Submitted:
22 February 2024
Posted:
23 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Establishing DNA Replication Forks—Origins of DNA Replication and Chromatin-Loading of Replication Proteins
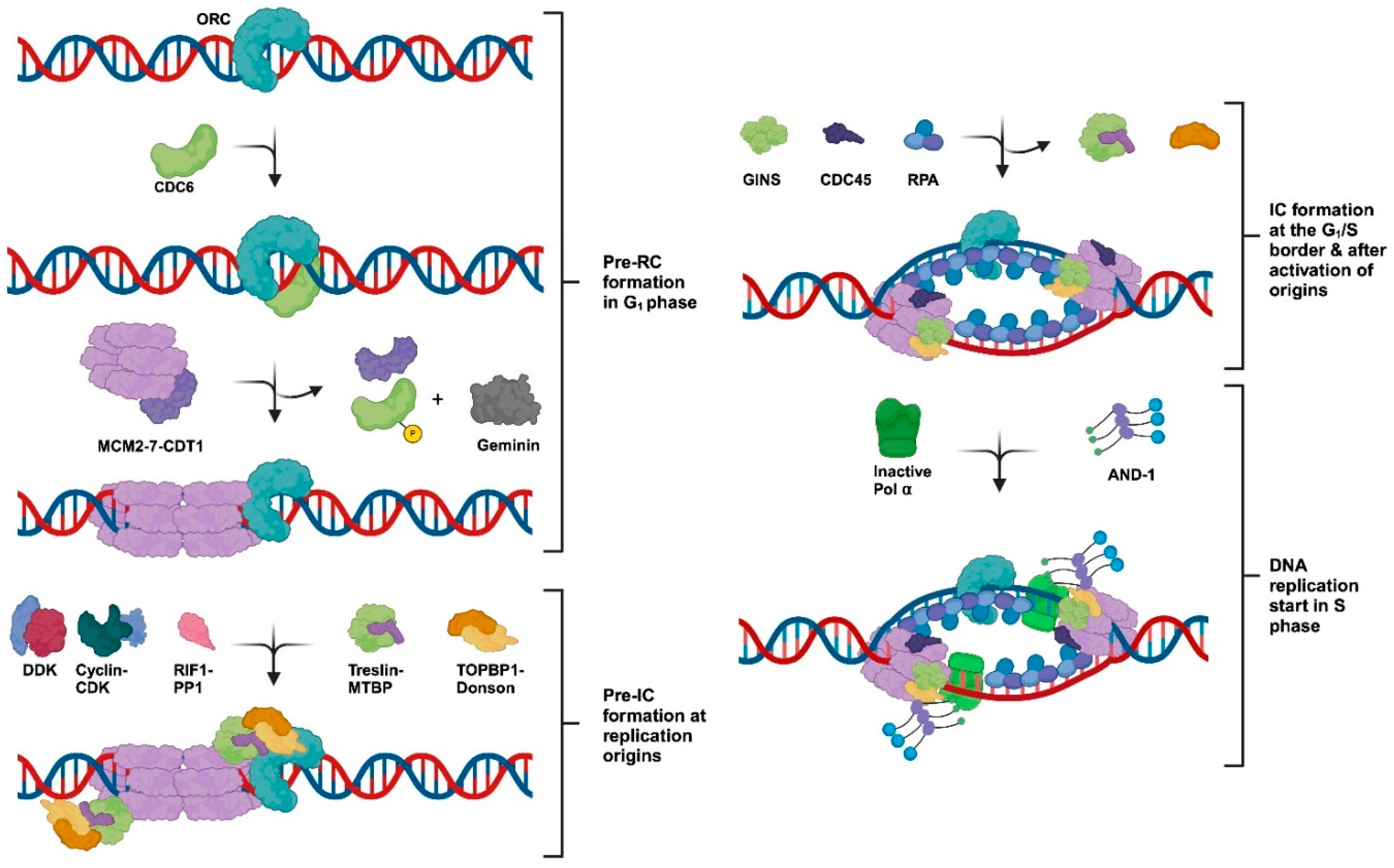
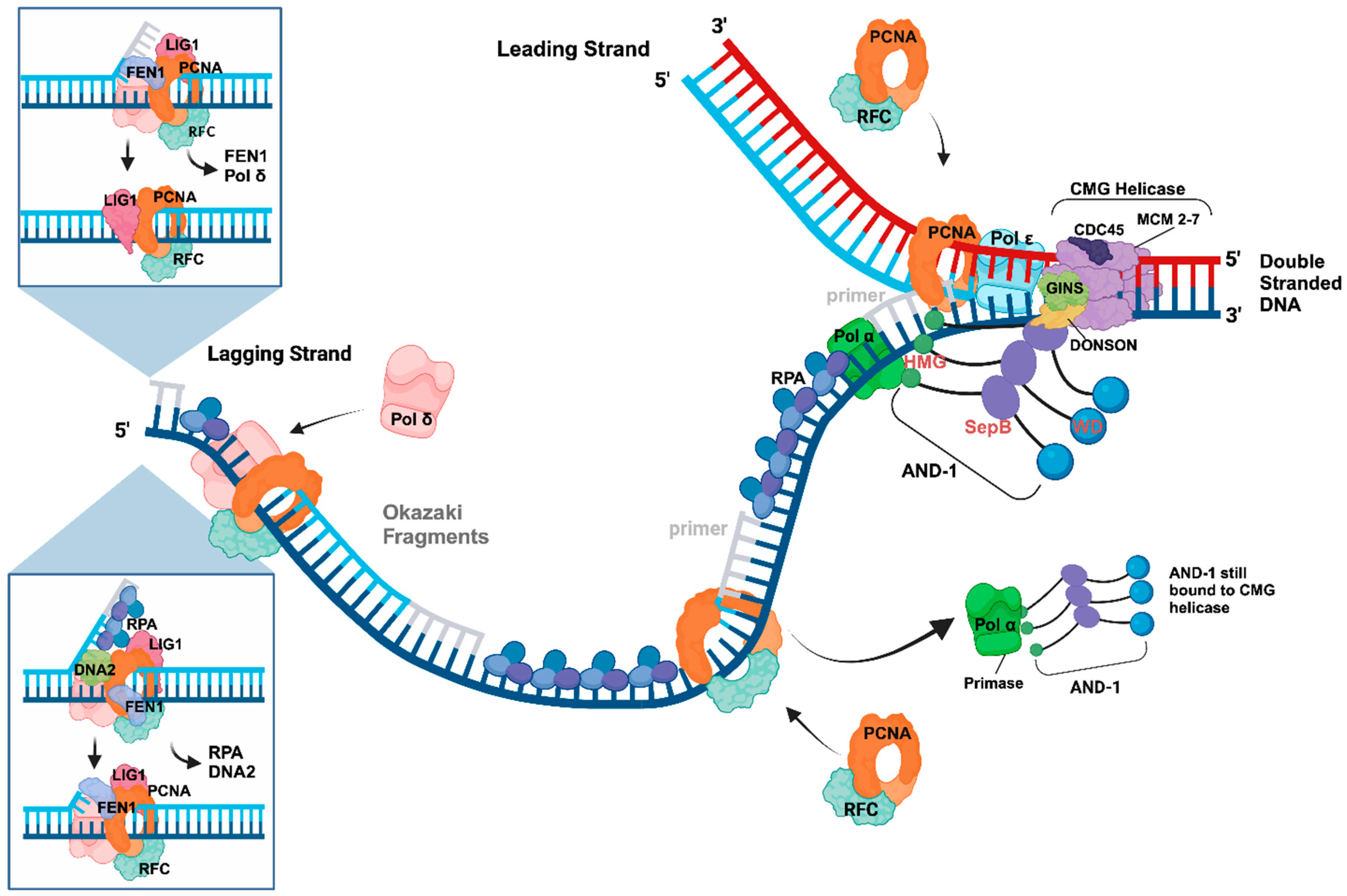
3. The Initiation of DNA Synthesis at Origins
4. Initiation Processes at DNA replication forks—Okazaki Fragment Synthesis
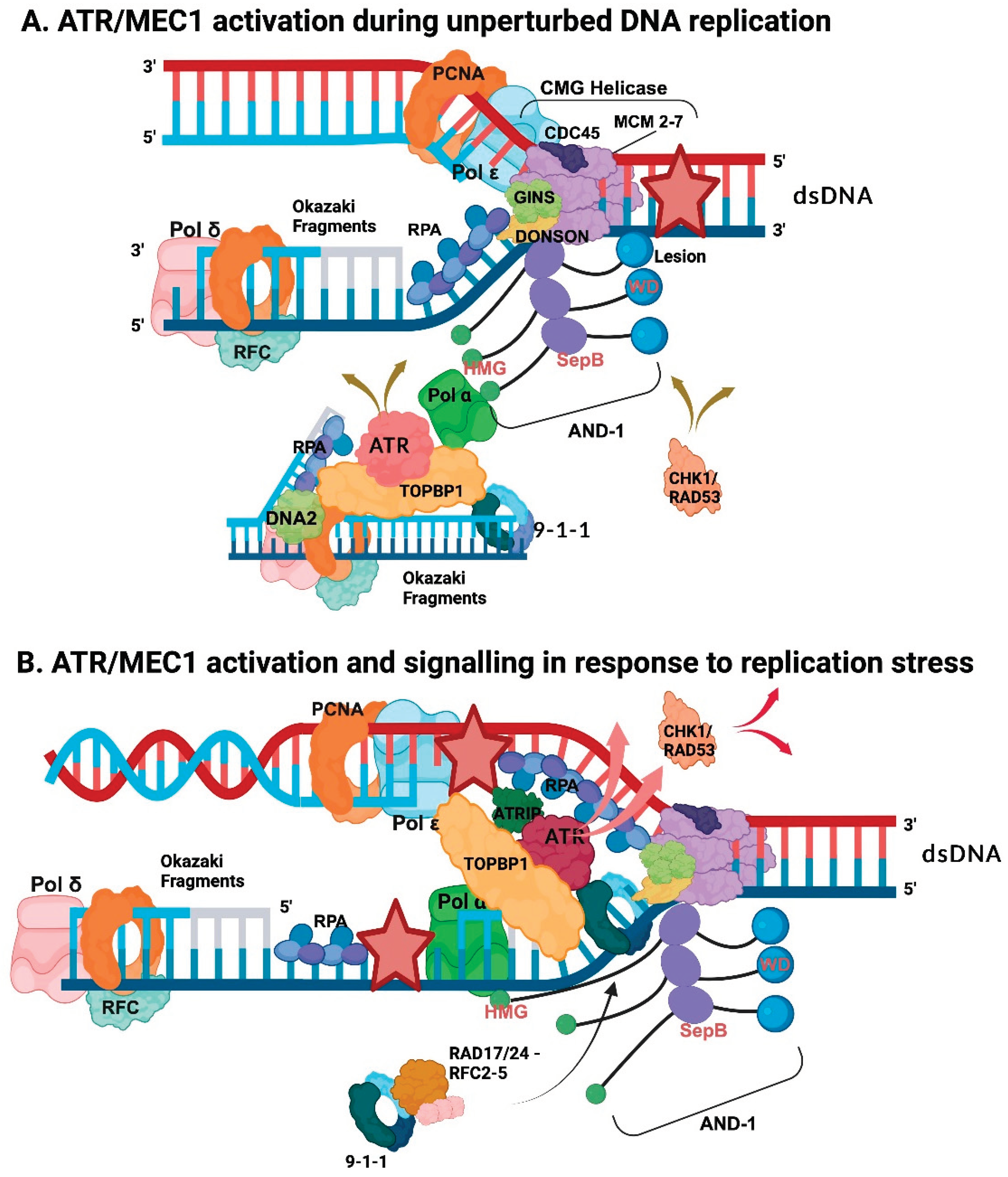
5. Challenges at Replication Forks—Replication Fork Stalling
6. Okazaki Fragment Synthesis and the End-Replication Problem at Telomeres
7. Outlook
Abbreviations
| 9-1-1 | Rad 9-Hus 1-Rad 1 complex (fission yeast and human) equivalent to the |
| complex | Rad 17-Mec3-Ddc1 complex in budding yeast |
| aa | amino acid |
| AND-1/CTF4/ | acidic and nucleoplasmic DNA- binding protein/Chromosome - |
| WDHD1 | Transmission Fidelity 4/WD repeat and HMG-box DNA-binding protein 1 |
| A-T | Ataxia telangiectasia |
| ATM | Ataxia telangiectasia-mutated |
| ATR | ATM-Rad 3-related protein kinase |
| ATRIP | ATR interacting protein |
| ba | bases |
| BLM | Bloom’s helicase |
| CDC | cell division cycle |
| CDK | cyclin-dependent kinase |
| CMG | CDC45-MCM2-7-GINS |
| CST | CTC1-STN1-TEN1 |
| DBD | DNA-binding domain |
| DDR | DNA damage response |
| DH | double hexamer |
| DSB | double-strand break |
| dsDNA | double-stranded DNA |
| ETAA1 | Ewing’s Tumor-Associated Antigen 1 |
| G4 | G-quadruplexes |
| IC | initiation complex |
| mt | mitochondrial |
| MCM | minichromosome maintenance |
| hPolprim1 | human primase-polymerase 1 |
| MCM | Minichromosome maintainance |
| MTBP | Mouse double minute 2-binding protein |
| nt | nucleotide |
| PCNA | proliferating cell nuclear antigen |
| PIKK | phosphotnositol-3 kinase-like protein kinase |
| Pol α | DNA polymerase α–primase |
| Pol δ | DNA polymerase δ |
| Pol ε | DNA polymerase ε |
| POT1 | protector of telomeres 1 |
| Pre-RC | pre-replicative complex |
| Pre-IC | pre-initiation complex |
| RAD | radiation-induced mutation |
| RFC | replication factor C |
| RPA | replication protein A |
| SSB | single-stranded DNA-binding protein |
| ssDNA | single-stranded DNA |
| S. cerevisiae | Saccharomyces cerevisiae |
| S. pombe | Schizosaccharomyces pombe |
| SV40 | simian virus 40 |
| Tp53 | tumour suppressor protein p53 |
| TOPBP1 | topoisomerase II-binding protein 1 |
References
- Burgers, P.M.J. and T.A. Kunkel. Eukaryotic DNA Replication Fork. Annu Rev Biochem, 2017. 86: p. 417-438. [CrossRef]
- Bleichert, F., M.R. Botchan, and J.M. Berger. Mechanisms for initiating cellular DNA replication. Science, 2017. 355: p. eaah6317. [CrossRef]
- Costa, A. and J.F.X. Diffley. The Initiation of Eukaryotic DNA Replication. Annu Rev Biochem, 2022. 91: p. 107-131. [CrossRef]
- Costa, A., I.V. Hood, and J.M. Berger. Mechanisms for initiating cellular DNA replication. Annu Rev Biochem, 2013. 82: p. 25-54. [CrossRef]
- Masai, H., S. Matsumoto, Z. You, N. Yoshizawa-Sugata, and M. Oda. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem, 2010. 79: p. 89-130. [CrossRef]
- Nasheuer, H.P.; Pospiech, H.; Syväoja, J. Progress towards the anatomy of the eukaryotic DNA replication fork. In Genome Integrity: Facets and Perspectives; Lankenau, D.H., Ed.; Springer: Berlin-Heidelberg-NewYork, 2007; Genome Dynamics & Stability; Volume 1, pp. 27–68. [Google Scholar] [CrossRef]
- O’Donnell, M., L. Langston, and B. Stillman. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol, 2013. 5. [CrossRef]
- González-Acosta, D. and M. Lopes. DNA replication and replication stress response in the context of nuclear architecture. Chromosoma, 2023. [CrossRef]
- Attali, I., M.R. Botchan, and J.M. Berger. Structural Mechanisms for Replicating DNA in Eukaryotes. Annu Rev Biochem, 2021. 90: p. 77-106. [CrossRef]
- Howard, A.; Pelc, S.R. Synthesis of deoxyribonucleic acid in normal and irradiated cells and its relation to chromosome breakage. Heredity 1953, 6, 261–273. [Google Scholar]
- Voorhees, J.J., E.A. Duell, D.A. Chambers, and C.L. Marcelo. Regulation of Cell Cycles. Journal of Investigative Dermatology, 1976. 67: p. 15-19. [CrossRef]
- Blow, J.J.; Dutta, A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005, 6, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Blow, J.J.; Laskey, R.A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature 1988, 332, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Mechali, M.; Kearsey, S. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell 1984, 38, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Stillman, B. Origins of DNA replication in eukaryotes. Mol. Cell 2023, 83, 352–372. [Google Scholar] [CrossRef] [PubMed]
- Diffley, J.F.X. Early events in eukaryotic DNA replication. Trends Cell Biol. 1992, 2, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Diffley, J.F.X.; Cocker, J.H. Protein-DNA interactions at a yeast replication origin. Nature 1992, 357, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Natale, D.A., C.J. Li, W.H. Sun, and M.L. DePamphilis. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. Embo J, 2000. 19: p. 2728-2738. [CrossRef]
- Sun, W.H., T.R. Coleman, and M.L. DePamphilis. Cell cycle-dependent regulation of the association between origin recognition proteins and somatic cell chromatin. Embo J, 2002. 21: p. 1437-46. [CrossRef]
- DePamphilis, M.L. The ‘ORC cycle’: a novel pathway for regulating eukaryotic DNA replication. Gene 2003, 310, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Saha, T., S. Ghosh, A. Vassilev, and M.L. Depamphilis. Ubiquitylation, phosphorylation and Orc2 modulate the subcellular location of Orc1 and prevent it from inducing apoptosis. J Cell Sci, 2006. [CrossRef]
- Cvetkovic, M.A., P. Passaretti, A. Butryn, A. Reynolds-Winczura, G. Kingsley, A. Skagia, C. Fernandez-Cuesta, D. Poovathumkadavil, R. George, A.S. Chauhan, S.S. Jhujh, G.S. Stewart, A. Gambus, and A. Costa. The structural mechanism of dimeric DONSON in replicative helicase activation. Mol Cell, 2023. 83: p. 4017-4031.e9. [CrossRef]
- Dhar, S.K., L. Delmolino, and A. Dutta. Architecture of the human origin recognition complex. J Biol Chem, 2001. 276: p. 29067-29071. [CrossRef]
- Evrin, C., V. Alvarez, J. Ainsworth, R. Fujisawa, C. Alabert, and K.P. Labib. DONSON is required for CMG helicase assembly in the mammalian cell cycle. EMBO Rep, 2023. 24: p. e57677. [CrossRef]
- Hashimoto, Y., K. Sadano, N. Miyata, H. Ito, and H. Tanaka. Novel role of DONSON in CMG helicase assembly during vertebrate DNA replication initiation. Embo j, 2023. 42: p. e114131. [CrossRef]
- Kingsley, G., A. Skagia, P. Passaretti, C. Fernandez-Cuesta, A. Reynolds-Winczura, K. Koscielniak, and A. Gambus. DONSON facilitates Cdc45 and GINS chromatin association and is essential for DNA replication initiation. Nucleic Acids Res, 2023. 51: p. 9748-9763. [CrossRef]
- Lim, Y., L. Tamayo-Orrego, E. Schmid, Z. Tarnauskaite, O.V. Kochenova, R. Gruar, S. Muramatsu, L. Lynch, A.V. Schlie, P.L. Carroll, G. Chistol, M.A.M. Reijns, M.T. Kanemaki, A.P. Jackson, and J.C. Walter. In silico protein interaction screening uncovers DONSON’s role in replication initiation. Science, 2023. 381: p. eadi3448. [CrossRef]
- Xia, Y., R. Sonneville, M. Jenkyn-Bedford, L. Ji, C. Alabert, Y. Hong, J.T.P. Yeeles, and K.P.M. Labib. DNSN-1 recruits GINS for CMG helicase assembly during DNA replication initiation in Caenorhabditis elegans. Science, 2023. 381: p. eadi4932. [CrossRef]
- Vashee, S., P. Simancek, M.D. Challberg, and T.J. Kelly. Assembly of the human origin recognition complex. J Biol Chem, 2001. 276: p. 26666-26673. [CrossRef]
- Pellegrini, L. The CMG DNA helicase and the core replisome. Curr Opin Struct Biol, 2023. 81: p. 102612. [CrossRef]
- Blow, J.J., P.J. Gillespie, D. Francis, and D.A. Jackson. Replication origins in Xenopus egg extract Are 5-15 kilobases apart and are activated in clusters that fire at different times. J Cell Biol, 2001. 152: p. 15-25. [CrossRef]
- Smith, D.J. and I. Whitehouse. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature, 2012. 483: p. 434-8. [CrossRef]
- Jones, M.L., Y. Baris, M.R. Taylor, and J.T.P. Yeeles. Structure of a human replisome shows the organisation and interactions of a DNA replication machine. Embo j, 2023. 42: p. e115685. [CrossRef]
- Baretić, D., M. Jenkyn-Bedford, V. Aria, G. Cannone, M. Skehel, and J.T.P. Yeeles. Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork. Mol Cell, 2020. 78: p. 926-940.e13. [CrossRef]
- Sriramachandran, A.M., G. Petrosino, M. Méndez-Lago, A.J. Schäfer, L.S. Batista-Nascimento, N. Zilio, and H.D. Ulrich. Genome-wide Nucleotide-Resolution Mapping of DNA Replication Patterns, Single-Strand Breaks, and Lesions by GLOE-Seq. Mol Cell, 2020. 78: p. 975-985.e7. [CrossRef]
- Blin, M., L. Lacroix, N. Petryk, Y. Jaszczyszyn, C.L. Chen, O. Hyrien, and B. Le Tallec. DNA molecular combing-based replication fork directionality profiling. Nucleic Acids Res, 2021. 49: p. e69. [CrossRef]
- Langston, L.D., R.E. Georgescu, and M.E. O’Donnell. Mechanism of eukaryotic origin unwinding is a dual helicase DNA shearing process. Proc Natl Acad Sci U S A, 2023. 120: p. e2316466120. [CrossRef]
- Yuan, Z., L. Bai, J. Sun, R. Georgescu, J. Liu, M.E. O’Donnell, and H. Li. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol, 2016. 23: p. 217-24. [CrossRef]
- Georgescu, R.E., G.D. Schauer, N.Y. Yao, L.D. Langston, O. Yurieva, D. Zhang, J. Finkelstein, and M.E. O’Donnell. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. Elife, 2015. 4: p. e04988. [CrossRef]
- Bell, S.P. and B. Stillman. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 1992. 357: p. 128-134. [CrossRef]
- Qiu, S., G. Jiang, L. Cao, and J. Huang. Replication Fork Reversal and Protection. Front Cell Dev Biol, 2021. 9: p. 670392. [CrossRef]
- Vipat, S. and T.N. Moiseeva. The TIMELESS Roles in Genome Stability and Beyond. J Mol Biol, 2024. 436: p. 168206. [CrossRef]
- Nasheuer, H.P. and N.O. Onwubiko. Lagging Strand Initiation Processes in DNA Replication of Eukaryotes-Strings of Highly Coordinated Reactions Governed by Multiprotein Complexes. Genes (Basel), 2023. 14. [CrossRef]
- Nasheuer, H.P., R. Smith, C. Bauerschmidt, F. Grosse, and K. Weisshart. Initiation of eukaryotic DNA replication: regulation and mechanisms. Prog Nucleic Acid Res Mol Biol, 2002. 72: p. 41-94. [CrossRef]
- Kilkenny, M.L., A.C. Simon, J. Mainwaring, D. Wirthensohn, S. Holzer, and L. Pellegrini. The human CTF4-orthologue AND-1 interacts with DNA polymerase α/primase via its unique C-terminal HMG box. Open Biol, 2017. 7. [CrossRef]
- Yeeles, J.T., T.D. Deegan, A. Janska, A. Early, and J.F. Diffley. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature, 2015. 519: p. 431-5. [CrossRef]
- On, K.F., F. Beuron, D. Frith, A.P. Snijders, E.P. Morris, and J.F. Diffley. Prereplicative complexes assembled in vitro support origin-dependent and independent DNA replication. Embo j, 2014. 33: p. 605-20. [CrossRef]
- Labib, K., J.F. Diffley, and S.E. Kearsey. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat Cell Biol, 1999. 1: p. 415-422. [CrossRef]
- Deegan, T.D. and J.F. Diffley. MCM: one ring to rule them all. Curr Opin Struct Biol, 2016. 37: p. 145-51. [CrossRef]
- Lewis, J.S., M.H. Gross, J. Sousa, S.S. Henrikus, J.F. Greiwe, A. Nans, J.F.X. Diffley, and A. Costa. Mechanism of replication origin melting nucleated by CMG helicase assembly. Nature, 2022. 606: p. 1007-1014. [CrossRef]
- Frigola, J., D. Remus, A. Mehanna, and J.F. Diffley. ATPase-dependent quality control of DNA replication origin licensing. Nature, 2013. 495: p. 339-43. [CrossRef]
- Romanowski, P., M.A. Madine, A. Rowles, J.J. Blow, and R.A. Laskey. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol, 1996. 6: p. 1416-25. [CrossRef]
- McGarry, T.J. and M.W. Kirschner. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell, 1998. 93: p. 1043-1053. [CrossRef]
- Ferenbach, A., A. Li, M. Brito-Martins, and J.J. Blow. Functional domains of the Xenopus replication licensing factor Cdt1. Nucleic Acids Res, 2005. 33: p. 316-24. [CrossRef]
- Pozo, P.N. and J.G. Cook. Regulation and Function of Cdt1; A Key Factor in Cell Proliferation and Genome Stability. Genes (Basel), 2016. 8. [CrossRef]
- Yuan, Z., R. Georgescu, L. Bai, D. Zhang, H. Li, and M.E. O’Donnell. DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat Commun, 2020. 11: p. 688. [CrossRef]
- Xu, Z., J. Feng, D. Yu, Y. Huo, X. Ma, W.H. Lam, Z. Liu, X.D. Li, T. Ishibashi, S. Dang, and Y. Zhai. Synergism between CMG helicase and leading strand DNA polymerase at replication fork. Nat Commun, 2023. 14: p. 5849. [CrossRef]
- Szambowska, A., I. Tessmer, P. Prus, B. Schlott, H. Pospiech, and F. Grosse. Cdc45-induced loading of human RPA onto single-stranded DNA. Nucleic Acids Res, 2017. [CrossRef]
- Chen, R. and M.S. Wold. Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays, 2014. 36: p. 1156-61. [CrossRef]
- Nasheuer, H.P., A.M. Meaney, T. Hulshoff, I. Thiele, and N.O. Onwubiko. Replication Protein A, the Main Eukaryotic Single-Stranded DNA Binding Protein, a Focal Point in Cellular DNA Metabolism. Int J Mol Sci, 2024. 25: p. 588. [CrossRef]
- Fousek-Schuller, V.J. and G.E.O. Borgstahl. The Intriguing Mystery of RPA Phosphorylation in DNA Double-Strand Break Repair. Genes, 2024. 15: p. 167. [CrossRef]
- Jones, M.L., V. Aria, Y. Baris, and J.T.P. Yeeles. How Pol α-primase is targeted to replisomes to prime eukaryotic DNA replication. Mol Cell, 2023. 83: p. 2911-2924.e16. [CrossRef]
- Villa, F., A.C. Simon, M.A. Ortiz Bazan, M.L. Kilkenny, D. Wirthensohn, M. Wightman, D. Matak-Vinkovíc, L. Pellegrini, and K. Labib. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol Cell, 2016. 63: p. 385-96. [CrossRef]
- Li, J., J. Dong, W. Wang, D. Yu, X. Fan, Y.C. Hui, C.S.K. Lee, W.H. Lam, N. Alary, Y. Yang, Y. Zhang, Q. Zhao, C.L. Chen, B.K. Tye, S. Dang, and Y. Zhai. The human pre-replication complex is an open complex. Cell, 2023. 186: p. 98-111.e21. [CrossRef]
- Jiang, X., V. Klimovich, A.I. Arunkumar, E.B. Hysinger, Y. Wang, R.D. Ott, G.D. Guler, B. Weiner, W.J. Chazin, and E. Fanning. Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. Embo J, 2006. 25: p. 5516-26. [CrossRef]
- Broderick, R., M.D. Rainey, C. Santocanale, and H.P. Nasheuer. Cell cycle-dependent formation of Cdc45-Claspin complexes in human cells are compromized by UV-mediated DNA damage. FEBS J, 2013. 280: p. 4888-4902. [CrossRef]
- Nasheuer, H.P. and F. Grosse. DNA polymerase α-primase from calf thymus. Determination of the polypeptide responsible for primase activity. J. Biol. Chem., 1988. 263: p. 8981-8988. [CrossRef]
- Zerbe, L.K. and R.D. Kuchta. The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry, 2002. 41: p. 4891-4900. [CrossRef]
- Baranovskiy, A.G., N.D. Babayeva, Y. Zhang, J. Gu, Y. Suwa, Y.I. Pavlov, and T.H. Tahirov. Mechanism of Concerted RNA-DNA Primer Synthesis by the Human Primosome. J Biol Chem, 2016. 291: p. 10006-20. [CrossRef]
- Baranovskiy, A.G. and T.H. Tahirov. Elaborated Action of the Human Primosome. Genes (Basel), 2017. 8: p. 62. [CrossRef]
- Taylor, M.R.G. and J.T.P. Yeeles. Dynamics of Replication Fork Progression Following Helicase-Polymerase Uncoupling in Eukaryotes. J Mol Biol, 2019. 431: p. 2040-2049. [CrossRef]
- Taylor, M.R.G. and J.T.P. Yeeles. The Initial Response of a Eukaryotic Replisome to DNA Damage. Mol Cell, 2018. 70: p. 1067-1080.e12. [CrossRef]
- Jones, M.L., Y. Baris, M.R.G. Taylor, and J.T.P. Yeeles. Structure of a human replisome shows the organisation and interactions of a DNA replication machine. Embo j, 2021. 40: p. e108819. [CrossRef]
- Guilliam, T.A. and J.T.P. Yeeles. An updated perspective on the polymerase division of labor during eukaryotic DNA replication. Crit Rev Biochem Mol Biol, 2020. 55: p. 469-481. [CrossRef]
- Barbour, A.T. and D.S. Wuttke. RPA-like single-stranded DNA-binding protein complexes including CST serve as specialized processivity factors for polymerases. Curr Opin Struct Biol, 2023. 81: p. 102611. [CrossRef]
- Jain, R., A.K. Aggarwal, and O. Rechkoblit. Eukaryotic DNA polymerases. Curr Opin Struct Biol, 2018. 53: p. 77-87. [CrossRef]
- Liu, L., J. Mo, E.M. Rodriguez-Belmonte, and M.Y. Lee. Identification of a fourth subunit of mammalian DNA polymerase delta. J Biol Chem, 2000. 275: p. 18739-18744. [CrossRef]
- Zhang, S., H.H. Chao, X. Wang, Z. Zhang, E.Y.C. Lee, and M. Lee. Loss of the p12 subunit of DNA polymerase delta leads to a defect in HR and sensitization to PARP inhibitors. DNA Repair (Amst), 2019. 73: p. 64-70. [CrossRef]
- Cali, F., S.K. Bharti, R. Di Perna, R.M. Brosh, Jr., and F.M. Pisani. Tim/Timeless, a member of the replication fork protection complex, operates with the Warsaw breakage syndrome DNA helicase DDX11 in the same fork recovery pathway. Nucleic Acids Res, 2016. 44: p. 705-17. [CrossRef]
- Prindle, M.J. and L.A. Loeb. DNA polymerase delta in DNA replication and genome maintenance. Environ Mol Mutagen, 2012. 53: p. 666-82. [CrossRef]
- Zhang, D. and M. O’Donnell. The Eukaryotic Replication Machine. Enzymes, 2016. 39: p. 191-229. [CrossRef]
- He, Q., X. Lin, B.L. Chavez, S. Agrawal, B.L. Lusk, and C.J. Lim. Structures of the human CST-Polα–primase complex bound to telomere templates. Nature, 2022. 608: p. 826-832. [CrossRef]
- Cai, S.W., J.C. Zinder, V. Svetlov, M.W. Bush, E. Nudler, T. Walz, and T. de Lange. Cryo-EM structure of the human CST–Polα/primase complex in a recruitment state. Nature Structural & Molecular Biology, 2022. 29: p. 813-819. [CrossRef]
- Thömmes, P., R. Fett, B. Schray, R. Burkhart, M. Barnes, C. Kennedy, N.C. Brown, and R. Knippers. Properties of the nuclear P1 protein, a mammalian homologue of the yeast Mcm3 replication protein. Nucleic Acids Res., 1992. 20: p. 1069-1074. [CrossRef]
- Maine, G.T., P. Sinha, and B.K. Tye. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics, 1984. 106: p. 365-85. [CrossRef]
- Tye, B.K. The MCM2-3-5 proteins: are they replication licensing factors? Trends Cell Biol, 1994. 4: p. 160-6. [CrossRef]
- Takayama, Y., Y. Kamimura, M. Okawa, S. Muramatsu, A. Sugino, and H. Araki. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev, 2003. 17: p. 1153-1165. [CrossRef]
- Baranovskiy, A.G., Y. Zhang, Y. Suwa, N.D. Babayeva, J. Gu, Y.I. Pavlov, and T.H. Tahirov. Crystal structure of the human primase. J Biol Chem, 2015. 290: p. 5635-46. [CrossRef]
- Arezi, B. and R.D. Kuchta. Eukaryotic DNA primase. Trends Biochem Sci, 2000. 25: p. 572-576. [CrossRef]
- Kirk, B.W. and R.D. Kuchta. Human DNA primase: anion inhibition, manganese stimulation, and their effects on in vitro start-site selection. Biochemistry, 1999. 38: p. 10126-10134. [CrossRef]
- Kirk, B.W. and R.D. Kuchta. Arg304 of human DNA primase is a key contributor to catalysis and NTP binding: primase and the family X polymerases share significant sequence homology. Biochemistry, 1999. 38: p. 7727-7736. [CrossRef]
- Grosse, F. and G. Krauss. The primase activity of DNA polymerase α from calf thymus. J Biol Chem, 1985. 260: p. 1881-1888.
- Vaithiyalingam, S., D.R. Arnett, A. Aggarwal, B.F. Eichman, E. Fanning, and W.J. Chazin. Insights into eukaryotic primer synthesis from structures of the p48 subunit of human DNA primase. J Mol Biol, 2014. 426: p. 558-69. [CrossRef]
- Cordoba, J.J., E.A. Mullins, L.E. Salay, B.F. Eichman, and W.J. Chazin. Flexibility and Distributive Synthesis Regulate RNA Priming and Handoff in Human DNA Polymerase α-Primase. J Mol Biol, 2023. 435: p. 168330. [CrossRef]
- He, Q., A.G. Baranovskiy, L.M. Morstadt, A.E. Lisova, N.D. Babayeva, B.L. Lusk, C.J. Lim, and T.H. Tahirov. Structures of human primosome elongation complexes. Nat Struct Mol Biol, 2023. 30: p. 579-583. [CrossRef]
- Zaug, A.J., K.J. Goodrich, J.J. Song, A.E. Sullivan, and T.R. Cech. Reconstitution of a telomeric replicon organized by CST. Nature, 2022. 608: p. 819-825. [CrossRef]
- Kuchta, R.D., B. Reid, and L.M. Chang. DNA primase. Processivity and the primase to polymerase alpha activity switch. J Biol Chem, 1990. 265: p. 16158-65.
- Kaguni, L.S., J.M. Rossignol, R.C. Conaway, and I.R. Lehman. Isolation of an intact DNA polymerase-primase from embryos of Drosophila melanogaster. Proc Natl Acad Sci U S A, 1983. 80: p. 2221-5. [CrossRef]
- Hübscher, U., G. Maga, and S. Spadari. Eukaryotic DNA polymerases. Annu Rev Biochem, 2002. 71: p. 133-63. [CrossRef]
- Donnianni, R.A., Z.X. Zhou, S.A. Lujan, A. Al-Zain, V. Garcia, E. Glancy, A.B. Burkholder, T.A. Kunkel, and L.S. Symington. DNA Polymerase Delta Synthesizes Both Strands during Break-Induced Replication. Mol Cell, 2019. 76: p. 371-381.e4. [CrossRef]
- Yeeles, J.T.P., A. Janska, A. Early, and J.F.X. Diffley. How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Mol Cell, 2017. 65: p. 105-116. [CrossRef]
- Raducanu, V.S., M. Tehseen, A. Al-Amodi, L.I. Joudeh, A. De Biasio, and S.M. Hamdan. Mechanistic investigation of human maturation of Okazaki fragments reveals slow kinetics. Nat Commun, 2022. 13: p. 6973. [CrossRef]
- Stodola, J.L. and P.M. Burgers. Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale. Nat Struct Mol Biol, 2016. 23: p. 402-8. [CrossRef]
- Kao, H.I., J.L. Campbell, and R.A. Bambara. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J Biol Chem, 2004. 279: p. 50840-9. [CrossRef]
- Kao, H.I. and R.A. Bambara. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit Rev Biochem Mol Biol, 2003. 38: p. 433-52. [CrossRef]
- Williams, J.S., P.P. Tumbale, M.E. Arana, J.A. Rana, R.S. Williams, and T.A. Kunkel. High-fidelity DNA ligation enforces accurate Okazaki fragment maturation during DNA replication. Nature Communications, 2021. 12: p. 482. [CrossRef]
- Beattie, T.R. and S.D. Bell. The role of the DNA sliding clamp in Okazaki fragment maturation in archaea and eukaryotes. Biochem Soc Trans, 2011. 39: p. 70-6. [CrossRef]
- Maga, G., G. Villani, V. Tillement, M. Stucki, G.A. Locatelli, I. Frouin, S. Spadari, and U. Hübscher. Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc Natl Acad Sci USA, 2001. 98: p. 14298-14303. [CrossRef]
- Lin, S.H., X. Wang, S. Zhang, Z. Zhang, E.Y. Lee, and M.Y.W.T. Lee. Dynamics of enzymatic interactions during short flap human Okazaki fragment processing by two forms of human DNA polymerase δ. DNA Repair (Amst), 2013. 12: p. 922-35. [CrossRef]
- Ayyagari, R., X.V. Gomes, D.A. Gordenin, and P.M. Burgers. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem, 2003. 278: p. 1618-25. [CrossRef]
- Bastos de Oliveira, F.M., D. Kim, J.R. Cussiol, J. Das, M.C. Jeong, L. Doerfler, K.H. Schmidt, H. Yu, and M.B. Smolka. Phosphoproteomics reveals distinct modes of Mec1/ATR signaling during DNA replication. Mol Cell, 2015. 57: p. 1124-1132. [CrossRef]
- Mutreja, K., J. Krietsch, J. Hess, S. Ursich, M. Berti, F.K. Roessler, R. Zellweger, M. Patra, G. Gasser, and M. Lopes. ATR-Mediated Global Fork Slowing and Reversal Assist Fork Traverse and Prevent Chromosomal Breakage at DNA Interstrand Cross-Links. Cell Rep, 2018. 24: p. 2629-2642.e5. [CrossRef]
- Lanz, M.C., S. Oberly, E.J. Sanford, S. Sharma, A. Chabes, and M.B. Smolka. Separable roles for Mec1/ATR in genome maintenance, DNA replication, and checkpoint signaling. Genes Dev, 2018. 32: p. 822-835. [CrossRef]
- Michael, W.M., R. Ott, E. Fanning, and J. Newport. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science, 2000. 289: p. 2133-2137. [CrossRef]
- Yan, S. Yan, S. and W.M. Michael. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle, 2009. 8: p. 2877-84. [CrossRef]
- Saldivar, J.C., D. Cortez, and K.A. Cimprich. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol, 2017. 18: p. 622-636. [CrossRef]
- Hadjicharalambous, A., A.J. Whale, G. Can, J.M. Skehel, J.M. Houseley, and P. Zegerman. Checkpoint kinase interaction with DNA polymerase alpha regulates replication progression during stress. Wellcome Open Res, 2023. 8: p. 327. [CrossRef]
- McClure, A.W., B. Canal, and J.F.X. Diffley. A DNA replication fork-centric view of the budding yeast DNA damage response. DNA Repair (Amst), 2022. 119: p. 103393. [CrossRef]
- Nam, E.A. and D. Cortez. ATR signalling: more than meeting at the fork. Biochem J, 2011. 436: p. 527-36. [CrossRef]
- Yan, S. and W.M. Michael. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol, 2009. 184: p. 793-804. [CrossRef]
- Labib, K. and A. Gambus. A key role for the GINS complex at DNA replication forks. Trends Cell Biol, 2007. 17: p. 271-8. [CrossRef]
- O’Donnell, M. and H. Li. The Eukaryotic Replisome Goes Under the Microscope. Curr Biol, 2016. 26: p. R247-56. [CrossRef]
- Grabarczyk, D.B. The Fork Protection Complex: A Regulatory Hub at the Head of the Replisome. Subcell Biochem, 2022. 99: p. 83-107. [CrossRef]
- Bell, S.P. and K. Labib. Chromosome Duplication in Saccharomyces cerevisiae. Genetics, 2016. 203: p. 1027-67. [CrossRef]
- Liao, H., F. Ji, T. Helleday, and S. Ying. Mechanisms for stalled replication fork stabilization: new targets for synthetic lethality strategies in cancer treatments. EMBO Rep, 2018. 19. [CrossRef]
- Simoneau, A. and L. Zou. An extending ATR-CHK1 circuitry: the replication stress response and beyond. Curr Opin Genet Dev, 2021. 71: p. 92-98. [CrossRef]
- Bainbridge, L.J., R. Teague, and A.J. Doherty. Repriming DNA synthesis: an intrinsic restart pathway that maintains efficient genome replication. Nucleic Acids Res, 2021. 49: p. 4831-4847. [CrossRef]
- Kieffer, S.R. and N.F. Lowndes. Immediate-Early, Early, and Late Responses to DNA Double Stranded Breaks. Front Genet, 2022. 13: p. 793884. [CrossRef]
- Hustedt, N., S.M. Gasser, and K. Shimada. Replication checkpoint: tuning and coordination of replication forks in s phase. Genes (Basel), 2013. 4: p. 388-434. [CrossRef]
- Tye, S., G.E. Ronson, and J.R. Morris. A fork in the road: Where homologous recombination and stalled replication fork protection part ways. Semin Cell Dev Biol, 2021. 113: p. 14-26. [CrossRef]
- Kondratick, C.M., M.T. Washington, and M. Spies. Making Choices: DNA Replication Fork Recovery Mechanisms. Semin Cell Dev Biol, 2021. 113: p. 27-37. [CrossRef]
- Petermann, E. and K.W. Caldecott. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle, 2006. 5: p. 2203-9. [CrossRef]
- Castaneda, J.C., M. Schrecker, D. Remus, and R.K. Hite. Mechanisms of loading and release of the 9-1-1 checkpoint clamp. Nat Struct Mol Biol, 2022. 29: p. 369-375. [CrossRef]
- Majka, J., S.K. Binz, M.S. Wold, and P.M. Burgers. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem, 2006. 281: p. 27855-61. [CrossRef]
- Majka, J. and P.M. Burgers. Function of Rad17/Mec3/Ddc1 and its partial complexes in the DNA damage checkpoint. DNA Repair (Amst), 2005. 4: p. 1189-94. [CrossRef]
- Majka, J. and P.M. Burgers. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol, 2004. 78: p. 227-60. [CrossRef]
- Green, C.M., H. Erdjument-Bromage, P. Tempst, and N.F. Lowndes. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol, 2000. 10: p. 39-42. [CrossRef]
- Van, C., S. Yan, W.M. Michael, S. Waga, and K.A. Cimprich. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J Cell Biol, 2010. 189: p. 233-46. [CrossRef]
- Mordes, D.A., G.G. Glick, R. Zhao, and D. Cortez. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev, 2008. 22: p. 1478-89. [CrossRef]
- Zabrady, K., A.W.H. Li, and A.J. Doherty. Mechanism of primer synthesis by Primase-Polymerases. Curr Opin Struct Biol, 2023. 82: p. 102652. [CrossRef]
- Wang, F., J.A. Stewart, C. Kasbek, Y. Zhao, W.E. Wright, and C.M. Price. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep, 2012. 2: p. 1096-103. [CrossRef]
- Stewart, J.A., F. Wang, M.F. Chaiken, C. Kasbek, P.D. Chastain, 2nd, W.E. Wright, and C.M. Price. Human CST promotes telomere duplex replication and general replication restart after fork stalling. Embo j, 2012. 31: p. 3537-49. [CrossRef]
- Blow, J.J., X.Q. Ge, and D.A. Jackson. How dormant origins promote complete genome replication. Trends Biochem Sci, 2011. 36: p. 405-14. [CrossRef]
- Blow, J.J. and X.Q. Ge. Replication forks, chromatin loops and dormant replication origins. Genome Biol, 2008. 9: p. 244. [CrossRef]
- Woodward, A.M., T. Göhler, M.G. Luciani, M. Oehlmann, X. Ge, A. Gartner, D.A. Jackson, and J.J. Blow. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol, 2006. 173: p. 673-83. [CrossRef]
- Pabla, N., S. Huang, Q.S. Mi, R. Daniel, and Z. Dong. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J Biol Chem, 2008. 283: p. 6572-83. [CrossRef]
- Toledo, L.I., M. Murga, P. Gutierrez-Martinez, R. Soria, and O. Fernandez-Capetillo. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev, 2008. 22: p. 297-302. [CrossRef]
- Olovnikov, A.M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol, 1973. 41: p. 181-90. [CrossRef]
- de Lange, T. Protection of mammalian telomeres. Oncogene, 2002. 21: p. 532-540. [CrossRef]
- Takai, H., V. Aria, P. Borges, J.T.P. Yeeles, and T. de Lange. CST-Polymeraseα-primase solves a second telomere end-replication problem. bioRxiv, 2023. [CrossRef]
- Lim, C.J. and T.R. Cech. Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization. Nat Rev Mol Cell Biol, 2021. 22: p. 283-298. [CrossRef]
- Greider, C.W. and E.H. Blackburn. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell, 1987. 51: p. 887-98. [CrossRef]
- Szostak, J.W. and E.H. Blackburn. Cloning yeast telomeres on linear plasmid vectors. Cell, 1982. 29: p. 245-55. [CrossRef]
- Ohki, R., T. Tsurimoto, and F. Ishikawa. In vitro reconstitution of the end replication problem. Mol Cell Biol, 2001. 21: p. 5753-66. [CrossRef]
- de Lange, T. Shelterin-Mediated Telomere Protection. Annu Rev Genet, 2018. 52: p. 223-247. [CrossRef]
- Ganduri, S. and N.F. Lue. STN1-POLA2 interaction provides a basis for primase-pol α stimulation by human STN1. Nucleic Acids Research, 2017. 45: p. 9455-9466. [CrossRef]
- Anderson, B.H., P.R. Kasher, J. Mayer, M. Szynkiewicz, E.M. Jenkinson, S.S. Bhaskar, J.E. Urquhart, S.B. Daly, J.E. Dickerson, J. O’Sullivan, E.O. Leibundgut, J. Muter, G.M. Abdel-Salem, R. Babul-Hirji, P. Baxter, A. Berger, L. Bonafé, J.E. Brunstom-Hernandez, J.A. Buckard, D. Chitayat, W.K. Chong, D.M. Cordelli, P. Ferreira, J. Fluss, E.H. Forrest, E. Franzoni, C. Garone, S.R. Hammans, G. Houge, I. Hughes, S. Jacquemont, P.Y. Jeannet, R.J. Jefferson, R. Kumar, G. Kutschke, S. Lundberg, C.M. Lourenço, R. Mehta, S. Naidu, K.K. Nischal, L. Nunes, K. Ounap, M. Philippart, P. Prabhakar, S.R. Risen, R. Schiffmann, C. Soh, J.B. Stephenson, H. Stewart, J. Stone, J.L. Tolmie, M.S. van der Knaap, J.P. Vieira, C.N. Vilain, E.L. Wakeling, V. Wermenbol, A. Whitney, S.C. Lovell, S. Meyer, J.H. Livingston, G.M. Baerlocher, G.C. Black, G.I. Rice, and Y.J. Crow. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet, 2012. 44: p. 338-42. [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov, 2022. 12: p. 31-46. [CrossRef]
- Hanahan, D. and R.A. Weinberg. Hallmarks of cancer: the next generation. Cell, 2011. 144: p. 646-74. [CrossRef]
- Hanahan, D. and R.A. Weinberg. The hallmarks of cancer. Cell, 2000. 100: p. 57-70. [CrossRef]
- Fenwick, A.L., M. Kliszczak, F. Cooper, J. Murray, L. Sanchez-Pulido, S.R. Twigg, A. Goriely, S.J. McGowan, K.A. Miller, I.B. Taylor, C. Logan, S. Bozdogan, S. Danda, J. Dixon, S.M. Elsayed, E. Elsobky, A. Gardham, M.J. Hoffer, M. Koopmans, D.M. McDonald-McGinn, G.W. Santen, R. Savarirayan, D. de Silva, O. Vanakker, S.A. Wall, L.C. Wilson, O.O. Yuregir, E.H. Zackai, C.P. Ponting, A.P. Jackson, A.O. Wilkie, W. Niedzwiedz, and L.S. Bicknell. Mutations in CDC45, Encoding an Essential Component of the Pre-initiation Complex, Cause Meier-Gorlin Syndrome and Craniosynostosis. Am J Hum Genet, 2016. 99: p. 125-38. [CrossRef]
- Piersimoni, L., P.L. Kastritis, C. Arlt, and A. Sinz. Cross-Linking Mass Spectrometry for Investigating Protein Conformations and Protein–Protein Interactions─A Method for All Seasons. Chemical Reviews, 2022. 122: p. 7500-7531. [CrossRef]
- Munden, A., M.T. Wright, D. Han, R. Tirgar, L. Plate, and J.T. Nordman. Identification of replication fork-associated proteins in Drosophila embryos and cultured cells using iPOND coupled to quantitative mass spectrometry. Sci Rep, 2022. 12: p. 6903. [CrossRef]
- Lenz, S., L.R. Sinn, F.J. O’Reilly, L. Fischer, F. Wegner, and J. Rappsilber. Reliable identification of protein-protein interactions by crosslinking mass spectrometry. Nature Communications, 2021. 12: p. 3564. [CrossRef]
- Ho, B., A. Baryshnikova, and G.W. Brown. Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces cerevisiae Proteome. Cell Syst, 2018. 6: p. 192-205.e3. [CrossRef]
- Petryk, N., M. Kahli, Y. d’Aubenton-Carafa, Y. Jaszczyszyn, Y. Shen, M. Silvain, C. Thermes, C.L. Chen, and O. Hyrien. Replication landscape of the human genome. Nat Commun, 2016. 7: p. 10208. [CrossRef]
- Tanigawa, K., Y. Tomioka, S. Misono, S. Asai, N. Kikkawa, Y. Hagihara, T. Suetsugu, H. Inoue, K. Mizuno, and N. Seki. Minichromosome maintenance proteins in lung adenocarcinoma: Clinical significance and therapeutic targets. FEBS Open Bio, 2023. 13: p. 1737-1755. [CrossRef]
- Mukherjee, G., B. Muralidhar, U.D. Bafna, R.A. Laskey, and N. Coleman. MCM immunocytochemistry as a first line cervical screening test in developing countries: a prospective cohort study in a regional cancer centre in India. Br J Cancer, 2007. 96: p. 1107-11. [CrossRef]
- Scarpini, C., V. White, B. Muralidhar, A. Patterson, N. Hickey, N. Singh, J. Mullerat, M. Winslet, R.J. Davies, M.L. Phillips, P. Stacey, R.A. Laskey, R. Miller, M. Nathan, and N. Coleman. Improved screening for anal neoplasia by immunocytochemical detection of minichromosome maintenance proteins. Cancer Epidemiol Biomarkers Prev, 2008. 17: p. 2855-64. [CrossRef]
- Simon, N.E. and A. Schwacha. The Mcm2-7 replicative helicase: a promising chemotherapeutic target. Biomed Res Int, 2014. 2014: p. 549719. [CrossRef]
- Montagnoli, A., P. Tenca, F. Sola, D. Carpani, D. Brotherton, C. Albanese, and C. Santocanale. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res, 2004. 64: p. 7110-7116. [CrossRef]
- Swords, R., D. Mahalingam, M. O’Dwyer, C. Santocanale, K. Kelly, J. Carew, and F. Giles. Cdc7 kinase—a new target for drug development. Eur J Cancer, 2010. 46: p. 33-40. [CrossRef]
- Menichincheri, M., A. Bargiotti, J. Berthelsen, J.A. Bertrand, R. Bossi, A. Ciavolella, A. Cirla, C. Cristiani, V. Croci, R. D’Alessio, M. Fasolini, F. Fiorentini, B. Forte, A. Isacchi, K. Martina, A. Molinari, A. Montagnoli, P. Orsini, F. Orzi, E. Pesenti, D. Pezzetta, A. Pillan, I. Poggesi, F. Roletto, A. Scolaro, M. Tatò, M. Tibolla, B. Valsasina, M. Varasi, D. Volpi, C. Santocanale, and E. Vanotti. First Cdc7 kinase inhibitors: pyrrolopyridinones as potent and orally active antitumor agents. 2. Lead discovery. J Med Chem, 2009. 52: p. 293-307. [CrossRef]
- Iwai, K., T. Nambu, R. Dairiki, M. Ohori, J. Yu, K. Burke, M. Gotou, Y. Yamamoto, S. Ebara, S. Shibata, R. Hibino, S. Nishizawa, T. Miyazaki, M. Homma, Y. Oguro, T. Imada, N. Cho, N. Uchiyama, A. Kogame, T. Takeuchi, O. Kurasawa, K. Yamanaka, H. Niu, and A. Ohashi. Molecular mechanism and potential target indication of TAK-931, a novel CDC7-selective inhibitor. Sci Adv, 2019. 5: p. eaav3660. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




