1. Introduction
Epilepsy is a common neurological condition, characterized by spontaneous seizures, and affecting about 1% of the global population [
1]. This results in a direct medical cost of
$24.4 billion in the U.S. annually [
2]. Despite the best efforts by healthcare providers towards early diagnosis and treatment of the symptoms of epilepsy, around 50% of patients continue to experience seizures, leading to a lower quality of life [
3]. Electroencephalography (EEG) is commonly used for diagnosing seizures in humans and for studying seizures in animal models. The low cost and easy collection of EEG allows its use in wearable devices for continuous monitoring. This allows the prediction of seizures, hence reducing its adverse effects [
4]. Better seizure prediction techniques increase the effectiveness of intervention devices that prevent the occurrence of a seizure by electrical stimulation or by administering acute medication. Electrical brain activity observed through EEG can vary depending on the subject’s condition and state, and is typically categorized into four main distinct phases. These include the interictal phase, which consists of normal brain activity and is also present in healthy individuals; the preictal phase, which precedes a seizure; the ictal phase, which starts with the seizure onset and concludes with the seizure’s end; and the postictal phase, which follows the ictal phase and lasts until the next interictal phase begins.
Animal models are often used for studying seizures and developing better models for seizure prediction in a research setting. This is because it is legal, ethical, and more affordable to induce neurological conditions in other animals and obtain continuous recordings spanning several hours. While traditionally, EEG is analyzed by human experts, it is infeasible and increases inter-observer variability for multi-hour recordings [
5,
6,
7].
Most studies focus on the automatic detection of seizures, however, few try to identify patterns in an EEG preceding a seizure. This is probably because there is no clear definition of how long before the occurrence of a seizure the preictal phase starts and there are several interpretations of this interval [
8,
9,
10]. The application of mathematical theories of non-linear dynamics initially allowed the beginning of preictal interval to be identified as the point beyond which brain activity develops deterministically, leading up to a seizure [
11,
12]. In other words, it is a point of no return, which once passed, a seizure will occur. However, the interval between that point and the occurrence of a seizure varies across patients [
11,
13].
Among automatic preictal phase-detection methods, statistical approaches compare the distribution of preictal and interictal EEG segments while some algorithmic approaches perform a grid search on preictal segments [
14]. Machine learning (ML) algorithms have widely been used for EEG analysis collected both from humans, mostly in clinical settings, and other animals, mostly in research settings [
15,
16,
17]. Advanced ML algorithms have proven effective in detecting seizure in both humans, usually scalp EEG [
18,
19,
20,
21,
22] and rodents [
23].
Very little work has been done on the detection of preictal phase in EEG. Among relevant literature, [
24] describes various feature extraction techniques before classifying ictal, preictal, and interictal phases of EEG using simple ML algorithms like a Support Vector Machine (SVM). Features like sharpness of the waves, expressed as kurtosis and spectral entropy, expressing the amount of information contained in the waves. On the other hand, [
25] uses a simple Convolutional Neural Network (CNN) to perform the same classification using frequency spectrogram. [
26] was the first to used unsupervised learning to identify preictal phase in EEG. They used clustering to group together interictal patterns. Later, [
27,
28,
29] to study a relationship between the patterns observed in the preictal phase and the type of seizure.
CNNs have been widely applied to image classification due to their ability to localize important features from data. Though not easily accessible, such features can still be investigated by methods such as Grad-CAM [
30]. Their widespread application allows easy access to large models, pre-trained on large datasets. Transfer learning allows these models to perform better on relatively smaller datasets, which is very relevant for medical data analysis. YOLOv8 is the latest version of the You Only Look Once (YOLO) architecture [
31]. It uses 50 layers of CNNs with cross-stage partial connections for better information flow. In this work, we propose to classify raw EEG data into preictal and interictal segments using a deep convolutional neural network (DCNN). While DCNNs have previously been applied to EEG data, to the best of our knowledge, our work is the first to use a DCNN to analyze raw EEG data. This allows explanation techniques to be applied to identify regions of interest in the raw EEG data.
2. Materials and Methods
2.1. Dataset
We used an EEG dataset, publicly available on Kaggle [
32]. It was a part of a competition organized by the National Institutes of Health (NINDS), the Epilepsy Foundation, and the American Epilepsy Society. Data was collected, using implantable leads, from 3 dogs with naturally occurring epilepsy. The voltages were recorded from 16 channels, at a sampling rate of 400Hz, and referenced to the group average. Recordings spanned from several months, up to a year. Preictal intervals were considered to be one hour long, ending 5 minutes before the beginning of a seizure. This was done to avoid contamination from any seizure activity before the start of the seizure, annotated by an epileptologist. Interictal intervals, of one hour duration, were considered as far apart, from seizures, as possible. Overall, there were 2744 interictal segments and 211 preictal segments.
2.2. Preprocessing
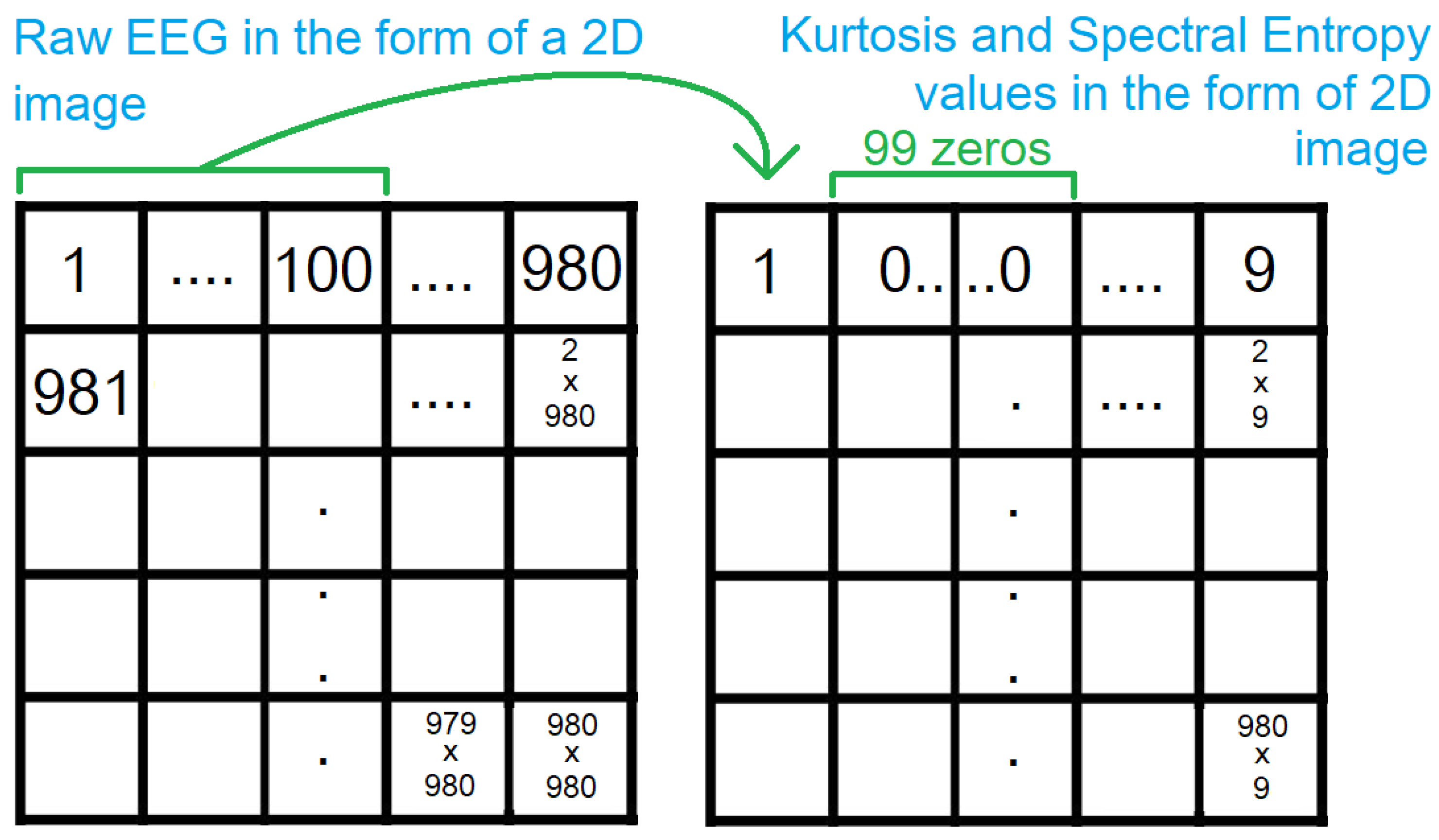
EEG is downsampled from 400Hz to 100Hz. Since the number of interictal segments is an order of magnitude greater than the number of preictal segments, we consider the 4 segments produced from each segment, during downsampling, for preictal segments but consider only the first one out of the 4 for interictal segments. This results in 844 preictal segments and 2744 interictal segments, out of which 1000 are considered for a better class balance. Raw EEG is expressed as time series data while CNNs accept image data. A common approach to representing EEG as image data is in the form of frequency spectrograms. However, such a representation does not allow the mapping of explanations, generated by methods such as Grad-CAM, back to the raw EEG data. Hence, instead of representing the raw EEG in an alternate form, we represent the voltage values as pixels in an image, in a row-major way, as shown in
Figure 1. The channels of a segment are concatenated before laying the voltage values on a 2D grid.
Most pre-trained CNNs use RGB images with 3 channels. The raw EEG laid out on a 2D grid is one channel, so we include kurtosis and spectral entropy as the two other channels. Both are calculated with a sliding window of size 100, without overlap. The consecutive values are padded by zeroes. This is then laid out on a 2D grid, as described above. This results in a sparse matrix, with 99 zeroes between 2 values as shown in
Figure 2. Due to the pooling layers in the CNN, the non-zero values, get focused on in the deeper layers because any trainable weight, multiplied by the zeroes, results in zeroes. Since the number of voltage values is not a perfect square, we append zeroes to the end, to obtain a square of dimensions 980x980. Thus, the resultant images have a dimension of 980x980x3.
2.3. Supervised Learning
Supervised learning algorithms depend on large annotated datasets to train well. However, given the cost of collecting and annotating medical data, it is often infeasible to train large supervised learning algorithms, from scratch, for medical applications. Thankfully, due to the popularity of DCNN models, several pre-trained DCNNs are publicly available. These were trained on real-life RGB images, from large-scale datasets like Imagenet [
33] and COCO datasets [
34]. For this work, we use a pre-trained YOLOv8, from Ultralytics [
35]. Due to memory restrictions, we use the medium version of the YOLOv8 model, with 50 CNN layers and 78.9 million trainable parameters. It was pre-trained on the COCO dataset [
34]. The model is trained for 100 epochs with the default hyperparameters. 80% of the data is used for training and 20% is used for testing. It is ensured that the copies of preictal segments, obtained during downsampling do not become part of two different sets. Weights are saved every 5 epochs and the weights corresponding to the best performance are used for the next step.
Due to memory restrictions during training, we added a padding of 10 on each side of a 980x980x3 image for all 3 channels, resulting in an image of dimensions 1000x1000x3. This was divided into 4 images of dimensions 250x250x3. The 4 images were provided as a batch to the model, with each image having the label of its parent image.
For comparison, the same model was trained and tested on a dataset containing only the raw EEG laid on a 2D grid (without the kurtosis and spectral entropy channels). To have 3 channels in these images, the single channel was copied twice and added to the image. This resulted in a 980x980x3 image, with each 980x980 slice being copies of one another. These were used for training in the same way as the images with raw eeg, kurtosis and spectral entropy (described in the previous paragraph). For comparison with the traditional approach, frequency spectrograms were generated for the EEG segments and used to train and test the model as well.
We also test the performance of the model by training it on data from one animal and testing it on 20% of the data from another animal.
2.4. Generating Explanation
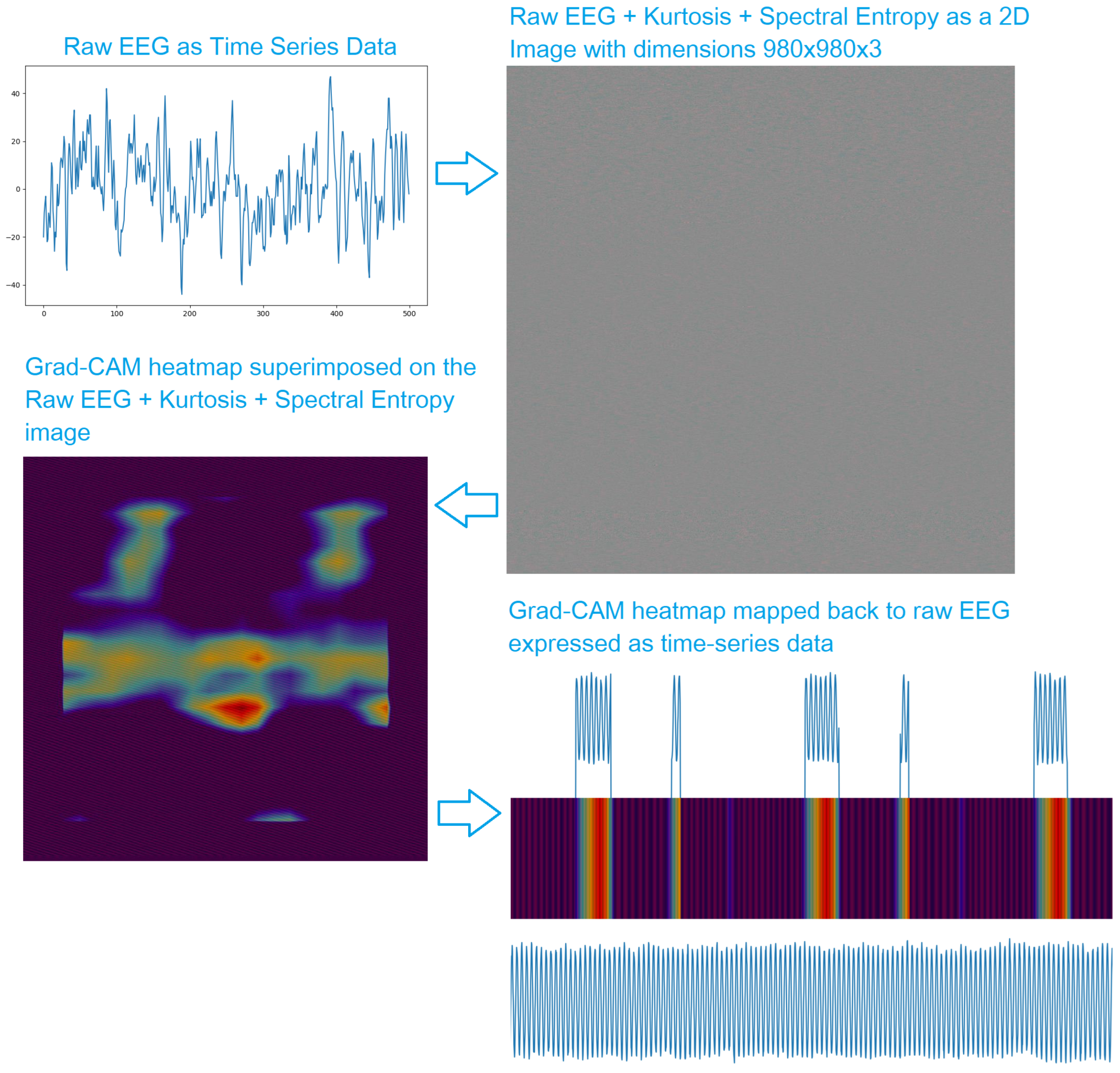
Once the model is trained, we use Grad-CAM [
30] to generate heatmaps. The heatmaps indicate how important each region in the image is to the model. Grad-CAM heatmaps are widely used to explain the performance of DCNNs on real-life RGB images, for example, if an image is classified as a cat, the heatmap, desirably, indicates the region of the image containing a cat is the most important. For our application, the heatmap tells us which regions of the image made the model "think" that it is a preictal segment. The most important regions in the raw EEG are then highlighted to observe relevant patterns in the preictal EEG. This is summarized in
Figure 3.
3. Results
As shown in
Table 1, the model performs best when trained on the images containing the raw EEG, kurtosis, and spectral entropy, with an accuracy of 95.75%, precision of 0.93, recall of 0.98, and F1 score of 0.95. The performances are close when the model is trained on only EEG and frequency spectrogram generated from EEG data, with a better performance for the EEG data.
As shown in
Table 2, the model performs better, when it is trained and tested on data from the same animal, than when it is trained on data from one animal and tested on data from another animal.
4. Discussion
In this study, we demonstrated that a pre-existing powerful DCNN can be repurposed to analyze EEG data in its original form. We observe that including additional features, namely kurtosis and spectral entropy improves the performance of the model. Kurtosis, broadly speaking, represents the sharpness of the waves. Seizures are known to contain more sharp spikes, suggesting their possible relevance for preictal phase identification as well. Spectral entropy represents the amount of information contained in a wave. It is known to be lower during a seizure. Hence, we hypothesized that this might be an important feature for identifying the preictal phase as well. Though we considered these two features, there are a lot of other features that remain unexplored. The same idea also applies to the DCNN model used. We used YOLOv8 because it is the state of the art, at the moment, for real-life object detection. However, it is not specialized for EEG analysis. A comprehensive study comparing the performance of various models trained on different features would be crucial to determine the best combination for preictal phase detection.
The use of raw EEG to train the model allows us to generate explanations, in the form of Grad-CAM heatmap, on the raw EEG. This allows us to localize patterns in the EEG that the model considers relevant to detecting the preictal phase. Such patterns might help experts look for consistency in the highlighted patterns and potentially discover biomarkers of specific conditions.
We observe that the model performs better when it is trained and tested on data obtained from the same animal. This might be explained by subtle differences in brain activity among individual animals with the same underlying condition. There are also minute differences in the position of the electrodes for different animals. The model seems to be able to pick up on these subtle differences. The wide disparity in performance, when the model is trained on data from different animals, can also be attributed to the difference in the number of segments from each animal. The original dataset had 42, 72, and 97 preictal and 500, 1440, and 804 interictal segments from dogs 1, 2, and 3 respectively. We obtained 4 segments from each preictal segment by downsampling, as described in the preprocessing section above. We also considered a proportional number of interictal segments such that the total number of interictal segments was equal to 1000. Thus, our final dataset contained 168, 288, and 388 preictal and 182, 525, and 293 interictal segments from dogs 1, 2, and 3 respectively.
5. Conclusions
In this work we demonstrated that (1) A pre-existing DCNN model i.e. YOLOv8 can be used for EEG analysis, (2) The model performs better when features such as Kurtosis and Spectral entropy are provided along with raw EEG, (3) The model performs better when it is trained and tested on data from the same animal, and (4) Grad-CAM heatmaps can be mapped to the raw EEG signal to localize regions of interest and potentially recognize important patterns. A comprehensive study comparing more features used to train more neural networks might help obtain better results.
Author Contributions
Conceptualization, S.B.; Investigation, S.B., A.B., K.K., C.A., and D.D.; Methodology, S.B., A.B., K.K., C.A., and D.D.; Project administration, D.D.; Supervision, D.D.; Validation, S.B., A.B., K.K., C.A., and D.D.; Visualization, S.B., A.B., K.K., C.A., and D.D.; Writing—original draft, S.B., A.B., K.K., C.A., and D.D.; Writing—review and editing, S.B., A.B., K.K., C.A., and D.D. All authors have read and agreed to the published version of the manuscript.
Funding
This study was conducted with the support of the National Institutes of Health (NIH) under award number R01NS111744.
Institutional Review Board Statement
Not applicable
Informed Consent Statement
Not applicable
Data Availability Statement
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| ML |
Machine Learning |
| CNN |
Convolutional Neural Network |
| DCNN |
Deep Convolutional Neural Network |
| YOLO |
You Only Look Once |
| EEG |
Electroencephalogram |
| SVM |
Support Vector Machine |
| NINDS |
National Institutes of Health |
References
- Kobau, R.; Zahran, H.; Thurman, D.J.; Zack, M.M.; Henry, T.R.; Schachter, S.C.; Price, P.H. Epilepsy surveillance among adults–19 states, behavioral risk factor surveillance system, 2005 2008.
- Begley, C.E.; Famulari, M.; Annegers, J.F.; Lairson, D.R.; Reynolds, T.F.; Coan, S.; Dubinsky, S.; Newmark, M.E.; Leibson, C.; So, E.; et al. The cost of epilepsy in the United States: an estimate from population-based clinical and survey data. Epilepsia 2000, 41, 342–351. [Google Scholar] [CrossRef]
- Loring, D.W.; Meador, K.J.; Lee, G.P. Determinants of quality of life in epilepsy. Epilepsy & Behavior 2004, 5, 976–980. [Google Scholar]
- Traynelis, S.F.; Dlugos, D.; Henshall, D.; Mefford, H.C.; Rogawski, M.A.; Staley, K.J.; Dacks, P.A.; Whittemore, V.; Poduri, A.; of Neurological Disorders, N.I.; et al. Epilepsy benchmarks area III: improved treatment options for controlling seizures and epilepsy-related conditions without side effects. Epilepsy currents 2020, 20, 23S–30S. [Google Scholar] [CrossRef] [PubMed]
- Abend, N.S.; Gutierrez-Colina, A.; Zhao, H.; Guo, R.; Marsh, E.; Clancy, R.R.; Dlugos, D.J. Interobserver reproducibility of electroencephalogram interpretation in critically ill children. Journal of clinical neurophysiology: official publication of the American Electroencephalographic Society 2011, 28, 15. [Google Scholar] [CrossRef]
- Gerber, P.A.; Chapman, K.E.; Chung, S.S.; Drees, C.; Maganti, R.K.; Ng, Y.t.; Treiman, D.M.; Little, A.S.; Kerrigan, J.F. Interobserver agreement in the interpretation of EEG patterns in critically ill adults. Journal of Clinical Neurophysiology 2008, 25, 241–249. [Google Scholar] [CrossRef]
- Williams, G.W.; Lüders, H.O.; Brickner, A.; Goormastic, M.; Klass, D.W. Interobserver variability in EEG interpretation. Neurology 1985, 35, 1714–1714. [Google Scholar] [CrossRef] [PubMed]
- Mormann, F.; Kreuz, T.; Rieke, C.; Andrzejak, R.G.; Kraskov, A.; David, P.; Elger, C.E.; Lehnertz, K. On the predictability of epileptic seizures. Clinical neurophysiology 2005, 116, 569–587. [Google Scholar] [CrossRef]
- Valderrama, M.; Alvarado, C.; Nikolopoulos, S.; Martinerie, J.; Adam, C.; Navarro, V.; Le Van Quyen, M. Identifying an increased risk of epileptic seizures using a multi-feature EEG–ECG classification. Biomedical Signal Processing and Control 2012, 7, 237–244. [Google Scholar] [CrossRef]
- Teixeira, C.A.; Direito, B.; Bandarabadi, M.; Le Van Quyen, M.; Valderrama, M.; Schelter, B.; Schulze-Bonhage, A.; Navarro, V.; Sales, F.; Dourado, A. Epileptic seizure predictors based on computational intelligence techniques: A comparative study with 278 patients. Computer methods and programs in biomedicine 2014, 114, 324–336. [Google Scholar] [CrossRef]
- Blauwblomme, T.; Jiruska, P.; Huberfeld, G. Mechanisms of ictogenesis. In International review of neurobiology; Elsevier, 2014; Vol. 114, pp. 155–185.
- Iasemidis, L.D. Epileptic seizure prediction and control. IEEE Transactions on Biomedical Engineering 2003, 50, 549–558. [Google Scholar] [CrossRef]
- Freestone, D.R.; Karoly, P.J.; Peterson, A.D.; Kuhlmann, L.; Lai, A.; Goodarzy, F.; Cook, M.J. Seizure prediction: science fiction or soon to become reality? Current neurology and neuroscience reports 2015, 15, 1–9. [Google Scholar] [CrossRef]
- Mormann, F.; Andrzejak, R.G.; Elger, C.E.; Lehnertz, K. Seizure prediction: the long and winding road. Brain 2007, 130, 314–333. [Google Scholar] [CrossRef]
- Gabor, A. Seizure detection using a self-organizing neural network: validation and comparison with other detection strategies. Electroencephalography and clinical Neurophysiology 1998, 107, 27–32. [Google Scholar] [CrossRef]
- Gotman, J. Automatic recognition of epileptic seizures in the EEG. Electroencephalography and clinical Neurophysiology 1982, 54, 530–540. [Google Scholar] [CrossRef]
- Ein Shoka, A.A.; Dessouky, M.M.; El-Sayed, A.; Hemdan, E.E.D. EEG seizure detection: concepts, techniques, challenges, and future trends. Multimedia Tools and Applications 2023, pp. 1–31.
- Farooq, M.S.; Zulfiqar, A.; Riaz, S. A review on Epileptic Seizure Detection using Machine Learning. arXiv preprint arXiv:2210.06292 2022.
- Abdelhameed, A.; Bayoumi, M. A deep learning approach for automatic seizure detection in children with epilepsy. Frontiers in Computational Neuroscience 2021, 15, 650050. [Google Scholar] [CrossRef]
- Siddiqui, M.K.; Morales-Menendez, R.; Huang, X.; Hussain, N. A review of epileptic seizure detection using machine learning classifiers. Brain informatics 2020, 7, 1–18. [Google Scholar] [CrossRef]
- Raghu, S.; Sriraam, N.; Temel, Y.; Rao, S.V.; Kubben, P.L. EEG based multi-class seizure type classification using convolutional neural network and transfer learning. Neural Networks 2020, 124, 202–212. [Google Scholar] [CrossRef]
- Gao, Y.; Gao, B.; Chen, Q.; Liu, J.; Zhang, Y. Deep convolutional neural network-based epileptic electroencephalogram (EEG) signal classification. Frontiers in neurology 2020, 11, 375. [Google Scholar] [CrossRef]
- Bergstrom, R.A.; Choi, J.H.; Manduca, A.; Shin, H.S.; Worrell, G.A.; Howe, C.L. Automated identification of multiple seizure-related and interictal epileptiform event types in the EEG of mice. Scientific reports 2013, 3, 1483. [Google Scholar] [CrossRef]
- Usman, S.M.; Usman, M.; Fong, S.; et al. Epileptic seizures prediction using machine learning methods. Computational and mathematical methods in medicine 2017, 2017. [Google Scholar] [CrossRef]
- Zhou, M.; Tian, C.; Cao, R.; Wang, B.; Niu, Y.; Hu, T.; Guo, H.; Xiang, J. Epileptic seizure detection based on EEG signals and CNN. Frontiers in neuroinformatics 2018, 12, 95. [Google Scholar] [CrossRef]
- Le Van Quyen, M.; Soss, J.; Navarro, V.; Robertson, R.; Chavez, M.; Baulac, M.; Martinerie, J. Preictal state identification by synchronization changes in long-term intracranial EEG recordings. Clinical Neurophysiology 2005, 116, 559–568. [Google Scholar] [CrossRef]
- Li, F.; Liang, Y.; Zhang, L.; Yi, C.; Liao, Y.; Jiang, Y.; Si, Y.; Zhang, Y.; Yao, D.; Yu, L.; et al. Transition of brain networks from an interictal to a preictal state preceding a seizure revealed by scalp EEG network analysis. Cognitive Neurodynamics 2019, 13, 175–181. [Google Scholar] [CrossRef]
- Quercia, A.; Frick, T.; Egli, F.E.; Pullen, N.; Dupanloup, I.; Tang, J.; Asif, U.; Harrer, S.; Brunschwiler, T. Preictal onset detection through unsupervised clustering for epileptic seizure prediction. In Proceedings of the 2021 IEEE International Conference on Digital Health (ICDH). IEEE, 2021, pp. 142–147.
- Leal, A.; Pinto, M.F.; Lopes, F.; Bianchi, A.M.; Henriques, J.; Ruano, M.G.; de Carvalho, P.; Dourado, A.; Teixeira, C.A. Heart rate variability analysis for the identification of the preictal interval in patients with drug-resistant epilepsy. Scientific reports 2021, 11, 5987. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Das, A.; Vedantam, R.; Cogswell, M.; Parikh, D.; Batra, D. Grad-CAM: Why did you say that? arXiv preprint arXiv:1611.07450 2016.
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 779–788.
- American Epilepsy Society Seizure Prediction Challenge — kaggle.com. https://www.kaggle.com/competitions/seizure-prediction. [Accessed 23-02-2024].
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE conference on computer vision and pattern recognition. Ieee; 2009; pp. 248–255. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6-12, 2014, Proceedings, Part V 13. Springer, 2014, pp. 740–755.
- Ultralytics. Home — docs.ultralytics.com. https://docs.ultralytics.com/. [Accessed 23-02-2024].
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).