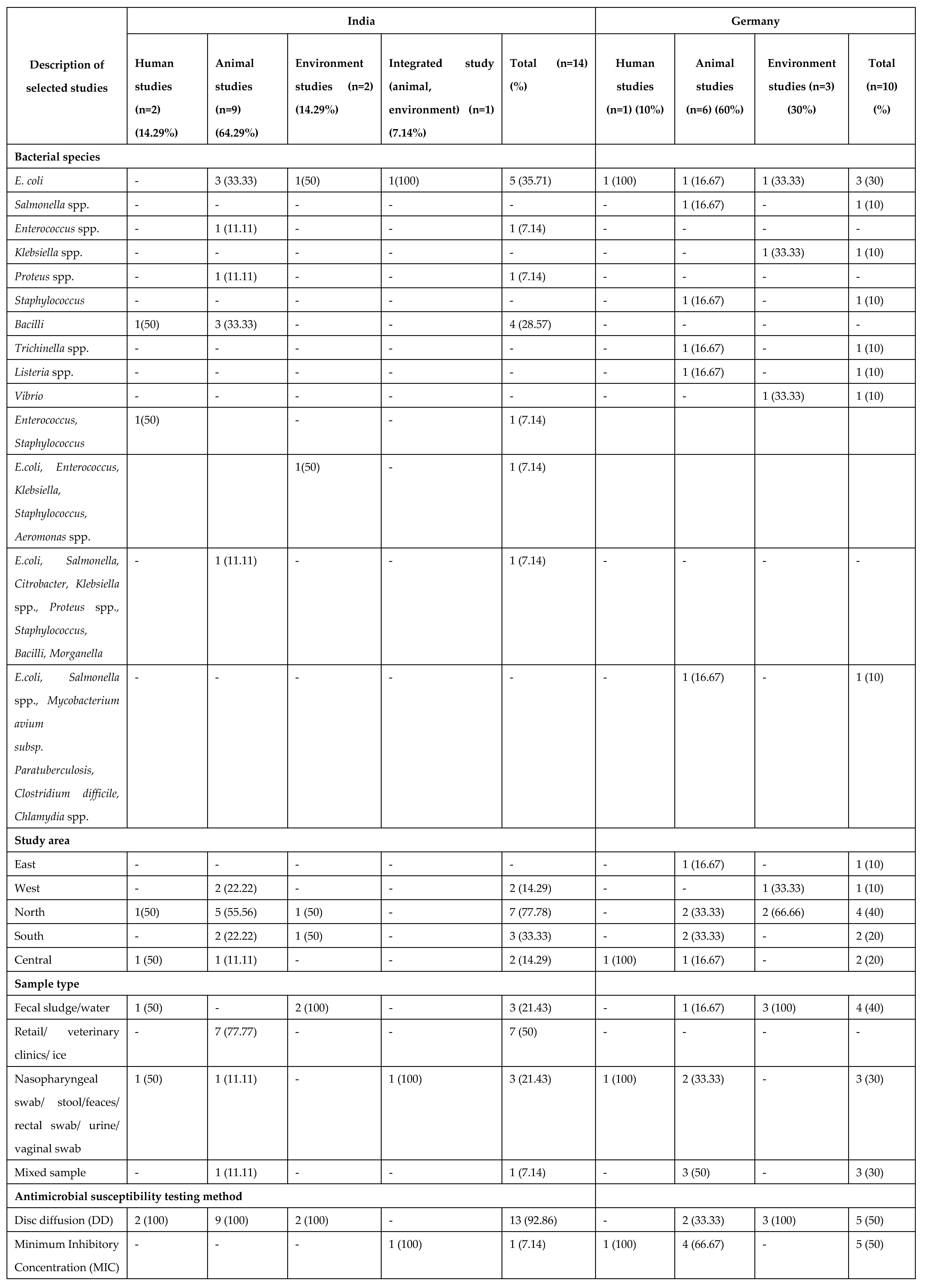
Table.
The total number of studies in each category.
Table.
The total number of studies in each category.
3.2.2. Microbial prevalence in India and Germany
In the selected research, In India, E. coli was the most prevalent species (5/14, 35.71%), followed by Bacilli (4/14, 28.57%), Enterococcus spp. (1/14, 7.14%), Proteus spp. (1/14, 7.14%), Moreover, there was one study covering more than one pathogen together, i.e. Enterococcus, Staphylococcus (1/14, 7.14%), and one covering E.coli, Enterococcus, Klebsiella, Staphylococcus, Aeromonas spp. (1/14, 7.14%) and one study that covers E.coli, Salmonella, Citrobacter, Klebsiella spp., Proteus spp., Staphylococcus, Bacilli, and Morganella (1/14, 7.14%). Retail/ veterinary clinics/ ice (7, 50%) were the most selected studies, followed by faecal sludge/water (3, 21.43%), nasopharyngeal swab/ stool/faeces/ rectal swab/ urine/ vaginal swab (3, 21.43%) and mixed sample (1, 7.14%). The agar disc diffusion (DD) method was employed by most of the selected research (13/14, 92.86%) for AST, whereas other studies (1/14, 7.14%) used agar/broth methods to assess the MIC of antimicrobial compounds.
E. coli (3/10) is also the most common species in Germany, followed by Salmonella spp. (1/10, 10%), Klebsiella spp. (1/10, 10%), Staphylococcus (1/10, 10%), Trichinella spp. (1/10, 10%), Listeria spp. (1/10, 10%), Vibrio (1/10, 10%), Furthermore, there was one study that covered more than one pathogen together, i.e. E.coli, Salmonella spp., Mycobacterium avium subsp. Paratuberculosis, Clostridium difficile, Chlamydia spp. (1/10, 10%). Faecal sludge/water were the most selected studies (4, 40%) along with nasopharyngeal swab/ stool/faeces/ rectal swab/ urine/ vaginal swab (3, 30%) and mixed sample (3, 30%). For AST, the agar DD method and MIC of antimicrobial agents (5/10, 50%) were used. (Table 1)
3.3. Quantitative syntheses of selected studies
3.3.1. AMR in India
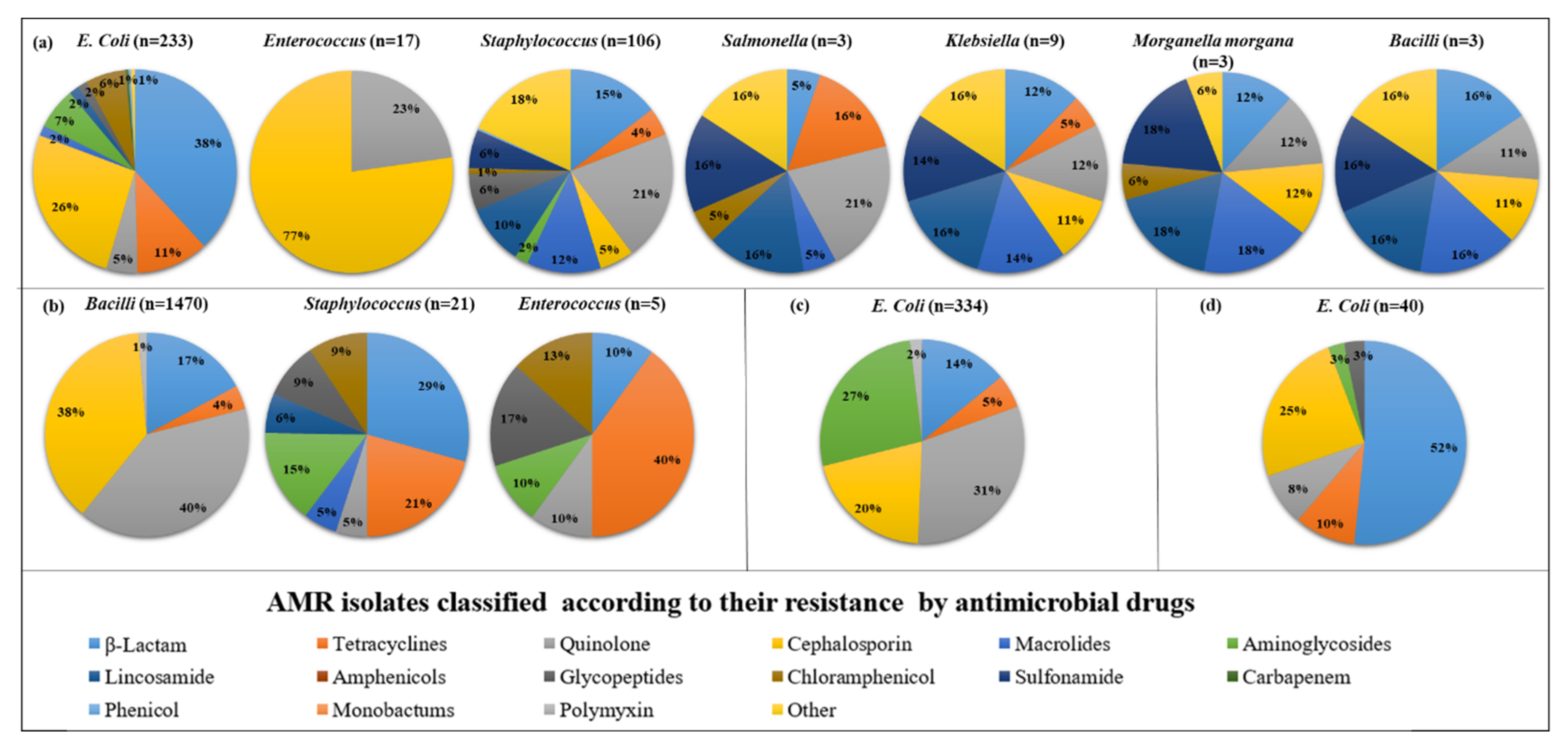
A total of 14 studies on AMR phenotypic susceptibility to 69 distinct antimicrobial agents (16 antimicrobial classes) among 10 Pathogens were considered. For ease of understanding, we categorized the studies into 4 groups: animals, humans, the environment, and integrated studies (animal and environment). In Animals, There are 7 bacteria which are found to be highly prevalent. The highest resistance shown by antibiotics in pathogens is as follows: in
E.coli, β-lactam found highly prevalent (38%), followed by cephalosporins (26%); in
Enterococcus, cephalosporins (77%); in
Staphylococcus, quinolones (21%) followed by other (18%); in
Salmonella, quinolones (20%); in
Klebsiella, lincosamide (16%) followed by macrolides (16%) and sulfonamides (16%);
Morganella morgana, lincosamide (18%) followed by sulfonamide (18%) and macrolides (18%);
Bacilli, β-lactam (16%), macrolides (16%), lincosamide (16%), sulfonamide (16%), other (16%). However, in humans 3 bacteria found high resistance to antibiotics, including
Bacilli, a quinolone (40%) followed by Cephalosporin (38%); in
Staphylococcus, β-lactam (29%) followed by tetracycline (21%); in
Enterococcus, tetracycline (40%). Moreover, in the environment, only
E. coli was highly resistant to antibiotics such as Quinolone (31%), followed by Aminoglycosides (27%). In addition, in integrated studies (animal and environment), only
E. coli was found to be highly resistant to antibiotics such as β-lactam (52%), followed by cephalosporin (25%). (
Figure 3)
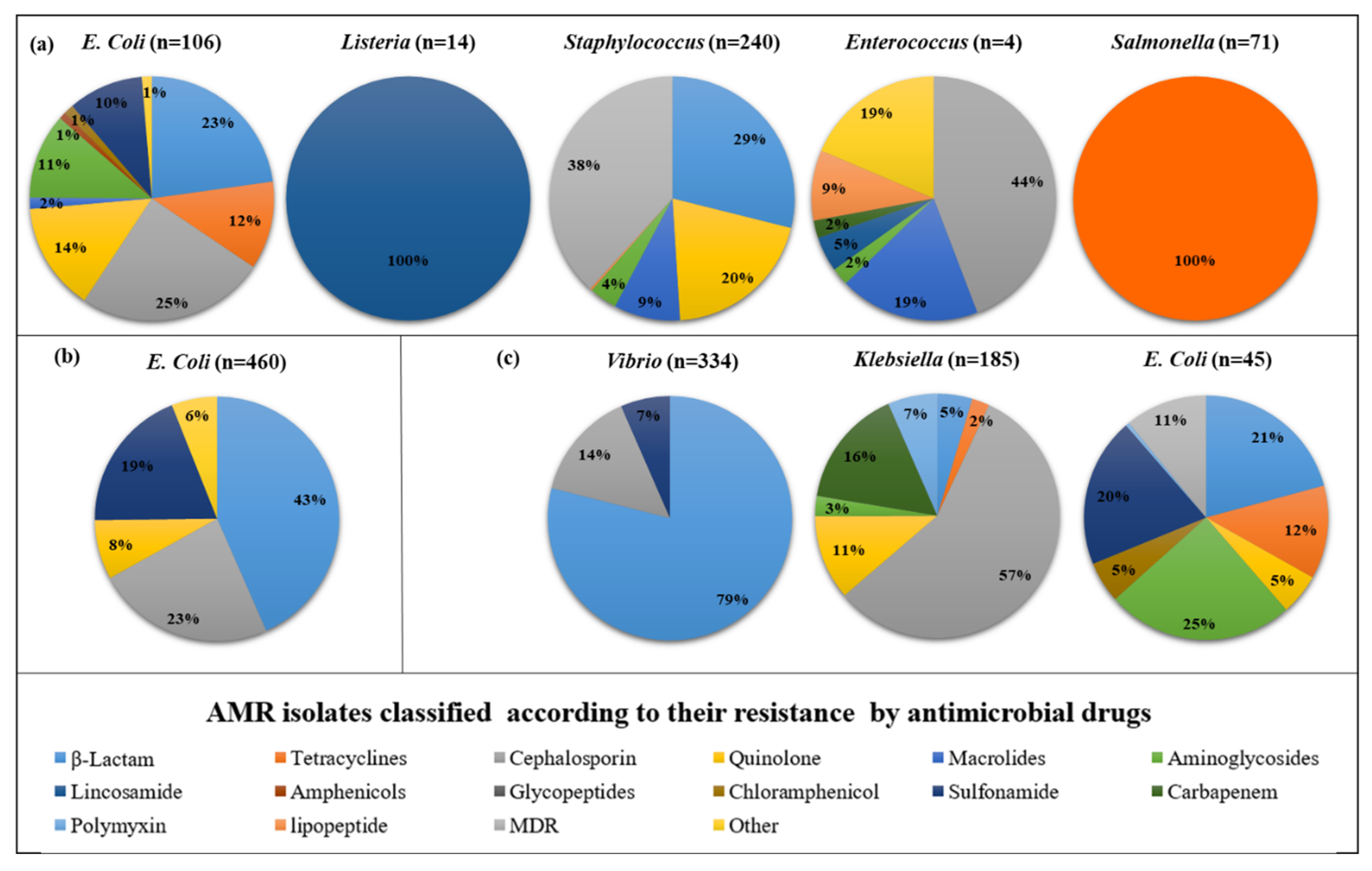
3.3.2. AMR in Germany
We analyzed 10 research on the AMR phenotypic susceptibility to 69 different antimicrobial drugs (16 antimicrobial classes) across 11 Pathogens. For clarity, we divided the studies into four categories: animals, humans, the environment, and integrated studies (animal and environment). In Animal, there are 5 bacteria found to be highly prevalent. The highest resistance showed by antibiotics in pathogens as follows: in
E. coli, Cephalosporin (25%) followed by β-lactam (23%);
Listeria, lincosamide (100%);
Staphylococcus, MDR (38%) followed by β-lactam (29%);
Enterococcus, Cephalosporin (44%) followed by macrolides (19%) and other (19%);
Salmonella, tetracyclines (100%). However, in humans, only
E. coli found high resistance to antibiotics such as β-lactam (43%) followed by Cephalosporin (23%). Moreover, in environment 3, bacteria found high resistance to antibiotics such as including
Vibrio, β-lactam (79%) followed by Cephalosporin (14%);
Klebsiella, Cephalosporin (57%) followed by carbapenem (16%);
E. coli, Aminoglycosides (25%) followed by β-lactam (20%). (
Figure 4)
4. Discussion
This study comprehensively reviews the prevalence of pathogens in various sectors, including humans, animals and the environment along with One Health perspective; the burden of AMR and the existence of AMR genes in India and Germany are also described. Data from 2022 were studied to establish a link between the occurrence of AMR and specific bacteria. In different sectors. Out of 532 papers, 24 were chosen as meeting the criterion. Human animals (including chicken, chicken meat, broiler, pig, pork, cattle, beef, fish, Octopus, lobster, shrimp, marine mammals, vulture, ostrich, camel, Dairy includes: milk and milk products) and environmental samples (including water, sewage water, soil) were all examined by an anticipated AMR risk group in a follow-up study.
AMR is a global problem that requires global action and nationally tailored responses. High AMR levels could be attributed to the spread of AMR bacteria and AMR-encoding genes as a result of intimate interaction between humans, animals, and the environment.[
36]. It was observed in animals that β-lactam is the antibiotic that is most frequent in the bacteria studied. Quinolone and cephalosporin’s, on the other hand, were highly resistant to the pathogens tested in humans. Furthermore, in the environment and integrated studies (which include animals and the environment), resistance to aminoglycosides and β-lactams is significant in India. While in Germany, a high rate of AMR resistance in animals, humans, and the environment was reported in β-lactam. Surprisingly, this data shows how different antibiotics interact with the food chain, and it is a serious concern if it is unchecked.
Implementing and establishing policies, legislation, and research to connect diverse sectors that can collaborate is part of the One Health concept[
37]. If proper methods are not implemented, the incidence of microbes developing resistant to various antimicrobial agents may continue to rise, posing a global danger[
38]. As a result, there is an urgent need to reduce antibiotic use in order to lower the burden of antimicrobial resistance. This can be accomplished using the One Health method, which takes into account hazards to the animal-human environment in number of different circumstances. In order to do that along with Global Action Plan (GAP) on AMR, India and Germany have launched several one-health strategies besides NAP. In India National Centre for Disease Control (NCDC), New Delhi is the focal point for implementing and coordinating the AMR program. (1) In April 2017, the Indian Ministry of Health and Family Welfare released the NAP to Combat AMR [
39]. The 12th Five-Year Plan (2012-17) is still in effect as the "National One Health Program for Zoonosis Prevention and Control" during the 15th Finance Commission (2021-26) term, Antimicrobial Resistance Surveillance & Research Network (AMRSN) established by the Indian Council of Medical Research (ICMR)[
40] started with six reference labs located in four tertiary care medical institutions, Infection prevention and control (IPC) programs[
41]. On the other hand in Germany, German Antimicrobial Resistance Strategy (DART) 2020[
42], Antibiotic Resistance Dynamics: the influence of geographic origin and management (ARDIG) [
43,
44,
45,
46,
47,
48], European Antimicrobial Resistance Surveillance Network (EARS-Net) and Global Antimicrobial Resistance Surveillance System (GLASS).
In terms of the study’s limitations, numerous publications were excluded from the meta-analysis because they did not provide enough information about the clinical impact of ABR in these two nations. Furthermore the variability in study results was thus demonstrated to be considerable in the meta-analysis because our research analyzed publications conducted in multiple geographic locations, namely the North and South in both nations, and with different sample types.
5. Conclusion
Our investigations have provided some data on the burden of AMR, which should be useful as a baseline for ongoing research into the development and transmission processes of AMR bacteria in all three compartments of humans, animals, and the environment in India and Germany in Given the significant frequency of resistance to the most vital antimicrobials, we suggest limiting their usage, particularly in Animals. A wide range of pathogens are involved, and resistance to important medicines, including beta-lactams and quinolones, is widespread. Finding strategies that can effectively lessen the burden of bacterial AMR is a top priority, whether they are applied in various settings or are precisely tailored to the resources available and the most effective pathogen-drug combinations in a given environment. In the future, broader adoption of One Health techniques will bring together various disciplines and data sources, resulting in considerably deeper insights. Multiple scales and levels under the One Health approach to tackle the AMR problem.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
References
- Tang, K.L., Caffrey, N.P., Nóbrega, D.B., Cork, S.C., Ronksley, P.E., Barkema, H.W., Polachek, A.J., Ganshorn, H., Sharma, N., Kellner, J.D., Ghali, W.A.: Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet. Heal. 1, e316–e327 (2017). [CrossRef]
- Gross, S., Müller, A., Seinige, D., Wohlsein, P., Oliveira, M., Steinhagen, D., Kehrenberg, C., Siebert, U.: Occurrence of Antimicrobial-Resistant Escherichia coli in Marine Mammals of the North and Baltic Seas: Sentinels for Human Health. Antibiotics. 11, 1–23 (2022). [CrossRef]
- Pormohammad, A., Nasiri, M.J., Azimi, T.: Prevalence of antibiotic resistance in escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: A systematic review and meta-analysis. Infect. Drug Resist. 12, 1181–1197 (2019). [CrossRef]
- Landers, T.F., Cohen, B., Wittum, T.E., Larson, E.L.: A Review of Antibiotic Use in Food Animals: Perspective, Policy, and Potential. Public Health Rep. 127, 4 (2012). [CrossRef]
- The review on antimicrobial resistance. Tackling drug-resistant infections globally: final report and recommendations. UK, 2016., https://amr-review.org/sites/default/files/160518_Final paper_with cover.pdf.
- de Kraker, M.E.A., Stewardson, A.J., Harbarth, S.: Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLOS Med. 13, e1002184 (2016). [CrossRef]
- Cassini, A., Högberg, L.D., Plachouras, D., Quattrocchi, A., Hoxha, A., Simonsen, G.S., Colomb-Cotinat, M., Kretzschmar, M.E., Devleesschauwer, B., Cecchini, M., Ouakrim, D.A., Oliveira, T.C., Struelens, M.J., Suetens, C., Monnet, D.L., Strauss, R., Mertens, K., Struyf, T., Catry, B., Latour, K., Ivanov, I.N., Dobreva, E.G., Tambic Andraševic, A., Soprek, S., Budimir, A., Paphitou, N., Žemlicková, H., Schytte Olsen, S., Wolff Sönksen, U., Märtin, P., Ivanova, M., Lyytikäinen, O., Jalava, J., Coignard, B., Eckmanns, T., Abu Sin, M., Haller, S., Daikos, G.L., Gikas, A., Tsiodras, S., Kontopidou, F., Tóth, Á., Hajdu, Á., Guólaugsson, Ó., Kristinsson, K.G., Murchan, S., Burns, K., Pezzotti, P., Gagliotti, C., Dumpis, U., Liuimiene, A., Perrin, M., Borg, M.A., de Greeff, S.C., Monen, J.C., Koek, M.B., Elstrøm, P., Zabicka, D., Deptula, A., Hryniewicz, W., Caniça, M., Nogueira, P.J., Fernandes, P.A., Manageiro, V., Popescu, G.A., Serban, R.I., Schréterová, E., Litvová, S., Štefkovicová, M., Kolman, J., Klavs, I., Korošec, A., Aracil, B., Asensio, A., Pérez-Vázquez, M., Billström, H., Larsson, S., Reilly, J.S., Johnson, A., Hopkins, S.: Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet. Infect. Dis. 19, 56–66 (2019). [CrossRef]
- Prestinaci, F., Pezzotti, P., Pantosti, A.: Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob. Health. 109, 309–318 (2015). [CrossRef]
- 2019 Antibiotic Resistance Threats Report | CDC, https://www.cdc.gov/drugresistance/biggest-threats.html.
- Antimicrobial resistance, https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
- Murray, C.J., Ikuta, K.S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., Han, C., Bisignano, C., Rao, P., Wool, E., Johnson, S.C., Browne, A.J., Chipeta, M.G., Fell, F., Hackett, S., Haines-Woodhouse, G., Kashef Hamadani, B.H., Kumaran, E.A.P., McManigal, B., Agarwal, R., Akech, S., Albertson, S., Amuasi, J., Andrews, J., Aravkin, A., Ashley, E., Bailey, F., Baker, S., Basnyat, B., Bekker, A., Bender, R., Bethou, A., Bielicki, J., Boonkasidecha, S., Bukosia, J., Carvalheiro, C., Castañeda-Orjuela, C., Chansamouth, V., Chaurasia, S., Chiurchiù, S., Chowdhury, F., Cook, A.J., Cooper, B., Cressey, T.R., Criollo-Mora, E., Cunningham, M., Darboe, S., Day, N.P.J., De Luca, M., Dokova, K., Dramowski, A., Dunachie, S.J., Eckmanns, T., Eibach, D., Emami, A., Feasey, N., Fisher-Pearson, N., Forrest, K., Garrett, D., Gastmeier, P., Giref, A.Z., Greer, R.C., Gupta, V., Haller, S., Haselbeck, A., Hay, S.I., Holm, M., Hopkins, S., Iregbu, K.C., Jacobs, J., Jarovsky, D., Javanmardi, F., Khorana, M., Kissoon, N., Kobeissi, E., Kostyanev, T., Krapp, F., Krumkamp, R., Kumar, A., Kyu, H.H., Lim, C., Limmathurotsakul, D., Loftus, M.J., Lunn, M., Ma, J., Mturi, N., Munera-Huertas, T., Musicha, P., Mussi-Pinhata, M.M., Nakamura, T., Nanavati, R., Nangia, S., Newton, P., Ngoun, C., Novotney, A., Nwakanma, D., Obiero, C.W., Olivas-Martinez, A., Olliaro, P., Ooko, E., Ortiz-Brizuela, E., Peleg, A.Y., Perrone, C., Plakkal, N., Ponce-de-Leon, A., Raad, M., Ramdin, T., Riddell, A., Roberts, T., Robotham, J.V., Roca, A., Rudd, K.E., Russell, N., Schnall, J., Scott, J.A.G., Shivamallappa, M., Sifuentes-Osornio, J., Steenkeste, N., Stewardson, A.J., Stoeva, T., Tasak, N., Thaiprakong, A., Thwaites, G., Turner, C., Turner, P., van Doorn, H.R., Velaphi, S., Vongpradith, A., Vu, H., Walsh, T., Waner, S., Wangrangsimakul, T., Wozniak, T., Zheng, P., Sartorius, B., Lopez, A.D., Stergachis, A., Moore, C., Dolecek, C., Naghavi, M.: Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 399, 629–655 (2022). [CrossRef]
- Jinks, T., Lee, N., Sharland, M., Rex, J., Gertler, N., Diver, M., Jones, I., Jones, K., Mathewson, S., Chiara, F., Farrar, J.: A time for action: antimicrobial resistance needs global response. Bull. World Health Organ. 94, 558-558A (2016). [CrossRef]
- Dahal, R., Upadhyay, A., Ewald, B.: One Health in South Asia and its challenges in implementation from stakeholder perspective. Vet. Rec. 181, 626–626 (2017). [CrossRef]
- Harris, J.D., Quatman, C.E., Manring, M.M., Siston, R.A., Flanigan, D.C.: How to write a systematic review. Am. J. Sports Med. 42, 2761–2768 (2014). [CrossRef]
- Grakh, K., Mittal, D., Prakash, A., Jindal, N.: Characterization and antimicrobial susceptibility of biofilm-producing Avian Pathogenic Escherichia coli from broiler chickens and their environment in India. Vet. Res. Commun. 46, 537–548 (2022). [CrossRef]
- Ronanki, S.P., Ramya, P., Babu, A.J., Sreedevi, B.: Multidrug resistant- Proteus mirabilis and Proteus vulgaris isolated from milk and meat samples in Tirupati, Andhra Pradesh: An emerging public health threat. 11, 1427–1434 (2022).
- Amreen, F.: Quarantine and antibiotic susceptibility of Enterobacteriaceae strains and other gram negative bacteria in dairy sweetmeat milk ( doodh ) peda. 1–17 (2022).
- Microbiological Contamination of Retail Meat from Mizoram ( India ) with Special Reference to Molecular Detection and Multi-Drug Resistance of Escherichia coli. 16–19 (2022).
- Panwar, K., Bhati, T., Ritod, S., Shringi, B.N.: Detection of antibiotic resistance in Escherichia coli isolates from Egyptian vultures from arid regions of India. Environ. Conserv. J. 23, 65–71 (2022). [CrossRef]
- Patel, N.M., Kumar, R., Savalia, C. V, Kalyani, I.H., Solanki, J.B.: Public Health risk of antibiotic resistant phenotypes and molecular confirmation of staphylococcus aureus isolated from bovine raw milk of South Gujarat, India. 11, 327–334 (2022).
- Charles, I., Boniface, A.: ANTI-MICROBIAL RESISTANCE PROFILE OF Escherichia coli ISOLATES. 2, 322–328 (2005).
- Singh, B.R., Karthikeyan, R., K Sinha, D., OR, V., Jaykumar, V., Yadav, A., Agri, H.: Potentially Pathogenic Bacteria in Water Bodies and Drinking Water Supplies in and Around Bareilly, India. Acta Sci. Microbiol. 113–126 (2022). [CrossRef]
- Sharma, M., Qurashi, M.R., Sharma, S.: Phenotypic and genotypic characterizations of antimicrobial resistance among gram-negative bacilli of clinical isolates. Microb. Biosyst. 7, 1–8 (2022). [CrossRef]
- Sivaraman, G.K., Vijayan, A., Visnuvinayagam, S., Muthulakshmi, T., Prasad, M.M., Ravishankar, C.N.: Incidence of multi drug resistant coagulase-negative Staphylococci from seafood samples, Veraval, Gujarat. Indian J. Anim. Heal. (2022). [CrossRef]
- Desai, D., Rajkumar, S.: ISOLATION AND CHARACTERIZATION OF ESBL PRODUCING ESCHERICHIA COLI FROM HEALTHY GOATS AND FROM THEIR ENVIRONMENT IN WEST COASTAL. (2023). [CrossRef]
- Moktan, J.B., Venkataraman, R., Shrestha, Y.: The Prevalence of Multidrug-Resistant Bacteria Detected in Poultry Products in Mandya, India. Arch. Pharm. Pract. 14, 35–39 (2023). [CrossRef]
- Ahamad Khan, J., Rathore, R.S., Ahmad, I., Gill, R., Husain, F.M., Akhtar, J.: High Prevalence of Multidrug Resistant Staphylococcus aureus from Buffalo Beef Sold at Retail Butcheries in Northern India. Acta Sci. Microbiol. 12–21 (2022). [CrossRef]
- Soundararajan, M., Marincola, G., Liong, O., Marciniak, T., Wencker, F.D.R., Hofmann, F., Schollenbruch, H., Kobusch, I., Linnemann, S., Wolf, S.A., Helal, M., Semmler, T., Walther, B., Schoen, C., Nyasinga, J., Revathi, G., Boelhauve, M., Ziebuhr, W.: Farming Practice Influences Antimicrobial Resistance Burden of Non-Aureus Staphylococci in Pig Husbandries. Microorganisms. 11, 31 (2022). [CrossRef]
- Klose, C., Scuda, N., Ziegler, T., Eisenberger, D., Hanczaruk, M., Riehm, J.M.: Whole-Genome Investigation of Salmonella Dublin Considering Mountain Pastures as Reservoirs in Southern Bavaria, Germany. Microorganisms. 10, (2022). [CrossRef]
- Oswaldi, V., Lüth, S., Dzierzon, J., Meemken, D., Schwarz, S., Feßler, A.T., Félix, B., Langforth, S.: Distribution and Characteristics of Listeria spp. in Pigs and Pork Production Chains in Germany. Microorganisms. 10, (2022). [CrossRef]
- Fleischmann, S., Herrig, I., Wesp, J., Stiedl, J., Reifferscheid, G., Strauch, E., Alter, T., Brennholt, N.: Prevalence and Distribution of Potentially Human Pathogenic Vibrio spp. on German North and Baltic Sea Coasts. Front. Cell. Infect. Microbiol. 12, 1–15 (2022). [CrossRef]
- González-Santamarina, B., Schnee, C., Köhler, H., Weber, M., Methner, U., Seyboldt, C., Berens, C., Menge, C.: Survey on shedding of selected patho-genic, zoonotic or antimicrobial resistant bacteria by South American camelids in Central Germany. Berl. Munch. Tierarztl. Wochenschr. 135, (2022). [CrossRef]
- Quality, G.M., Beindorf, P., Kovalenko, O., Ulrich, S., Geißler, H., Korbel, R., Schwaiger, K., Dorn-in, S., Esteban-cuesta, I.: Resistance. (2022).
- Kresken, M., Pfeifer, Y., Wagenlehner, F., Werner, G., Wohlfarth, E.: Resistance to Mecillinam and Nine Other Antibiotics for Oral Use in Escherichia coli Isolated from Urine Specimens of Primary Care Patients in Germany, 2019/Antibiotics. 11, (2022). [CrossRef]
- Savin, M., Bierbaum, G., Mutters, N.T., Schmithausen, R.M., Kreyenschmidt, J., García-Meniño, I., Schmoger, S., Käsbohrer, A., Hammerl, J.A.: Genetic Characterization of Carbapenem-Resistant Klebsiella spp. from Municipal and Slaughterhouse Wastewater. Antibiotics. 11, 1–14 (2022). [CrossRef]
- Vale, F.F., Lehours, P., Yamaoka, Y.: Editorial: The Role of Mobile Genetic Elements in Bacterial Evolution and Their Adaptability. Front. Microbiol. 13, 198 (2022). [CrossRef]
- Gunjan, Vidic, J., Manzano, M., Raj, V.S., Pandey, R.P., Chang, C.M.: Comparative meta-analysis of antimicrobial resistance from different food sources along with one health approach in Italy and Thailand. One Heal. 16, 100477 (2023). [CrossRef]
- Von Wintersdorff, C.J.H., Penders, J., Van Niekerk, J.M., Mills, N.D., Majumder, S., Van Alphen, L.B., Savelkoul, P.H.M., Wolffs, P.F.G.: Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7, 173 (2016). [CrossRef]
- India: National action plan on antimicrobial resistance (NAP-AMR) 2017 – 2021, https://www.who.int/publications/m/item/india-national-action-plan-on-antimicrobial-resistance-(nap-amr)-2017-2021.
- AMRSN, https://iamrsn.icmr.org.in/index.php/amrsn/amrsn.
- National Guidelines for Infection Prevention and Control in Healthcare Facilities, MoHFW 2020 Training Modules:: National Centre for Disease Control (NCDC), https://ncdc.gov.in/index1.php?lang=1&level=2&sublinkid=1019&lid=794.
- BMEL - Animal health - “DART 2020” continues the German Antibiotic Resistance Strategy, https://www.bmel.de/EN/topics/animals/animal-health/DART2020.html.
- Antibiotic Resistance Dynamics: the influence of geographic origin and management systems on resistance gene flows within humans, animals and the environment (ARDIG), https://www.fisaonline.de/en/find-institutions/international-research-institutions/?tx_fisaresearch_internationalresearchinstitutions%5Bp_id%5D=11522&tx_fisaresearch_internationalresearchinstitutions%5Baction%5D=projectDetails&tx_fisaresearch_internationalresearchinstitutions%5Bcontroller%5D=Projects&cHash=3f3470d77b190d844235809dbf1953e7.
- D. Shailendiran, N. Pawar, A. Chanchal, R. P. Pandey, H. B. Bohidar and A. K. Verma, "Characterization and Antimicrobial Activity of Nanocurcumin and Curcumin," 2011 International Conference on Nanoscience, Technology and Societal Implications, Bhubaneswar, India, 2011, pp. 1-7. [CrossRef]
- Tyagi R, Srivastava M, Jain P, Pandey RP, Asthana S, Kumar D, Raj VS. Development of potential proteasome inhibitors against Mycobacterium tuberculosis. J Biomol Struct Dyn. 2022 Mar;40(5):2189-2203. [CrossRef]
- Mukherjee S, Mishra AK, Peer GDG, Bagabir SA, Haque S, Pandey RP, Raj VS, Jain N, Pandey A, Kar SK. The Interplay of the Unfolded Protein Response in Neurodegenerative Diseases: A Therapeutic Role of Curcumin. Front Aging Neurosci. 2021 Nov 19;13:767493. [CrossRef]
- Mishra, P. K., and R. Pandey. "Physico-chemical properties of starch and protein and their relation to grain quality and nutritional value of rice." Rice Breed. Pub. IRRI, Los Banos, Phillippines (1998): 389-405.
- do Socorro Fôro Ramos E, de Oliveira Ribeiro G, Villanova F, de Padua Milagres FA, Brustulin R, Araújo ELL, Pandey RP, Raj VS, Deng X, Delwart E, Luchs A, da Costa AC, Leal É. Composition of Eukaryotic Viruses and Bacteriophages in Individuals with Acute Gastroenteritis. Viruses. 2021 Nov 25;13(12). [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).