Submitted:
25 February 2024
Posted:
26 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction: STAT transcription factors
2. Inappropriate activation of STATs in cancer
2.2. STAT3
2.3. STAT5
2.4. Other STATs
3. Targeting STATs for cancer therapy
4. Strategies to directly target STATs
4.1. Direct STAT binding molecules
4.2. STAT degraders
5. Targeting upstream kinases: STAT phosphorylation as a biomarker for on-target effects
5.1. Inhibiting STAT5 in chronic myeloid leukemia
5.2. Targeting kinases upstream of STATs in AML
6. Novel ways to identify STAT transcriptional inhibitors
6.1. Chemical Biology approaches
6.2. Computational approaches leveraging transcriptional signatures
7. Conclusions and future directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- “Global Cancer Facts & Figures.” Accessed: Aug. 24, 2023. [Online]. Available: https://www.cancer.org/research/cancer-facts-statistics/global.html.
- H. B. Sadowski, K. Shuai, J. E. Darnell, and M. Z. Gilman, “A common nuclear signal transduction pathway activated by growth factor and cytokine receptors,” Science, vol. 261, no. 5129, pp. 1739–1744, Sep. 1993. [CrossRef]
- U. M. Wegenka et al., “The interleukin-6-activated acute-phase response factor is antigenically and functionally related to members of the signal transducer and activator of transcription (STAT) family,” Mol Cell Biol, vol. 14, no. 5, pp. 3186–3196, May 1994. [CrossRef]
- J. E. Darnell, I. M. Kerr, and G. R. Stark, “Jak-STAT Pathways and Transcriptional Activation in Response to IFNs and Other Extracellular Signaling Proteins,” Science, vol. 264, no. 5164, pp. 1415–1421, 1994. [CrossRef]
- A. Orlova et al., “Direct Targeting Options for STAT3 and STAT5 in Cancer,” Cancers (Basel), vol. 11, no. 12, p. 1930, Dec. 2019. [CrossRef]
- W. Wang et al., “The complementary roles of STAT3 and STAT1 in cancer biology: insights into tumor pathogenesis and therapeutic strategies,” Front Immunol, vol. 14, p. 1265818, 2023. [CrossRef]
- T. J. Mitchell and S. John, “Signal transducer and activator of transcription (STAT) signalling and T-cell lymphomas,” Immunology, vol. 114, no. 3, pp. 301–312, Mar. 2005. [CrossRef]
- A. Yoshimura, T. Naka, and M. Kubo, “SOCS proteins, cytokine signalling and immune regulation,” Nat Rev Immunol, vol. 7, no. 6, pp. 454–465, Jun. 2007. [CrossRef]
- C. D. Chung et al., “Specific Inhibition of Stat3 Signal Transduction by PIAS3,” Science, vol. 278, no. 5344, pp. 1803–1805, Dec. 1997. [CrossRef]
- L. N. Heppler and D. A. Frank, “Rare mutations provide unique insight into oncogenic potential of STAT transcription factors,” J Clin Invest, vol. 128, no. 1, pp. 113–115, Jan. 2018. [CrossRef]
- M. Bar-Natan, E. A. Nelson, M. Xiang, and D. A. Frank, “STAT signaling in the pathogenesis and treatment of myeloid malignancies,” JAKSTAT, vol. 1, no. 2, pp. 55–64, Apr. 2012. [CrossRef]
- D. A. Frank, “STAT signaling in the pathogenesis and treatment of cancer,” Mol Med, vol. 5, no. 7, pp. 432–456, Jul. 1999. [CrossRef]
- V. Boudny and J. Kovarik, “JAK/STAT signaling pathways and cancer. Janus kinases/signal transducers and activators of transcription,” Neoplasma, vol. 49, no. 6, pp. 349–355, 2002.
- R. Catlett-Falcone et al., “Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells,” Immunity, vol. 10, no. 1, pp. 105–115, Jan. 1999. [CrossRef]
- J. F. Bromberg et al., “Stat3 as an Oncogene,” Cell, vol. 98, no. 3, pp. 295–303, Aug. 1999. [CrossRef]
- H. Lee, A. J. Jeong, and S.-K. Ye, “Highlighted STAT3 as a potential drug target for cancer therapy,” BMB Rep, vol. 52, no. 7, pp. 415–423, Jul. 2019. [CrossRef]
- T. Xia et al., “Advances in the role of STAT3 in macrophage polarization,” Frontiers in Immunology, vol. 14, 2023, Accessed: Nov. 29, 2023. [Online]. Available: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1160719.
- J. R. Grandis et al., “Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo,” Proceedings of the National Academy of Sciences, vol. 97, no. 8, pp. 4227–4232, Apr. 2000. [CrossRef]
- J. Bromberg, “Stat proteins and oncogenesis,” J Clin Invest, vol. 109, no. 9, pp. 1139–1142, May 2002. [CrossRef]
- H. Lee et al., “Persistently Activated Stat3 Maintains Constitutive NF-κB Activity in Tumors,” Cancer Cell, vol. 15, no. 4, pp. 283–293, Apr. 2009. [CrossRef]
- K. Berthenet et al., “HSP110 promotes colorectal cancer growth through STAT3 activation,” Oncogene, vol. 36, no. 16, pp. 2328–2336, Apr. 2017. [CrossRef]
- K.-W. Hsu et al., “Activation of the Notch1/STAT3/Twist signaling axis promotes gastric cancer progression,” Carcinogenesis, vol. 33, no. 8, pp. 1459–1467, Aug. 2012. [CrossRef]
- Y. Niwa et al., “Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma,” Oncogene, vol. 24, no. 42, pp. 6406–6417, Sep. 2005. [CrossRef]
- H. Isomoto et al., “Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing,” Gastroenterology, vol. 132, no. 1, pp. 384–396, Jan. 2007. [CrossRef]
- H. Ogata et al., “Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta1 production,” Oncogene, vol. 25, no. 17, pp. 2520–2530, Apr. 2006. [CrossRef]
- M. Z. Kamran, P. Patil, and R. P. Gude, “Role of STAT3 in Cancer Metastasis and Translational Advances,” Biomed Res Int, vol. 2013, p. 421821, 2013. [CrossRef]
- M. Tolomeo and A. Cascio, “The Multifaced Role of STAT3 in Cancer and Its Implication for Anticancer Therapy,” Int J Mol Sci, vol. 22, no. 2, p. 603, Jan. 2021. [CrossRef]
- L. Lin et al., “STAT3 is necessary for proliferation and survival in colon cancer-initiating cells,” Cancer Res, vol. 71, no. 23, pp. 7226–7237, Dec. 2011. [CrossRef]
- G. Niu et al., “Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis,” Oncogene, vol. 21, no. 13, pp. 2000–2008, Mar. 2002. [CrossRef]
- N. Priego et al., “STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis,” Nat Med, vol. 24, no. 7, pp. 1024–1035, Jul. 2018. [CrossRef]
- Q. Xu et al., “Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways,” Oncogene, vol. 24, no. 36, pp. 5552–5560, Aug. 2005. [CrossRef]
- S. K. Tripathi et al., “Genome-wide Analysis of STAT3-Mediated Transcription during Early Human Th17 Cell Differentiation,” Cell Reports, vol. 19, no. 9, pp. 1888–1901, May 2017. [CrossRef]
- J. Odajima et al., “Full oncogenic activities of v-Src are mediated by multiple signaling pathways. Ras as an essential mediator for cell survival,” J Biol Chem, vol. 275, no. 31, pp. 24096–24105, Aug. 2000. [CrossRef]
- J. Luo, R. Yan, X. He, and J. He, “Constitutive activation of STAT3 and cyclin D1 overexpression contribute to proliferation, migration and invasion in gastric cancer cells,” Am J Transl Res, vol. 9, no. 12, pp. 5671–5677, Dec. 2017.
- C. Yue et al., “STAT3 in CD8+ T Cells Inhibits Their Tumor Accumulation by Downregulating CXCR3/CXCL10 Axis,” Cancer Immunol Res, vol. 3, no. 8, pp. 864–870, Aug. 2015. [CrossRef]
- T. Wang et al., “Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells,” Nat Med, vol. 10, no. 1, pp. 48–54, Jan. 2004. [CrossRef]
- J.-P. Herbeuval, E. Lelievre, C. Lambert, M. Dy, and C. Genin, “Recruitment of STAT3 for production of IL-10 by colon carcinoma cells induced by macrophage-derived IL-6,” J Immunol, vol. 172, no. 7, pp. 4630–4636, Apr. 2004. [CrossRef]
- T.-X. Xie et al., “Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis,” Oncogene, vol. 23, no. 20, pp. 3550–3560, Apr. 2004. [CrossRef]
- H.-W. Lo et al., “Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression,” Cancer Res, vol. 67, no. 19, pp. 9066–9076, Oct. 2007. [CrossRef]
- T. N. Dechow et al., “Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C,” Proc Natl Acad Sci U S A, vol. 101, no. 29, pp. 10602–10607, Jul. 2004. [CrossRef]
- Y. Wu, I. Diab, X. Zhang, E. S. Izmailova, and Z. E. Zehner, “Stat3 enhances vimentin gene expression by binding to the antisilencer element and interacting with the repressor protein, ZBP-89,” Oncogene, vol. 23, no. 1, pp. 168–178, Jan. 2004. [CrossRef]
- R. Garcia et al., “Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells,” Oncogene, vol. 20, no. 20, pp. 2499–2513, May 2001. [CrossRef]
- J. V. Alvarez et al., “A STAT3 Gene Expression Signature in Gliomas is Associated with a Poor Prognosis,” Transl Oncogenomics, vol. 2, pp. 99–105, 2007. [CrossRef]
- R. L. Carpenter and H.-W. Lo, “STAT3 Target Genes Relevant to Human Cancers,” Cancers (Basel), vol. 6, no. 2, pp. 897–925, Apr. 2014. [CrossRef]
- S. H. Swerdlow et al., “The 2016 revision of the World Health Organization classification of lymphoid neoplasms,” Blood, vol. 127, no. 20, pp. 2375–2390, May 2016. [CrossRef]
- A. Moignet and T. Lamy, “Latest Advances in the Diagnosis and Treatment of Large Granular Lymphocytic Leukemia,” Am Soc Clin Oncol Educ Book, vol. 38, pp. 616–625, May 2018. [CrossRef]
- T. Lamy, A. Moignet, and T. P. Loughran, “LGL leukemia: from pathogenesis to treatment,” Blood, vol. 129, no. 9, pp. 1082–1094, Mar. 2017. [CrossRef]
- A. Fasan, W. Kern, V. Grossmann, C. Haferlach, T. Haferlach, and S. Schnittger, “STAT3 mutations are highly specific for large granular lymphocytic leukemia,” Leukemia, vol. 27, no. 7, pp. 1598–1600, Jul. 2013. [CrossRef]
- H. L. M. Koskela et al., “Somatic STAT3 Mutations in Large Granular Lymphocytic Leukemia,” N Engl J Med, vol. 366, no. 20, pp. 1905–1913, May 2012. [CrossRef]
- E. Andersson et al., “Activating somatic mutations outside the SH2-domain of STAT3 in LGL-Leukemia,” Leukemia, vol. 30, no. 5, pp. 1204–1208, May 2016. [CrossRef]
- G. Semenzato, A. Teramo, G. Calabretto, V. R. Gasparini, and R. Zambello, “All that glitters is not LGL Leukemia,” Leukemia, vol. 36, no. 11, pp. 2551–2557, Nov. 2022. [CrossRef]
- A. Dutta, D. Yan, R. E. Hutchison, and G. Mohi, “STAT3 mutations are not sufficient to induce large granular lymphocytic leukaemia in mice,” Br J Haematol, vol. 180, no. 6, pp. 911–915, Mar. 2018. [CrossRef]
- A. Teramo et al., “STAT3 mutation impacts biological and clinical features of T-LGL leukemia,” Oncotarget, vol. 8, no. 37, pp. 61876–61889, Jun. 2017. [CrossRef]
- T. P. Loughran et al., “Immunosuppressive therapy of LGL leukemia: prospective multicenter phase II study by the Eastern Cooperative Oncology Group (E5998),” Leukemia, vol. 29, no. 4, pp. 886–894, Apr. 2015. [CrossRef]
- E. Masle-Farquhar et al., “STAT3 gain-of-function mutations connect leukemia with autoimmune disease by pathological NKG2Dhi CD8+ T cell dysregulation and accumulation,” Immunity, vol. 55, no. 12, pp. 2386-2404.e8, Dec. 2022. [CrossRef]
- S. Grivennikov et al., “IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer,” Cancer Cell, vol. 15, no. 2, pp. 103–113, Feb. 2009. [CrossRef]
- H. Kitamura et al., “Interleukin-6/STAT3 signaling as a promising target to improve the efficacy of cancer immunotherapy,” Cancer Science, vol. 108, no. 10, pp. 1947–1952, 2017. [CrossRef]
- F. Cheng et al., “A critical role for Stat3 signaling in immune tolerance,” Immunity, vol. 19, no. 3, pp. 425–436, Sep. 2003. [CrossRef]
- M. L. McLemore et al., “STAT-3 activation is required for normal G-CSF-dependent proliferation and granulocytic differentiation,” Immunity, vol. 14, no. 2, pp. 193–204, Feb. 2001. [CrossRef]
- H. Zhang, H. Nguyen-Jackson, A. D. Panopoulos, H. S. Li, P. J. Murray, and S. S. Watowich, “STAT3 controls myeloid progenitor growth during emergency granulopoiesis,” Blood, vol. 116, no. 14, pp. 2462–2471, Oct. 2010. [CrossRef]
- C. Lee et al., “STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation,” Immunity, vol. 17, no. 1, pp. 63–72, Jul. 2002. [CrossRef]
- W.-C. Chou, D. E. Levy, and C.-K. Lee, “STAT3 positively regulates an early step in B-cell development,” Blood, vol. 108, no. 9, pp. 3005–3011, Nov. 2006. [CrossRef]
- S. A. Diehl, H. Schmidlin, M. Nagasawa, B. Blom, and H. Spits, “IL-6 triggers IL-21 production by human CD4+ T cells to drive STAT3-dependent plasma cell differentiation in B cells,” Immunol Cell Biol, vol. 90, no. 8, pp. 802–811, Sep. 2012. [CrossRef]
- L. Durant et al., “Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis,” Immunity, vol. 32, no. 5, pp. 605–615, May 2010. [CrossRef]
- S. M. Kaech and W. Cui, “Transcriptional control of effector and memory CD8+ T cell differentiation,” Nat Rev Immunol, vol. 12, no. 11, pp. 749–761, Nov. 2012. [CrossRef]
- X. Y. Fu, “STAT3 in immune responses and inflammatory bowel diseases,” Cell Res, vol. 16, no. 2, Art. no. 2, Feb. 2006. [CrossRef]
- Z. Wang and K. D. Bunting, “STAT5 in hematopoietic stem cell biology and transplantation,” JAKSTAT, vol. 2, no. 4, p. e27159, Oct. 2013. [CrossRef]
- K. D. Bunting, H. L. Bradley, T. S. Hawley, R. Moriggl, B. P. Sorrentino, and J. N. Ihle, “Reduced lymphomyeloid repopulating activity from adult bone marrow and fetal liver of mice lacking expression of STAT5,” Blood, vol. 99, no. 2, pp. 479–487, Jan. 2002. [CrossRef]
- Z. Wang, G. Li, W. Tse, and K. D. Bunting, “Conditional deletion of STAT5 in adult mouse hematopoietic stem cells causes loss of quiescence and permits efficient nonablative stem cell replacement,” Blood, vol. 113, no. 20, pp. 4856–4865, May 2009. [CrossRef]
- G. Li, Z. Wang, K. L. Miskimen, Y. Zhang, W. Tse, and K. D. Bunting, “Gab2 Promotes Hematopoietic Stem Cell Maintenance and Self-Renewal Synergistically with STAT5,” PLoS One, vol. 5, no. 2, p. e9152, Feb. 2010. [CrossRef]
- J.-X. Lin et al., “Critical functions for STAT5 tetramers in the maturation and survival of natural killer cells,” Nat Commun, vol. 8, no. 1, p. 1320, Nov. 2017. [CrossRef]
- L. M. Heltemes-Harris et al., “Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia,” J Exp Med, vol. 208, no. 6, pp. 1135–1149, Jun. 2011. [CrossRef]
- M. A. Burchill et al., “Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells,” J Immunol, vol. 171, no. 11, pp. 5853–5864, Dec. 2003. [CrossRef]
- S. L. Nutt and B. L. Kee, “The Transcriptional Regulation of B Cell Lineage Commitment,” Immunity, vol. 26, no. 6, pp. 715–725, Jun. 2007. [CrossRef]
- T. W. Hand et al., “Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival,” Proceedings of the National Academy of Sciences, vol. 107, no. 38, pp. 16601–16606, Sep. 2010. [CrossRef]
- P. Tripathi et al., “STAT5 IS CRITICAL TO MAINTAIN EFFECTOR CD8+ T CELL RESPONSES,” J Immunol, vol. 185, no. 4, pp. 2116–2124, Aug. 2010. [CrossRef]
- J.-H. Park et al., “Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells,” Nat Immunol, vol. 11, no. 3, pp. 257–264, Mar. 2010. [CrossRef]
- T. Kanai et al., “Identification of STAT5A and STAT5B Target Genes in Human T Cells,” PLoS One, vol. 9, no. 1, p. e86790, Jan. 2014. [CrossRef]
- C. E. Halim, S. Deng, M. S. Ong, and C. T. Yap, “Involvement of STAT5 in Oncogenesis,” Biomedicines, vol. 8, no. 9, p. 316, Aug. 2020. [CrossRef]
- D. A. Frank, “StAT signaling in cancer: insights into pathogenesis and treatment strategies,” Cancer Treat Res, vol. 115, pp. 267–291, 2003. [CrossRef]
- E. A. Nelson, S. R. Walker, J. V. Alvarez, and D. A. Frank, “Isolation of Unique STAT5 Targets by Chromatin Immunoprecipitation-based Gene Identification *,” Journal of Biological Chemistry, vol. 279, no. 52, pp. 54724–54730, Dec. 2004. [CrossRef]
- Y. Verhoeven et al., “The potential and controversy of targeting STAT family members in cancer,” Seminars in Cancer Biology, vol. 60, pp. 41–56, Feb. 2020. [CrossRef]
- W. Lin, J. W. Schmidt, B. A. Creamer, A. A. Triplett, and K.-U. Wagner, “Gain-of-Function of Stat5 Leads to Excessive Granulopoiesis and Lethal Extravasation of Granulocytes to the Lung,” PLOS ONE, vol. 8, no. 4, p. e60902, Apr. 2013. [CrossRef]
- J. C. G. van der Zwet et al., “STAT5 does not drive steroid resistance in T-cell acute lymphoblastic leukemia despite the activation of BCL and BCLXL following glucocorticoid treatment,” Haematologica, vol. 108, no. 3, pp. 732–746, Jun. 2022. [CrossRef]
- I. Barash, “Stat5 in breast cancer: potential oncogenic activity coincides with positive prognosis for the disease,” Carcinogenesis, vol. 33, no. 12, pp. 2320–2325, Dec. 2012. [CrossRef]
- K.-U. Wagner and H. Rui, “Jak2/Stat5 Signaling in Mammogenesis, Breast Cancer Initiation and Progression,” J Mammary Gland Biol Neoplasia, vol. 13, no. 1, pp. 93–103, Mar. 2008. [CrossRef]
- “Stat5a is tyrosine phosphorylated and nuclear localized in a high proportion of human breast cancers - Cotarla - 2004 - International Journal of Cancer - Wiley Online Library.” Accessed: Jan. 07, 2024. [Online]. Available: https://onlinelibrary.wiley.com/doi/10.1002/ijc.11619.
- M. Lin et al., “STAT5 confers lactogenic properties in breast tumorigenesis and restricts metastatic potential,” Oncogene, vol. 41, no. 48, pp. 5214–5222, Nov. 2022. [CrossRef]
- S. R. Walker et al., “Reciprocal Effects of STAT5 and STAT3 in Breast Cancer,” Molecular Cancer Research, vol. 7, no. 6, pp. 966–976, Jun. 2009. [CrossRef]
- B. Wingelhofer et al., “Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer,” Leukemia, vol. 32, no. 8, Art. no. 8, Aug. 2018. [CrossRef]
- K. Basso and R. Dalla-Favera, “Germinal centres and B cell lymphomagenesis,” Nat Rev Immunol, vol. 15, no. 3, pp. 172–184, Mar. 2015. [CrossRef]
- S. R. Walker, E. A. Nelson, J. E. Yeh, L. Pinello, G.-C. Yuan, and D. A. Frank, “STAT5 Outcompetes STAT3 To Regulate the Expression of the Oncogenic Transcriptional Modulator BCL6,” Mol Cell Biol, vol. 33, no. 15, pp. 2879–2890, Aug. 2013. [CrossRef]
- “STAT1 signal transducer and activator of transcription 1 [Homo sapiens (human)] - Gene - NCBI.” Accessed: Jan. 07, 2024. [Online]. Available: https://www.ncbi.nlm.nih.gov/gene/6772.
- S. E. Dovhey, N. S. Ghosh, and K. L. Wright, “Loss of interferon-gamma inducibility of TAP1 and LMP2 in a renal cell carcinoma cell line,” Cancer Res, vol. 60, no. 20, pp. 5789–5796, Oct. 2000.
- B. Kovacic et al., “STAT1 acts as a tumor promoter for leukemia development,” Cancer Cell, vol. 10, no. 1, pp. 77–87, Jul. 2006. [CrossRef]
- C. Park, S. Li, E. Cha, and C. Schindler, “Immune Response in Stat2 Knockout Mice,” Immunity, vol. 13, no. 6, pp. 795–804, Dec. 2000. [CrossRef]
- S. Hambleton et al., “STAT2 deficiency and susceptibility to viral illness in humans,” Proceedings of the National Academy of Sciences, vol. 110, no. 8, pp. 3053–3058, Feb. 2013. [CrossRef]
- J. Canar, kennedy Darling, R. Dadey, and A. M. Gamero, “The duality of STAT2 mediated type I interferon signaling in the tumor microenvironment and chemoresistance,” Cytokine, vol. 161, p. 156081, Jan. 2023. [CrossRef]
- C.-J. Lee et al., “FBXW7-mediated stability regulation of signal transducer and activator of transcription 2 in melanoma formation,” Proceedings of the National Academy of Sciences, vol. 117, no. 1, pp. 584–594, Jan. 2020. [CrossRef]
- L. Zhou, Y. Li, Z. Li, and Q. Huang, “Mining therapeutic and prognostic significance of STATs in renal cell carcinoma with bioinformatics analysis,” Genomics, vol. 112, no. 6, pp. 4100–4114, Nov. 2020. [CrossRef]
- X.-Y. Zhou, H.-Y. Dai, H. Zhang, J.-L. Zhu, and H. Hu, “Signal transducer and activator of transcription family is a prognostic marker associated with immune infiltration in endometrial cancer,” Journal of Clinical Laboratory Analysis, vol. 36, no. 4, p. e24315, 2022. [CrossRef]
- Y.-C. Huang, J.-L. Huang, L.-C. Tseng, P.-H. Yu, S.-Y. Chen, and C.-S. Lin, “High Expression of Interferon Pathway Genes CXCL10 and STAT2 Is Associated with Activated T-Cell Signature and Better Outcome of Oral Cancer Patients,” J Pers Med, vol. 12, no. 2, p. 140, Jan. 2022. [CrossRef]
- K. B. Nguyen et al., “Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection,” Science, vol. 297, no. 5589, pp. 2063–2066, Sep. 2002. [CrossRef]
- C. Parham et al., “A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R,” J Immunol, vol. 168, no. 11, pp. 5699–5708, Jun. 2002. [CrossRef]
- C. M. Bacon et al., “Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes.,” Proc Natl Acad Sci U S A, vol. 92, no. 16, pp. 7307–7311, Aug. 1995. [CrossRef]
- L. Zhao et al., “An integrated analysis identifies STAT4 as a key regulator of ovarian cancer metastasis,” Oncogene, vol. 36, no. 24, pp. 3384–3396, Jun. 2017. [CrossRef]
- M. Nishi et al., “High STAT4 Expression Indicates Better Disease-free Survival in Patients with Gastric Cancer,” Anticancer Research, vol. 37, no. 12, pp. 6723–6729, Dec. 2017. [CrossRef]
- K. Anderson, N. Ryan, G. Volpedo, S. Varikuti, A. R. Satoskar, and S. Oghumu, “Immune Suppression Mediated by STAT4 Deficiency Promotes Lymphatic Metastasis in HNSCC,” Frontiers in Immunology, vol. 10, 2020, Accessed: Jan. 07, 2024. [Online]. Available: https://www.frontiersin.org/articles/10.3389/fimmu.2019.03095.
- S. Goenka and M. H. Kaplan, “Transcriptional regulation by STAT6,” Immunol Res, vol. 50, no. 1, pp. 87–96, May 2011. [CrossRef]
- J. Zhu and W. E. Paul, “CD4 T cells: fates, functions, and faults,” Blood, vol. 112, no. 5, pp. 1557–1569, Sep. 2008. [CrossRef]
- K. Binnemars-Postma, R. Bansal, G. Storm, and J. Prakash, “Targeting the Stat6 pathway in tumor-associated macrophages reduces tumor growth and metastatic niche formation in breast cancer,” FASEB J, vol. 32, no. 2, pp. 969–978, Feb. 2018. [CrossRef]
- Y. Delgado-Ramirez, V. Colly, G. V. Gonzalez, and S. Leon-Cabrera, “Signal transducer and activator of transcription 6 as a target in colon cancer therapy,” Oncol Lett, vol. 20, no. 1, pp. 455–464, Jul. 2020. [CrossRef]
- A. B. Di Stefano et al., “Survivin is regulated by interleukin-4 in colon cancer stem cells,” J Cell Physiol, vol. 225, no. 2, pp. 555–561, Nov. 2010. [CrossRef]
- A. Jayakumar and A. L. M. Bothwell, “Stat6 Promotes Intestinal Tumorigenesis in a Mouse Model of Adenomatous Polyposis by Expansion of MDSCs and Inhibition of Cytotoxic CD8 Response,” Neoplasia, vol. 19, no. 8, pp. 595–605, Aug. 2017. [CrossRef]
- J. Iqbal et al., “Nanomedicines for developing cancer nanotherapeutics: from benchtop to bedside and beyond,” Appl Microbiol Biotechnol, vol. 102, no. 22, pp. 9449–9470, Nov. 2018. [CrossRef]
- D. Hong et al., “AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer,” Sci Transl Med, vol. 7, no. 314, p. 314ra185, Nov. 2015. [CrossRef]
- M. J. Reilley et al., “STAT3 antisense oligonucleotide AZD9150 in a subset of patients with heavily pretreated lymphoma: results of a phase 1b trial,” J Immunother Cancer, vol. 6, no. 1, p. 119, Nov. 2018. [CrossRef]
- L. Bai et al., “A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo,” Cancer Cell, vol. 36, no. 5, pp. 498-511.e17, Nov. 2019. [CrossRef]
- S. M. Holland et al., “STAT3 mutations in the hyper-IgE syndrome,” N Engl J Med, vol. 357, no. 16, pp. 1608–1619, Oct. 2007. [CrossRef]
- Y. Minegishi et al., “Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome,” Nature, vol. 448, no. 7157, Art. no. 7157, Aug. 2007. [CrossRef]
- D. E. Levy and C. A. Loomis, “STAT3 Signaling and the Hyper-IgE Syndrome,” New England Journal of Medicine, vol. 357, no. 16, pp. 1655–1658, Oct. 2007. [CrossRef]
- R. T. Rhodes, “Understanding Hyper IgE Syndrome,” 2020.
- M. Furqan, A. Akinleye, N. Mukhi, V. Mittal, Y. Chen, and D. Liu, “STAT inhibitors for cancer therapy,” Journal of Hematology & Oncology, vol. 6, no. 1, p. 90, Dec. 2013. [CrossRef]
- U. Bharadwaj, M. M. Kasembeli, and D. J. Tweardy, “STAT3 Inhibitors in Cancer: A Comprehensive Update,” in STAT Inhibitors in Cancer, A. C. Ward, Ed., in Cancer Drug Discovery and Development., Cham: Springer International Publishing, 2016, pp. 95–161. [CrossRef]
- K. S. Bhullar et al., “Kinase-targeted cancer therapies: progress, challenges and future directions,” Molecular Cancer, vol. 17, no. 1, p. 48, Feb. 2018. [CrossRef]
- P. Cohen, D. Cross, and P. A. Jänne, “Kinase drug discovery 20 years after imatinib: progress and future directions,” Nat Rev Drug Discov, vol. 20, no. 7, Art. no. 7, Jul. 2021. [CrossRef]
- C. M. Lovly and A. T. Shaw, “Molecular Pathways: Resistance to Kinase Inhibitors and Implications for Therapeutic Strategies,” Clin Cancer Res, vol. 20, no. 9, pp. 2249–2256, May 2014. [CrossRef]
- S. A. Rosenzweig, “Acquired Resistance to Drugs Targeting Tyrosine Kinases,” Adv Cancer Res, vol. 138, pp. 71–98, 2018. [CrossRef]
- E. Benedettini et al., “Met activation in non-small cell lung cancer is associated with de novo resistance to EGFR inhibitors and the development of brain metastasis,” Am J Pathol, vol. 177, no. 1, pp. 415–423, Jul. 2010. [CrossRef]
- S. Zou, Q. Tong, B. Liu, W. Huang, Y. Tian, and X. Fu, “Targeting STAT3 in Cancer Immunotherapy,” Molecular Cancer, vol. 19, no. 1, p. 145, Sep. 2020. [CrossRef]
- P. Demela, N. Pirastu, and B. Soskic, “Cross-disorder genetic analysis of immune diseases reveals distinct gene associations that converge on common pathways,” Nat Commun, vol. 14, no. 1, Art. no. 1, May 2023. [CrossRef]
- A. Sarapultsev, E. Gusev, M. Komelkova, I. Utepova, S. Luo, and D. Hu, “JAK-STAT signaling in inflammation and stress-related diseases: implications for therapeutic interventions,” Mol Biomed, vol. 4, p. 40, Nov. 2023. [CrossRef]
- A. N. Koehler, “A complex task? Direct modulation of transcription factors with small molecules,” Curr Opin Chem Biol, vol. 14, no. 3, pp. 331–340, Jun. 2010. [CrossRef]
- X. Xie et al., “Recent advances in targeting the ‘undruggable’ proteins: from drug discovery to clinical trials,” Sig Transduct Target Ther, vol. 8, no. 1, Art. no. 1, Sep. 2023. [CrossRef]
- X. Xu, M. M. Kasembeli, X. Jiang, B. J. Tweardy, and D. J. Tweardy, “Chemical probes that competitively and selectively inhibit Stat3 activation,” PLoS One, vol. 4, no. 3, p. e4783, 2009. [CrossRef]
- H. Chen et al., “Targeting STAT3 by a small molecule suppresses pancreatic cancer progression,” Oncogene, vol. 40, no. 8, p. 1440, 2021. [CrossRef]
- E. A. Nelson, S. V. Sharma, J. Settleman, and D. A. Frank, “A chemical biology approach to developing STAT inhibitors: molecular strategies for accelerating clinical translation,” Oncotarget, vol. 2, no. 6, pp. 518–524, Jun. 2011. [CrossRef]
- P. J. M. Jackson, S. Jamshidi, and D. B. Farag, “Computational Approaches in the Development of Small-molecule Transcription Factor Inhibitors,” Sep. 2018. [CrossRef]
- P. Yue and J. Turkson, “Targeting STAT3 in cancer: how successful are we?,” Expert Opin Investig Drugs, vol. 18, no. 1, pp. 45–56, Jan. 2009. [CrossRef]
- U. Hemmann et al., “Differential Activation of Acute Phase Response Factor/Stat3 and Stat1 via the Cytoplasmic Domain of the Interleukin 6 Signal Transducer gp130,” Journal of Biological Chemistry, vol. 271, no. 22, pp. 12999–13007, May 1996. [CrossRef]
- A. Diop et al., “SH2 Domains: Folding, Binding and Therapeutical Approaches,” Int J Mol Sci, vol. 23, no. 24, p. 15944, Dec. 2022. [CrossRef]
- U. Bharadwaj et al., “Small-molecule inhibition of STAT3 in radioresistant head and neck squamous cell carcinoma,” Oncotarget, vol. 7, no. 18, pp. 26307–26330, Mar. 2016. [CrossRef]
- R. A. Cerulli and J. A. Kritzer, “Phosphotyrosine Isosteres: Past, Present and Future,” Org Biomol Chem, vol. 18, no. 4, pp. 583–605, Jan. 2020. [CrossRef]
- Z. Shi et al., “Silibinin inhibits endometrial carcinoma via blocking pathways of STAT3 activation and SREBP1-mediated lipid accumulation,” Life Sci, vol. 217, pp. 70–80, Jan. 2019. [CrossRef]
- “BP-1-102 exerts antitumor effects on T-cell acute lymphoblastic leukemia cells by suppressing the JAK2/STAT3/c-Myc signaling pathway - PubMed.” Accessed: Feb. 17, 2024. [Online]. Available: https://pubmed.ncbi.nlm.nih.gov/37020528/.
- R. Zhang, X. Chen, S. Fu, L. Xu, and J. Lin, “A small molecule STAT3 inhibitor, LLL12, enhances cisplatin- and paclitaxel-mediated inhibition of cell growth and migration in human ovarian cancer cells,” Oncol Rep, vol. 44, no. 3, pp. 1224–1232, Sep. 2020. [CrossRef]
- J. R. Brown et al., “Targeting constitutively active STAT3 in chronic lymphocytic leukemia: A clinical trial of the STAT3 inhibitor pyrimethamine with pharmacodynamic analyses,” Am J Hematol, vol. 96, no. 4, pp. E95–E98, Apr. 2021. [CrossRef]
- “STAT3 Inhibitor OPB-51602 Is Cytotoxic to Tumor Cells Through Inhibition of Complex I and ROS Induction - PubMed.” Accessed: Feb. 17, 2024. [Online]. Available: https://pubmed.ncbi.nlm.nih.gov/33305182/.
- M. Skwarski et al., “Mitochondrial Inhibitor Atovaquone Increases Tumor Oxygenation and Inhibits Hypoxic Gene Expression in Patients with Non-Small Cell Lung Cancer,” Clin Cancer Res, vol. 27, no. 9, pp. 2459–2469, May 2021. [CrossRef]
- A. M. Stevens et al., “Atovaquone is active against AML by upregulating the integrated stress pathway and suppressing oxidative phosphorylation,” Blood Adv, vol. 3, no. 24, pp. 4215–4227, Dec. 2019. [CrossRef]
- X. Qi et al., “Trichothecin Inhibits Cancer-Related Features in Colorectal Cancer Development by Targeting STAT3,” Molecules, vol. 25, no. 10, p. 2306, May 2020. [CrossRef]
- L. H et al., “Design, synthesis, and biological characterization of a potent STAT3 degrader for the treatment of gastric cancer,” Frontiers in pharmacology, vol. 13, Aug. 2022. [CrossRef]
- A. Kaneshige et al., “A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo,” Nat Chem Biol, vol. 19, no. 6, pp. 703–711, Jun. 2023. [CrossRef]
- M. Roschewski et al., “Phase I Study of Acalabrutinib Plus Danvatirsen (AZD9150) in Relapsed/Refractory Diffuse Large B-Cell Lymphoma Including Circulating Tumor DNA Biomarker Assessment,” Clin Cancer Res, vol. 29, no. 17, pp. 3301–3312, Sep. 2023. [CrossRef]
- T. Nishina, T. Fujita, N. Yoshizuka, K. Sugibayashi, K. Murayama, and Y. Kuboki, “Safety, tolerability, pharmacokinetics and preliminary antitumour activity of an antisense oligonucleotide targeting STAT3 (danvatirsen) as monotherapy and in combination with durvalumab in Japanese patients with advanced solid malignancies: a phase 1 study,” BMJ Open, vol. 12, no. 10, p. e055718, Oct. 2022. [CrossRef]
- G. Casas, F. Perche, P. Midoux, C. Pichon, and J.-M. Malinge, “DNA minicircles as novel STAT3 decoy oligodeoxynucleotides endowed with anticancer activity in triple-negative breast cancer,” Mol Ther Nucleic Acids, vol. 29, pp. 162–175, Sep. 2022. [CrossRef]
- M. Aftabizadeh et al., “Potent antitumor effects of cell-penetrating peptides targeting STAT3 axis,” JCI Insight, vol. 6, no. 2, p. e136176. [CrossRef]
- P. Oleksak et al., “Design, synthesis, and in vitro evaluation of BP-1-102 analogs with modified hydrophobic fragments for STAT3 inhibition,” J Enzyme Inhib Med Chem, vol. 36, no. 1, pp. 410–424. [CrossRef]
- Y. Gu, I. S. Mohammad, and Z. Liu, “Overview of the STAT-3 signaling pathway in cancer and the development of specific inhibitors (Review),” Oncology Letters, vol. 19, no. 4, pp. 2585–2594, Apr. 2020. [CrossRef]
- D. Wang, M. Xu, F. Li, Y. Gao, and H. Sun, “Target Identification-Based Analysis of Mechanism of Betulinic Acid-Induced Cells Apoptosis of Cervical Cancer SiHa,” Natural Product Communications, vol. 17, no. 7, p. 1934578X221115528, Jul. 2022. [CrossRef]
- L. Zhao, J. Zhao, K. Zhong, A. Tong, and D. Jia, “Targeted protein degradation: mechanisms, strategies and application,” Sig Transduct Target Ther, vol. 7, no. 1, Art. no. 1, Apr. 2022. [CrossRef]
- K. M. Sakamoto, K. B. Kim, A. Kumagai, F. Mercurio, C. M. Crews, and R. J. Deshaies, “Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation,” Proceedings of the National Academy of Sciences, vol. 98, no. 15, pp. 8554–8559, Jul. 2001. [CrossRef]
- F. Potjewyd et al., “Degradation of Polycomb Repressive Complex 2 with an EED-Targeted Bivalent Chemical Degrader,” Cell Chem Biol, vol. 27, no. 1, pp. 47-56.e15, Jan. 2020. [CrossRef]
- S.-M. Qi, J. Dong, Z.-Y. Xu, X.-D. Cheng, W.-D. Zhang, and J.-J. Qin, “PROTAC: An Effective Targeted Protein Degradation Strategy for Cancer Therapy,” Front Pharmacol, vol. 12, p. 692574, May 2021. [CrossRef]
- M. He, W. Lv, and Y. Rao, “Opportunities and Challenges of Small Molecule Induced Targeted Protein Degradation,” Frontiers in Cell and Developmental Biology, vol. 9, 2021, Accessed: Nov. 29, 2023. [Online]. Available: https://www.frontiersin.org/articles/10.3389/fcell.2021.685106.
- H. Zhou et al., “Structure-Based Discovery of SD-36 as a Potent, Selective, and Efficacious PROTAC Degrader of STAT3 Protein,” J. Med. Chem., vol. 62, no. 24, pp. 11280–11300, Dec. 2019. [CrossRef]
- S. Kong et al., “SD-36 promotes growth inhibition and induces apoptosis via suppression of Mcl-1 in glioma,” J Cell Mol Med, vol. 25, no. 17, pp. 8261–8270, Sep. 2021. [CrossRef]
- H. Zhou et al., “SD-91 as A Potent and Selective STAT3 Degrader Capable of Achieving Complete and Long-Lasting Tumor Regression,” ACS Med Chem Lett, vol. 12, no. 6, pp. 996–1004, Jun. 2021. [CrossRef]
- A. Shastri, “Preliminary Safety, Pharmacokinetics, Pharmacodynamics and Clinical Activity of KT-333, a Targeted Protein Degrader of STAT3, in Patients with Relapsed or Refractory Lymphomas, Large Granular Lymphocytic Leukemia, and Solid Tumors,” presented at the 65th ASH Annual Meeting & Exposition, ASH, Dec. 2023. Accessed: Jan. 04, 2024. [Online]. Available: https://ash.confex.com/ash/2023/webprogram/Paper181130.html.
- J. M. Sasso, R. Tenchov, D. Wang, L. S. Johnson, X. Wang, and Q. A. Zhou, “Molecular Glues: The Adhesive Connecting Targeted Protein Degradation to the Clinic,” Biochemistry, vol. 62, no. 3, pp. 601–623, Jul. 2022. [CrossRef]
- G. Dong, Y. Ding, S. He, and C. Sheng, “Molecular Glues for Targeted Protein Degradation: From Serendipity to Rational Discovery,” J. Med. Chem., vol. 64, no. 15, pp. 10606–10620, Aug. 2021. [CrossRef]
- W. den Besten and J. R. Lipford, “Prospecting for molecular glues,” Nat Chem Biol, vol. 16, no. 11, pp. 1157–1158, Nov. 2020. [CrossRef]
- D. E. Johnson, R. A. O’Keefe, and J. R. Grandis, “Targeting the IL-6/JAK/STAT3 signalling axis in cancer,” Nat Rev Clin Oncol, vol. 15, no. 4, pp. 234–248, Apr. 2018. [CrossRef]
- Y. Ohsugi and N. Tsuchimoto, “[Pharmacological and clinical profile of anti-human IL-6 receptor antibody (tocilizumab, ACTEMRA), a novel therapeutic drug for Castleman’s disease],” Nihon Yakurigaku Zasshi, vol. 126, no. 6, pp. 419–425, Dec. 2005. [CrossRef]
- M. Sheppard, F. Laskou, P. P. Stapleton, S. Hadavi, and B. Dasgupta, “Tocilizumab (Actemra),” Hum Vaccin Immunother, vol. 13, no. 9, pp. 1972–1988, Sep. 2017. [CrossRef]
- R. Salgado et al., “Circulating interleukin-6 predicts survival in patients with metastatic breast cancer,” Int J Cancer, vol. 103, no. 5, pp. 642–646, Feb. 2003. [CrossRef]
- B. E. Lippitz and R. A. Harris, “Cytokine patterns in cancer patients: A review of the correlation between interleukin 6 and prognosis,” Oncoimmunology, vol. 5, no. 5, p. e1093722, May 2016. [CrossRef]
- X. Hu, J. Li, M. Fu, X. Zhao, and W. Wang, “The JAK/STAT signaling pathway: from bench to clinic,” Sig Transduct Target Ther, vol. 6, no. 1, Art. no. 1, Nov. 2021. [CrossRef]
- R. Morris, N. J. Kershaw, and J. J. Babon, “The molecular details of cytokine signaling via the JAK/STAT pathway,” Protein Science, vol. 27, no. 12, pp. 1984–2009, 2018. [CrossRef]
- D. M. Schwartz, Y. Kanno, A. Villarino, M. Ward, M. Gadina, and J. J. O’Shea, “JAK inhibition as a therapeutic strategy for immune and inflammatory diseases,” Nat Rev Drug Discov, vol. 17, no. 1, p. 78, Dec. 2017. [CrossRef]
- K. L. Winthrop and S. B. Cohen, “Oral surveillance and JAK inhibitor safety: the theory of relativity,” Nat Rev Rheumatol, vol. 18, no. 5, Art. no. 5, May 2022. [CrossRef]
- M. von Locquenghien, C. Rozalén, and T. Celià-Terrassa, “Interferons in cancer immunoediting: sculpting metastasis and immunotherapy response,” J Clin Invest, vol. 131, no. 1, p. e143296. [CrossRef]
- A. Ribas, W. N. Haining, and T. N. M. Schumacher, “When Cancer Cells Become the Enablers of an Antitumor Immune Response,” Cancer Discovery, vol. 12, no. 10, pp. 2244–2248, Oct. 2022. [CrossRef]
- J. V. Melo, “The molecular biology of chronic myeloid leukaemia,” Leukemia, vol. 10, no. 5, pp. 751–756, May 1996.
- D. A. Frank and L. Varticovski, “BCR/abl leads to the constitutive activation of Stat proteins, and shares an epitope with tyrosine phosphorylated Stats,” Leukemia, vol. 10, no. 11, pp. 1724–1730, Nov. 1996.
- N. Carlesso, D. A. Frank, and J. D. Griffin, “Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl,” J Exp Med, vol. 183, no. 3, pp. 811–820, Mar. 1996. [CrossRef]
- K. Yanagisawa, H. Yamauchi, M. Kaneko, H. Kohno, H. Hasegawa, and S. Fujita, “Suppression of cell proliferation and the expression of a bcr-abl fusion gene and apoptotic cell death in a new human chronic myelogenous leukemia cell line, KT-1, by interferon-alpha,” Blood, vol. 91, no. 2, pp. 641–648, Jan. 1998. [CrossRef]
- A. Hoelbl et al., “Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia,” EMBO Mol Med, vol. 2, no. 3, pp. 98–110, Mar. 2010. [CrossRef]
- O. Hantschel et al., “BCR-ABL uncouples canonical JAK2-STAT5 signaling in chronic myeloid leukemia,” Nat Chem Biol, vol. 8, no. 3, Art. no. 3, Mar. 2012. [CrossRef]
- F. Rossari, F. Minutolo, and E. Orciuolo, “Past, present, and future of Bcr-Abl inhibitors: from chemical development to clinical efficacy,” Journal of Hematology & Oncology, vol. 11, no. 1, p. 84, Jun. 2018. [CrossRef]
- E. A. Nelson et al., “The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors,” Blood, vol. 117, no. 12, pp. 3421–3429, Mar. 2011. [CrossRef]
- M. Bar-Natan, E. A. Nelson, S. R. Walker, Y. Kuang, R. J. Distel, and D. A. Frank, “Dual inhibition of Jak2 and STAT5 enhances killing of myeloproliferative neoplasia cells,” Leukemia, vol. 26, no. 6, pp. 1407–1410, Jun. 2012. [CrossRef]
- A. Kentsis et al., “Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia,” Nat Med, vol. 18, no. 7, pp. 1118–1122, Jul. 2012. [CrossRef]
- L. Naldini et al., “Scatter factor and hepatocyte growth factor are indistinguishable ligands for the MET receptor,” EMBO J, vol. 10, no. 10, pp. 2867–2878, Oct. 1991. [CrossRef]
- S. McGee et al., “Biological properties of ligand-dependent activation of the MET receptor kinase in acute myeloid leukemia,” Leukemia, vol. 29, no. 5, pp. 1218–1221, May 2015. [CrossRef]
- B. Kim et al., “Synthetic lethal screening reveals FGFR as one of the combinatorial targets to overcome resistance to Met-targeted therapy,” Oncogene, vol. 34, no. 9, pp. 1083–1093, Feb. 2015. [CrossRef]
- E. C. Chen et al., “Targeting MET and FGFR in Relapsed or Refractory Acute Myeloid Leukemia: Preclinical and Clinical Findings, and Signal Transduction Correlates,” Clinical Cancer Research, vol. 29, no. 5, pp. 878–887, Mar. 2023. [CrossRef]
- C. Zhao, H. Li, H.-J. Lin, S. Yang, J. Lin, and G. Liang, “Feedback Activation of STAT3 as a Cancer Drug-Resistance Mechanism,” Trends Pharmacol Sci, vol. 37, no. 1, pp. 47–61, Jan. 2016. [CrossRef]
- L. N. Heppler and D. A. Frank, “Targeting Oncogenic Transcription Factors: Therapeutic Implications of Endogenous STAT Inhibitors,” Trends Cancer, vol. 3, no. 12, pp. 816–827, Dec. 2017. [CrossRef]
- C. L. Verlinde and W. G. Hol, “Structure-based drug design: progress, results and challenges,” Structure, vol. 2, no. 7, pp. 577–587, Jul. 1994. [CrossRef]
- A. Chen and A. N. Koehler, “Transcription Factor Inhibition: Lessons Learned and Emerging Targets,” Trends Mol Med, vol. 26, no. 5, pp. 508–518, May 2020. [CrossRef]
- E. A. Nelson et al., “Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3,” Blood, vol. 112, no. 13, pp. 5095–5102, Dec. 2008. [CrossRef]
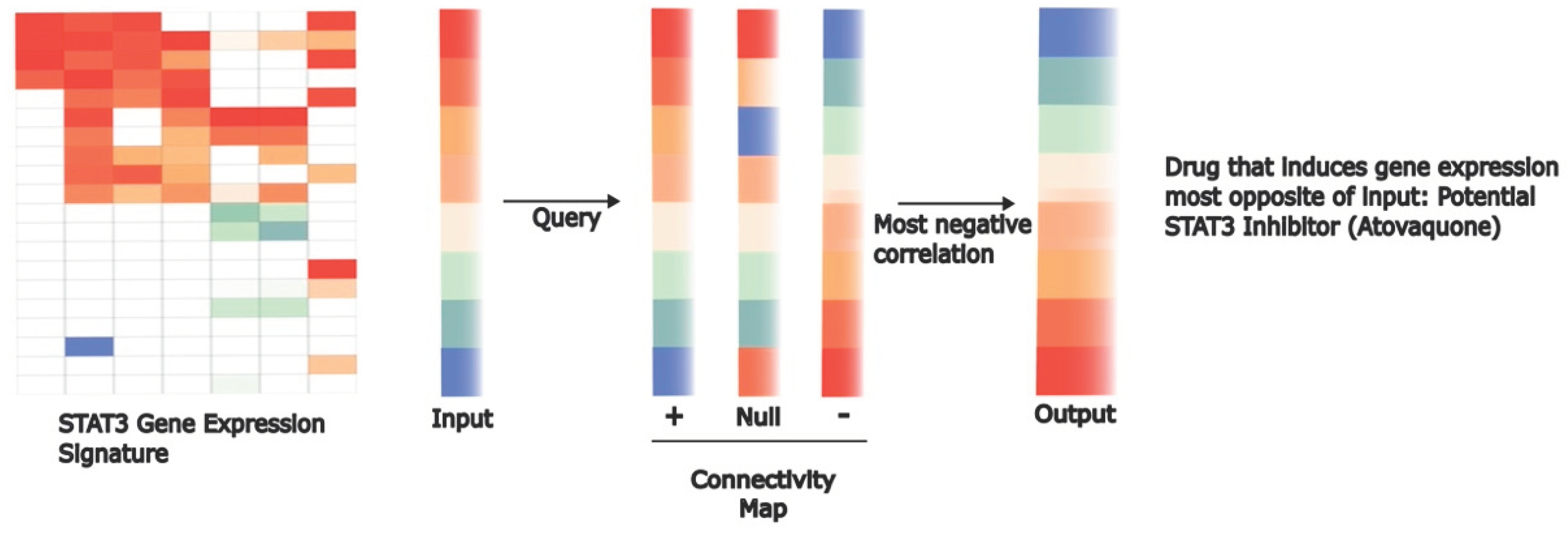
- M. Xiang et al., “Gene expression-based discovery of atovaquone as a STAT3 inhibitor and anticancer agent,” Blood, vol. 128, no. 14, pp. 1845–1853, Oct. 2016. [CrossRef]
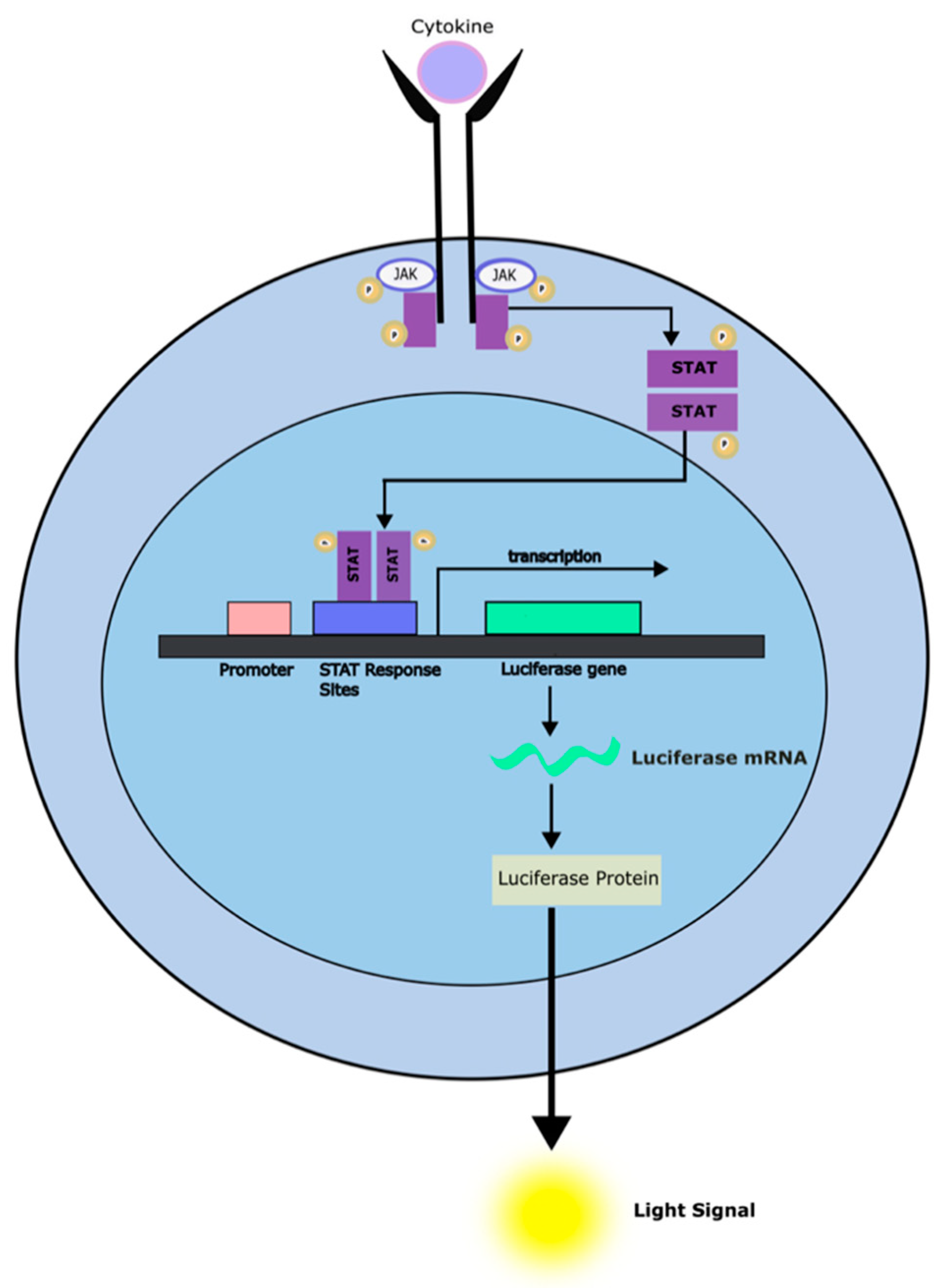
- M. Hirschenberger, M. Hayn, A. Laliberté, L. Koepke, F. Kirchhoff, and K. M. J. Sparrer, “Luciferase reporter assays to monitor interferon signaling modulation by SARS-CoV-2 proteins,” STAR Protoc, vol. 2, no. 4, p. 100781, Aug. 2021. [CrossRef]
- S. R. Walker and D. A. Frank, “Screening approaches to generating STAT inhibitors,” JAKSTAT, vol. 1, no. 4, pp. 292–299, Oct. 2012. [CrossRef]
- E. A. Nelson et al., “The STAT5 Inhibitor Pimozide Displays Efficacy in Models of Acute Myelogenous Leukemia Driven by FLT3 Mutations,” Genes Cancer, vol. 3, no. 7–8, pp. 503–511, Jul. 2012. [CrossRef]
- A. Takakura et al., “Pyrimethamine inhibits adult polycystic kidney disease by modulating STAT signaling pathways,” Hum Mol Genet, vol. 20, no. 21, pp. 4143–4154, Nov. 2011. [CrossRef]
- M. W. Khan et al., “The STAT3 inhibitor pyrimethamine displays anti-cancer and immune stimulatory effects in murine models of breast cancer,” Cancer Immunol Immunother, vol. 67, no. 1, pp. 13–23, Jan. 2018. [CrossRef]
- L. N. Heppler et al., “The antimicrobial drug pyrimethamine inhibits STAT3 transcriptional activity by targeting the enzyme dihydrofolate reductase,” J Biol Chem, vol. 298, no. 2, p. 101531, Feb. 2022. [CrossRef]
- J. I. Brown et al., “Investigating the anti-cancer potential of pyrimethamine analogues through a modern chemical biology lens,” Eur J Med Chem, vol. 264, p. 115971, Jan. 2024. [CrossRef]
- C.-C. for D. C. and Prevention, “CDC - Toxoplasmosis - Resources for Health Professionals.” Accessed: Jan. 09, 2024. [Online]. Available: https://www.cdc.gov/parasites/toxoplasmosis/health_professionals/index.html.
- D. A. Frank, S. Mahajan, and J. Ritz, “B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues,” J Clin Invest, vol. 100, no. 12, pp. 3140–3148, Dec. 1997. [CrossRef]
- I. Hazan-Halevy et al., “STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells,” Blood, vol. 115, no. 14, pp. 2852–2863, Apr. 2010. [CrossRef]
- M. Tolomeo, A. Cavalli, and A. Cascio, “STAT1 and Its Crucial Role in the Control of Viral Infections,” Int J Mol Sci, vol. 23, no. 8, p. 4095, Apr. 2022. [CrossRef]
- T. E. Battle, R. A. Lynch, and D. A. Frank, “Signal transducer and activator of transcription 1 activation in endothelial cells is a negative regulator of angiogenesis,” Cancer Res, vol. 66, no. 7, pp. 3649–3657, Apr. 2006. [CrossRef]
- D. A. Frank, S. Mahajan, and J. Ritz, “Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling,” Nat Med, vol. 5, no. 4, pp. 444–447, Apr. 1999. [CrossRef]
- R. A. Lynch, J. Etchin, T. E. Battle, and D. A. Frank, “A small-molecule enhancer of signal transducer and activator of transcription 1 transcriptional activity accentuates the antiproliferative effects of IFN-gamma in human cancer cells,” Cancer Res, vol. 67, no. 3, pp. 1254–1261, Feb. 2007. [CrossRef]
- J. V. Alvarez, P. G. Febbo, S. Ramaswamy, M. Loda, A. Richardson, and D. A. Frank, “Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors,” Cancer Res, vol. 65, no. 12, pp. 5054–5062, Jun. 2005. [CrossRef]
- J. Lamb, “The Connectivity Map: a new tool for biomedical research,” Nat Rev Cancer, vol. 7, no. 1, Art. no. 1, Jan. 2007. [CrossRef]
- J. Lamb et al., “The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease,” Science, vol. 313, no. 5795, pp. 1929–1935, Sep. 2006. [CrossRef]
- W. T. Hughes, “The Role of Atovaquone Tablets in Treating Pneumocystis carinii Pneumonia,” JAIDS Journal of Acquired Immune Deficiency Syndromes, vol. 8, no. 3, p. 247, Mar. 1995. [CrossRef]
- Z. Lv, X. Yan, L. Lu, C. Su, and Y. He, “Atovaquone enhances doxorubicin’s efficacy via inhibiting mitochondrial respiration and STAT3 in aggressive thyroid cancer,” J Bioenerg Biomembr, vol. 50, no. 4, pp. 263–270, Aug. 2018. [CrossRef]
- A. M. Stevens et al., “Repurposing Atovaquone as a Therapeutic against Acute Myeloid Leukemia (AML): Combination with Conventional Chemotherapy Is Feasible and Well Tolerated,” Cancers, vol. 15, no. 4, Art. no. 4, Jan. 2023. [CrossRef]
- S. S. Bashraheel, A. Domling, and S. K. Goda, “Update on targeted cancer therapies, single or in combination, and their fine tuning for precision medicine,” Biomed Pharmacother, vol. 125, p. 110009, May 2020. [CrossRef]
- K. A. Z. Siddiquee and J. Turkson, “STAT3 as a target for inducing apoptosis in solid and hematological tumors,” Cell Res, vol. 18, no. 2, pp. 254–267, Feb. 2008. [CrossRef]
- N. Jin, Y. Xia, and Q. Gao, “Combined PARP inhibitors and small molecular inhibitors in solid tumor treatment (Review),” Int J Oncol, vol. 62, no. 2, p. 28, Jan. 2023. [CrossRef]
- L. Chen et al., “Inflammatory responses and inflammation-associated diseases in organs,” Oncotarget, vol. 9, no. 6, pp. 7204–7218, Dec. 2017. [CrossRef]
- KB. Megha, X. Joseph, V. Akhil, and PV. Mohanan, “Cascade of immune mechanism and consequences of inflammatory disorders,” Phytomedicine, vol. 91, p. 153712, Oct. 2021. [CrossRef]
- W. Wang, F. Burton, S. Herter, L. Codarri Deak, C. Klein, and D. Frank, “Pharmacologic Inhibitors of STAT3 or BCL6 Transcriptional Function Sensitize Lymphoma Cells to the Novel PD-1 Cis-Targeted PD1-IL2v Immunocytokine in a Murine Model,” Blood, vol. 140, no. Supplement 1, pp. 8835–8836, Nov. 2022. [CrossRef]
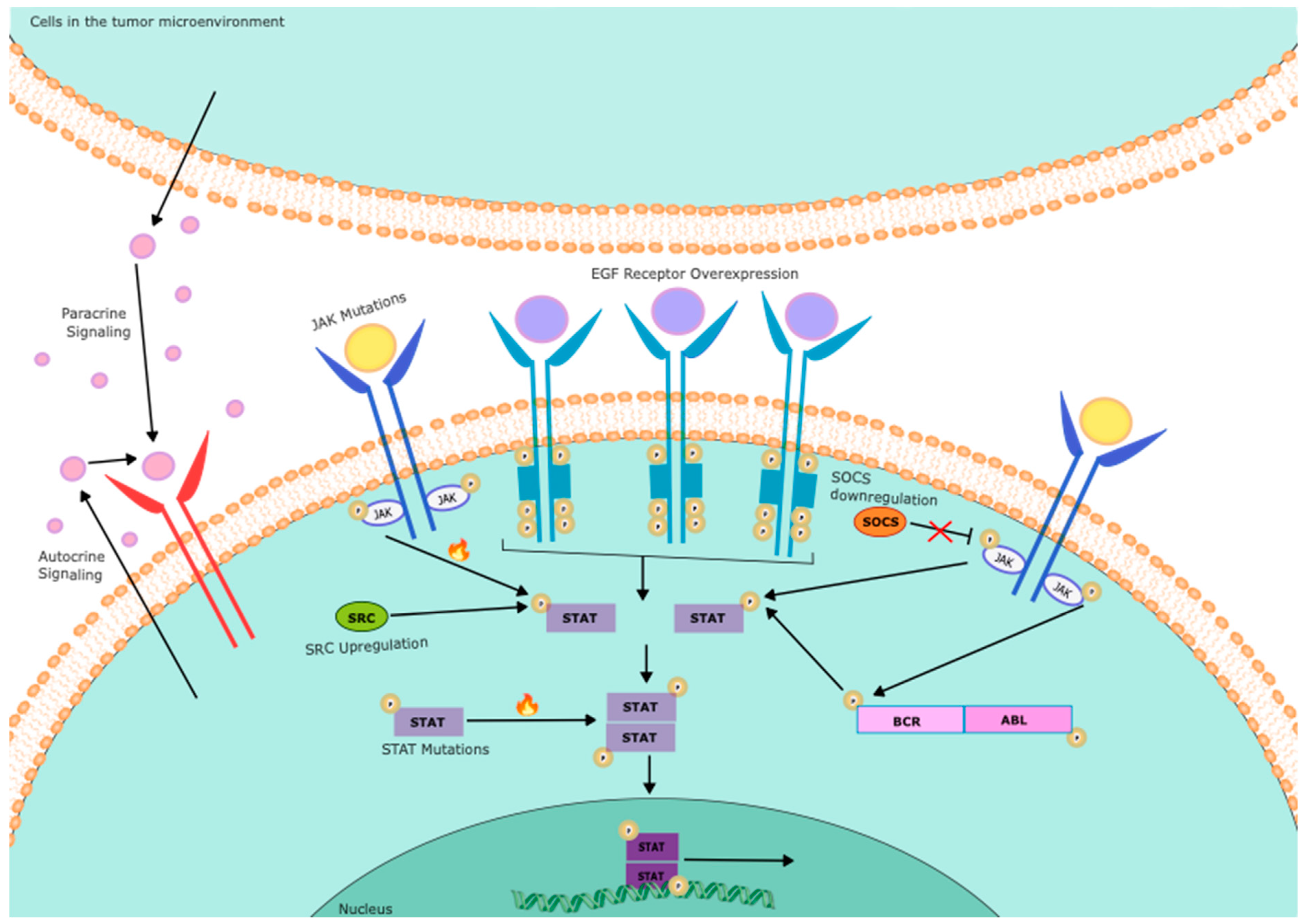
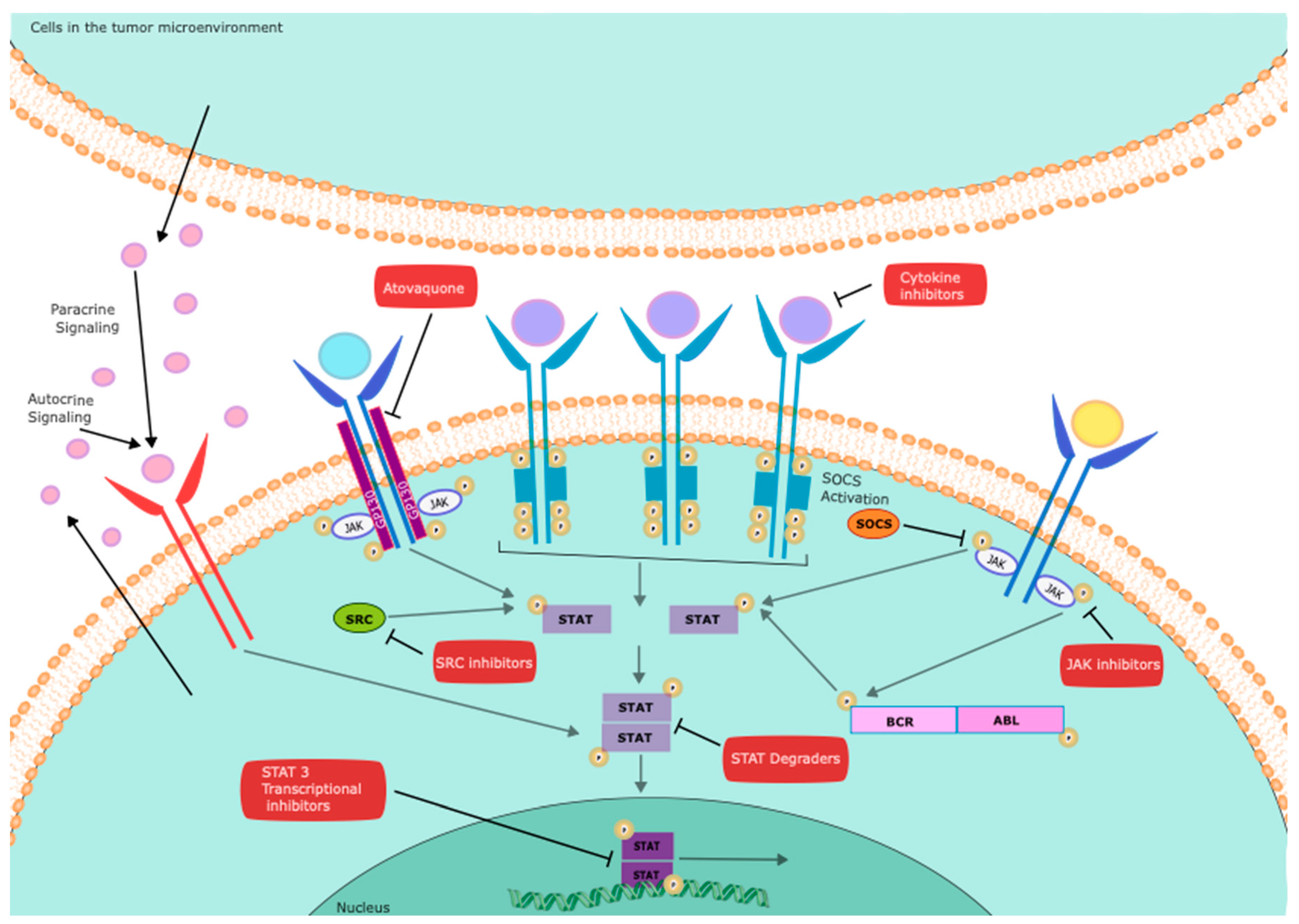
| Gene | Function | Status | Cell source | References |
|---|---|---|---|---|
| AKT1 | Proliferation | Upregulated | Various human cancer cells | [31] |
| BATF | Differentiation | Upregulated | Human Th17 cells | [32] |
| Bcl-xL | Anti-apoptosis | Upregulated | Human U266 cells | [14] |
| BCL6 | Proliferation | Upregulated | Human Th17 cells | [32] |
| MYC | Proliferation | Upregulated | Murine Ba/F3 cells | [33] |
| CCND1 | Proliferation | Upregulated | Human gastric cancer cells | [34] |
| CDKN2C | Cell cycle inhibition | Downregulated | Human Th17 cells | [32] |
| CREM | Spermatogenesis | Downregulated | Human Th17 cells | [32] |
| CXCL10 | Angiogenesis, Immune escape | Downregulated | Human CD8+ T cells | [35] |
| FOSL2 | Differentiation | Upregulated | Human Th17 cells | [32] |
| IKZF2 | Lymphocyte development | Downregulated | Human Th17 cells | [32] |
| IL6 | Immune escape | Upregulated | Murine melanoma cells | [36] |
| IL10 | Immune escape | Upregulated | Human colon Carcinoma | [37] |
| MMP2 | Immune escape | Upregulated | Murine melanoma cells | [38] |
| MMP9 | Immune escape | Upregulated | Murine fibroblasts | [39] |
| Ccl5 | Immune escape | Downregulated | Murine melanoma cells | [36] |
| RBPJ | Differentiation | Upregulated | Human Th17 cells | [32] |
| SMAD7 | Differentiation | Downregulated | Human Th17 cells | [32] |
| STAT1 | Differentiation | Downregulated | Human Th17 cells | [32] |
| STAT2 | Antiviral activity | Downregulated | Human Th17 cells | [32] |
| STAT3 | Differentiation | Upregulated | Human Th17 cells | [32] |
| TWIST | Immune escape | Upregulated | Human breast carcinomas | [40] |
| VEGF | Angiogenesis, immune escape | Upregulated | Murine fibroblasts | [39] |
| VIM | Immune escape | Upregulated | Monkey kidney cells | [41] |
| Gene | Function | Status | Cell source | References |
|---|---|---|---|---|
| Arnt | Protein sumoylation | Downregulated | Mouse proB cells | [81] |
| BCL2 | Anti-apoptosis | Upregulated | Human T cells | [84] |
| Bcl2l1 | Apoptosis | Upregulated | Mouse proB cells | [81] |
| BCLXL | Anti-apoptosis | Upregulated | Human T cells | [84] |
| C3ar1 | Chemotaxis | Upregulated | Murine proB cells | [78] |
| CISH | STAT inhibitor | Upregulated | Human T cells | [84] |
| Dusp1 | Anti-inflammation | Upregulated | Murine proB cells | [78] |
| DUSP5 | Anti-proliferation | No change | Human T cells | [84] |
| GTF2H5 | DNA repair | Downregulated | Human T cells | [84] |
| MBP | Inflammation | No change | Human T cells | [84] |
| Myc | Proliferation | Upregulated | Murine proB cells | [78] |
| OSM1 | Metabolic process | Upregulated | Human T cells | [84] |
| PIM1 | Proliferation, survival | Upregulated | Human T cells | [84] |
| Pim2 | Cell survival | Upregulated | Murine proB cells | [78] |
| Ro60 | Sperm antigen | Downregulated | Mouse proB cells | [81] |
| Ryk | Proliferation | Upregulated | Murine proB cells | [78] |
| Serpina3g | Proliferation | Upregulated | Murine proB cells | [78] |
| SGK1 | Proliferation | Downregulated | Human T cells | [84] |
| SLC22A5 | Carnitine uptake | Downregulated | Human T cells | [84] |
| Socs1 | Apoptosis | Upregulated | Murine proB cells | [78] |
| SOCS2 | inflammation | Upregulated | Human T cells | [84] |
| Srp9 | RNA binding | Upregulated | Mouse proB cells | [81] |
| Tnfrsf13b | B cell homeostasis | Upregulated | Murine proB cells | [78] |
| Type | Agent | Target | Cancer type | ClinicalTrial.gov identifier | Phase | References |
|---|---|---|---|---|---|---|
| Small molecules | Silibinin | STAT3 | Endometrial carcinoma | Preclinical | [144] | |
| SD-36 | STAT3 | Acute myeloid leukemia and anaplastic large cell lymphoma | Preclinical | [118] | ||
| BP-1-102 | STAT3 | Acute lymphoblastic leukemia | Preclinical | [145] | ||
| LLL12 | STAT3 | Ovarian cancer | Preclinical | [146] | ||
| Pyrimethamine | STAT3 | Chronic lymphocytic leukemia | NCT01066663 | Phase 1/2 | [147] | |
| OPB-51602 | STAT3 | Nasopharyngeal carcinoma | NCT01184807 | Phase 1 | [148] | |
| N4 | STAT3 | Pancreatic cancer | Preclinical | [136] | ||
| Atovaquone | STAT3 | Non-small cell lung cancer | NCT02628080 | Phase 1 | [149] | |
| STAT3 | Acute myeloid leukemia | Preclinical | [150] | |||
| Trichothecin | STAT3 | Colorectal Cancer | Preclinical | [151] | ||
| SDL-1 | STAT3 | Gastric cancer | Preclinical | [152] | ||
| AK-2292 | STAT5 | Chronic myeloid leukemia | Preclinical | [153] | ||
| Oligonucleotides | Danvatirsen | STAT3 | Diffuse large B-cell lymphoma | NCT03527147 | Phase 1 | [154] |
| STAT3 | Myelodysplastic syndromes, acute myeloid leukemia | NCT05986240 | Phase 1 | [155] | ||
| Double-stranded minicircles | STAT3 | Triple-negative breast cancer | Preclinical | [156] | ||
| Peptides | OPB-31121 | STAT3 | Hepatocellular carcinoma | NCT01406574 | Phase 1/2 | [154] |
| PS-acet.-STAT3 peptide | STAT3 | Melanoma | Preclinical | [157] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




