Submitted:
26 February 2024
Posted:
27 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
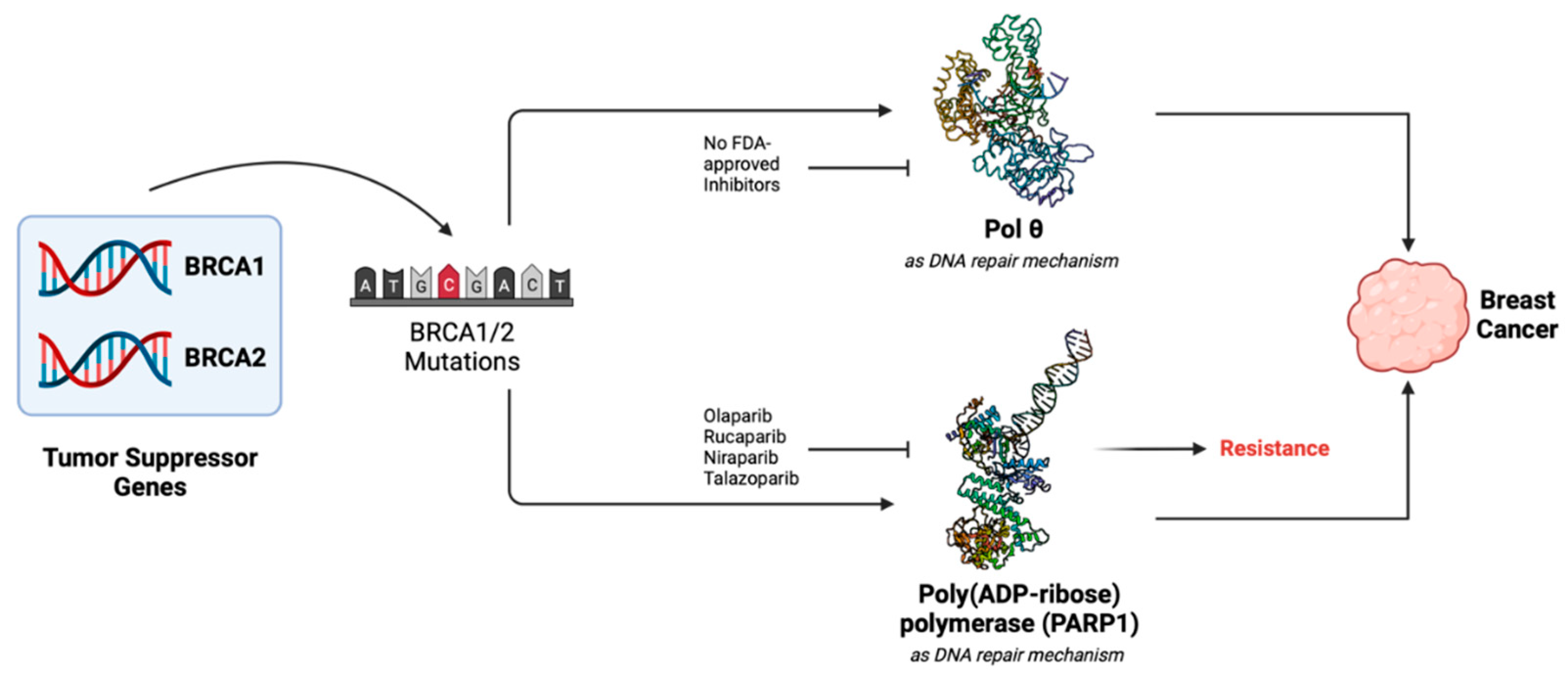
2. Pol θ as Drug-Development Target in HR-Deficient Cancers
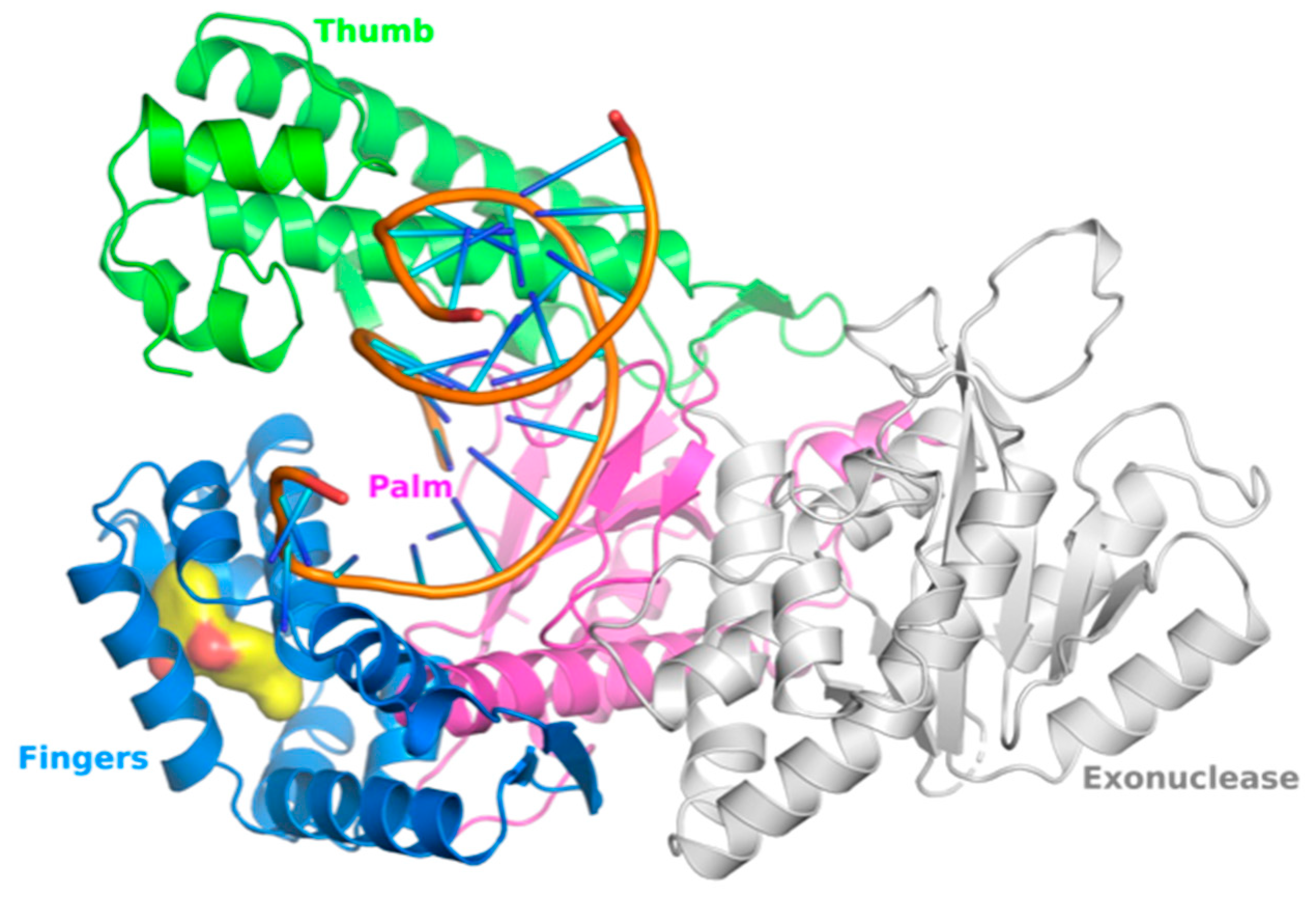
2.1. Human DNA pol θ
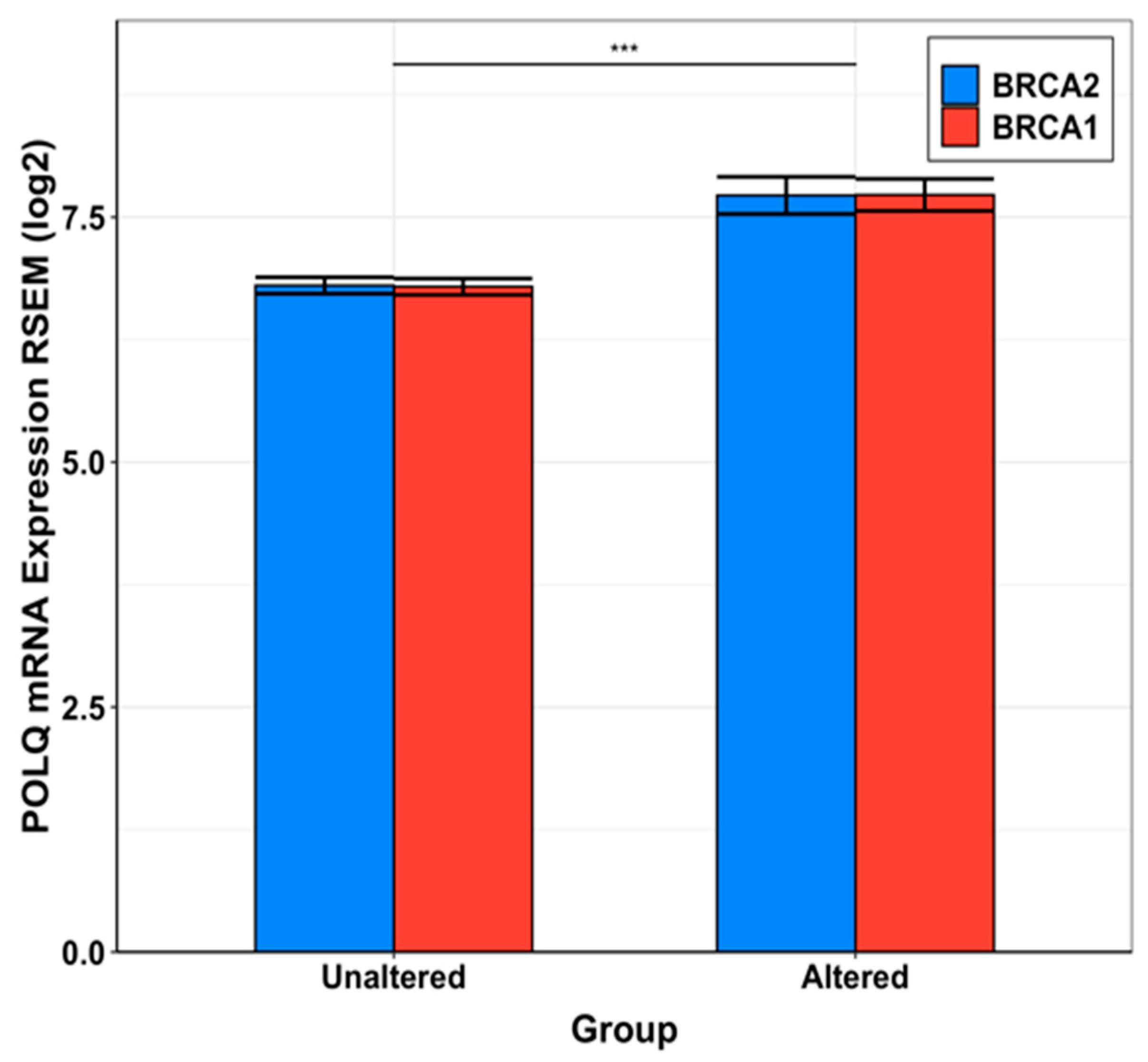
2.2. Human DNA pol θ as Double Strand Break (DSB) Repair Enzyme
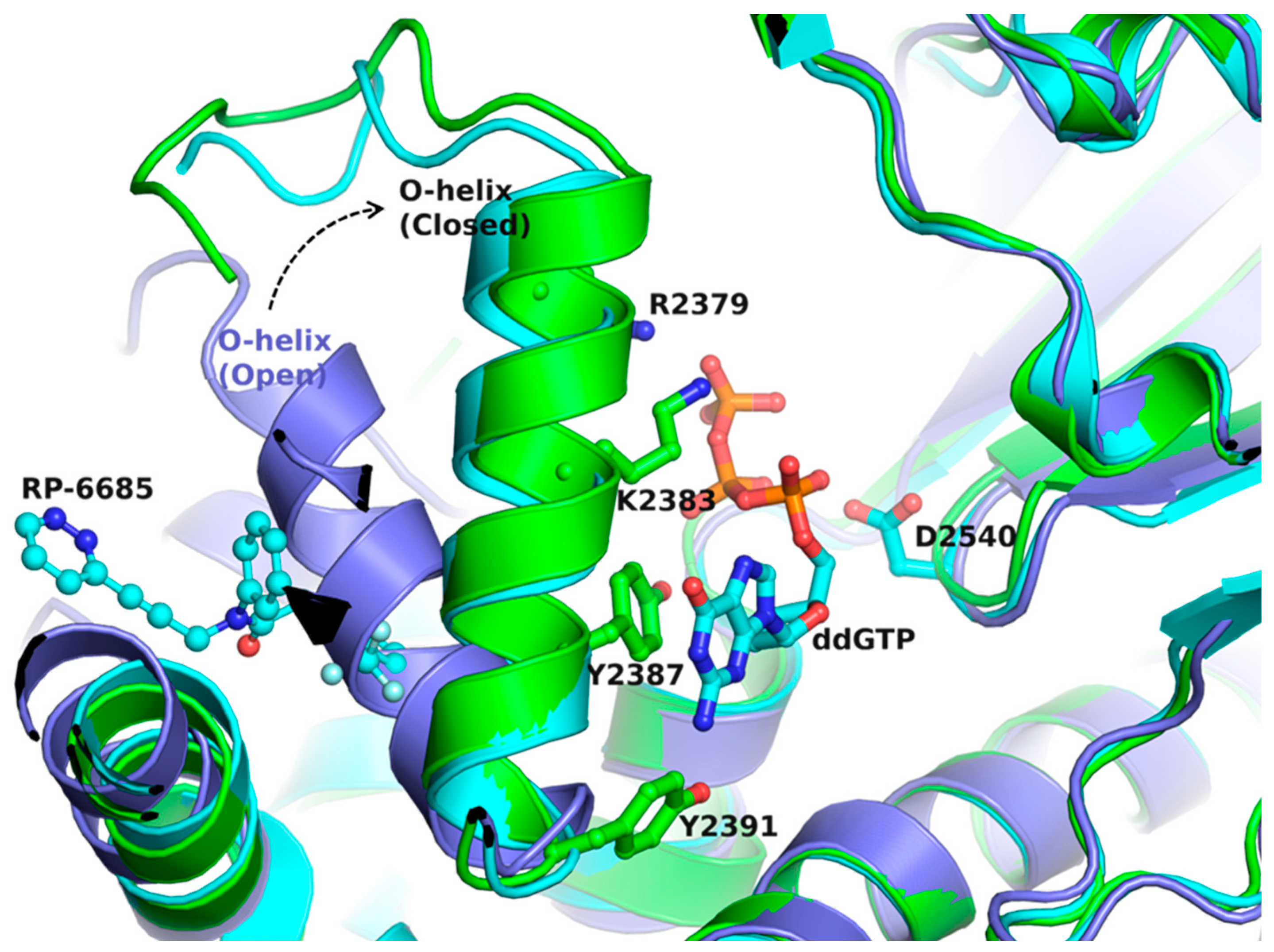
2.3. Status of Drug-Development against Human DNA pol θ
3. Apicoplast DNA Polymerase (apPOL) as Antimalaria Target
3.1. Status of Drug-Development against apPOL
3.2. Apicoplast as Antimalaria Drug Target
4. Pol I as Target for Combating Antimicrobial Resistance
4.1. Antimicrobial Resistance: A Global Health Concern
4.2. Family A Polymerase as a Novel Antibacterial Target
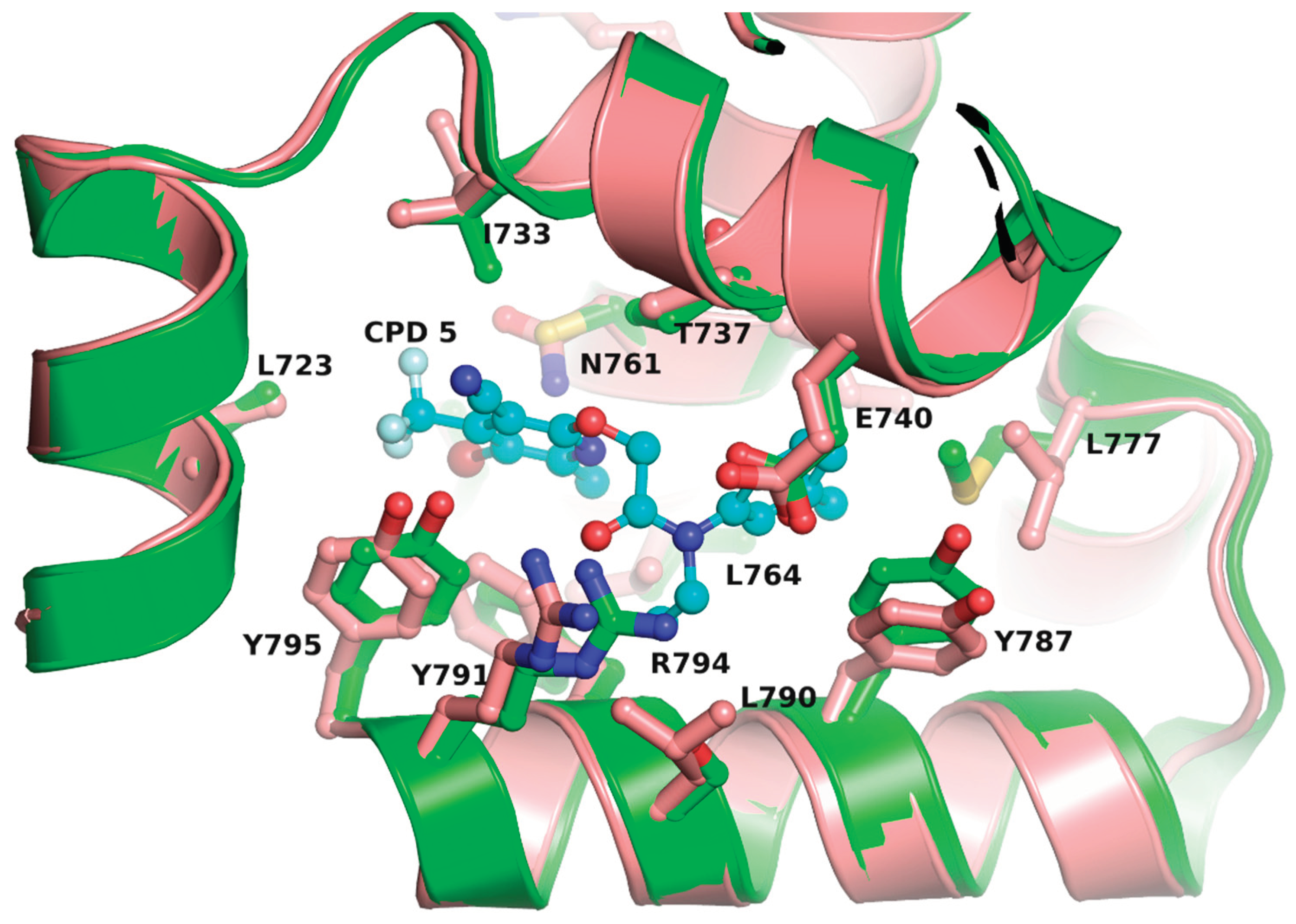
4.3. Pol I Inhibitors Acting through a Mechanism Analogous to NNRTI Inhibition of HIV-1 RT
4.4. Comparison of Allosteric Inhibitor Binding Site in pol I Enzymes from Diverse Species
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindahl, T.; Barnes, D.E.2000 Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol, 65, 127-133. [CrossRef]
- Modak, M.J.1978 Biochemistry of terminal deoxynucleotidyltransferase: Mechanism of inhibition by adenosine 5’-triphosphate. Biochemistry, 17, 3116-3120. [CrossRef]
- Delarue, M.; Poch, O.; Tordo, N.; Moras, D.; Argos, P.1990 An attempt to unify the structure of polymerases. Protein Eng, 3, 461-467. [CrossRef]
- Ishino, Y.; Komori, K.; Cann, I.K.; Koga, Y.1998 A novel DNA polymerase family found in archaea. J Bacteriol, 180, 2232-2236. [CrossRef]
- Braithwaite, D.K.; Ito, J.1993 Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res, 21, 787-802. [CrossRef]
- Cann, I.K.; Ishino, Y.1999 Archaeal DNA replication: Identifying the pieces to solve a puzzle. Genetics, 152, 1249-1267. [CrossRef]
- Ohmori, H.; Friedberg, E.C.; Fuchs, R.P.; Goodman, M.F.; Hanaoka, F.; Hinkle, D.; Kunkel, T.A.; Lawrence, C.W.; Livneh, Z.; Nohmi, T.; Prakash, L.; Prakash, S.; Todo, T.; Walker, G.C.; Wang, Z.; Woodgate, R.2001 The y-family of DNA polymerases. Mol Cell, 8, 7-8.
- Iyer, L.M.; Koonin, E.V.; Leipe, D.D.; Aravind, L.2005 Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: Structural insights and new members. Nucleic Acids Res, 33, 3875-3896. [CrossRef]
- Ito, J.; Braithwaite, D.K.1991 Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res, 19, 4045-4057. [CrossRef]
- Czernecki, D.; Nourisson, A.; Legrand, P.; Delarue, M.2023 Reclassification of family a DNA polymerases reveals novel functional subfamilies and distinctive structural features. Nucleic Acids Res, 51, 4488-4507. [CrossRef]
- Kornberg, A.; Baker, T.A. DNA replication. W.H. Freeman: New York, 1992.
- Klenow, H.; Henningsen, I.1970 Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from escherichia coli b by limited proteolysis. Proc Natl Acad Sci U S A, 65, 168-175. [CrossRef]
- Ollis, D.L.; Brick, P.; Hamlin, R.; Xuong, N.G.; Steitz, T.A.1985 Structure of large fragment of escherichia coli DNA polymerase i complexed with dtmp. Nature, 313, 762-766. [CrossRef]
- Kim, Y.; Eom, S.H.; Wang, J.; Lee, D.S.; Suh, S.W.; Steitz, T.A.1995 Crystal structure of thermus aquaticus DNA polymerase. Nature, 376, 612-616. [CrossRef]
- Carrodeguas, J.A.; Kobayashi, R.; Lim, S.E.; Copeland, W.C.; Bogenhagen, D.F.1999 The accessory subunit of xenopus laevis mitochondrial DNA polymerase gamma increases processivity of the catalytic subunit of human DNA polymerase gamma and is related to class ii aminoacyl-tRNA synthetases. Mol Cell Biol, 19, 4039-4046. [CrossRef]
- Malaby, A.W.; Martin, S.K.; Wood, R.D.; Doublie, S.2017 Expression and structural analyses of human DNA polymerase theta (polq). Methods Enzymol, 592, 103-121. [CrossRef]
- Leonhardt, E.A.; Henderson, D.S.; Rinehart, J.E.; Boyd, J.B.1993 Characterization of the mus308 gene in drosophila melanogaster. Genetics, 133, 87-96. [CrossRef]
- Kent, T.; Mateos-Gomez, P.A.; Sfeir, A.; Pomerantz, R.T.2016 Polymerase theta is a robust terminal transferase that oscillates between three different mechanisms during end-joining. Elife, 5. [CrossRef]
- Marini, F.; Kim, N.; Schuffert, A.; Wood, R.D.2003 Poln, a nuclear pola family DNA polymerase homologous to the DNA cross-link sensitivity protein mus308. J Biol Chem, 278, 32014-32019. [CrossRef]
- Takata, K.; Shimizu, T.; Iwai, S.; Wood, R.D.2006 Human DNA polymerase n (poln) is a low fidelity enzyme capable of error-free bypass of 5s-thymine glycol. J Biol Chem, 281, 23445-23455. [CrossRef]
- Sharief, F.S.; Vojta, P.J.; Ropp, P.A.; Copeland, W.C.1999 Cloning and chromosomal mapping of the human DNA polymerase theta (polq), the eighth human DNA polymerase. Genomics, 59, 90-96. [CrossRef]
- Inagaki, S.; Suzuki, T.; Ohto, M.A.; Urawa, H.; Horiuchi, T.; Nakamura, K.; Morikami, A.2006 Arabidopsis tebichi, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell, 18, 879-892. [CrossRef]
- Schoenfeld, T.W.; Murugapiran, S.K.; Dodsworth, J.A.; Floyd, S.; Lodes, M.; Mead, D.A.; Hedlund, B.P.2013 Lateral gene transfer of family a DNA polymerases between thermophilic viruses, aquificae, and apicomplexa. Mol Biol Evol, 30, 1653-1664. [CrossRef]
- Kent, T.; Rusanov, T.D.; Hoang, T.M.; Velema, W.A.; Krueger, A.T.; Copeland, W.C.; Kool, E.T.; Pomerantz, R.T.2016 DNA polymerase theta specializes in incorporating synthetic expanded-size (xdna) nucleotides. Nucleic Acids Res, 44, 9381-9392. [CrossRef]
- Zatreanu, D.; Robinson, H.M.R.; Alkhatib, O.; Boursier, M.; Finch, H.; Geo, L.; Grande, D.; Grinkevich, V.; Heald, R.A.; Langdon, S.; Majithiya, J.; McWhirter, C.; Martin, N.M.B.; Moore, S.; Neves, J.; Rajendra, E.; Ranzani, M.; Schaedler, T.; Stockley, M.; Wiggins, K.; Brough, R.; Sridhar, S.; Gulati, A.; Shao, N.; Badder, L.M.; Novo, D.; Knight, E.G.; Marlow, R.; Haider, S.; Callen, E.; Hewitt, G.; Schimmel, J.; Prevo, R.; Alli, C.; Ferdinand, A.; Bell, C.; Blencowe, P.; Bot, C.; Calder, M.; Charles, M.; Curry, J.; Ekwuru, T.; Ewings, K.; Krajewski, W.; MacDonald, E.; McCarron, H.; Pang, L.; Pedder, C.; Rigoreau, L.; Swarbrick, M.; Wheatley, E.; Willis, S.; Wong, A.C.; Nussenzweig, A.; Tijsterman, M.; Tutt, A.; Boulton, S.J.; Higgins, G.S.; Pettitt, S.J.; Smith, G.C.M.; Lord, C.J.2021 Poltheta inhibitors elicit brca-gene synthetic lethality and target parp inhibitor resistance. Nat Commun, 12, 3636. [CrossRef]
- Bubenik, M.; Mader, P.; Mochirian, P.; Vallee, F.; Clark, J.; Truchon, J.F.; Perryman, A.L.; Pau, V.; Kurinov, I.; Zahn, K.E.; Leclaire, M.E.; Papp, R.; Mathieu, M.C.; Hamel, M.; Duffy, N.M.; Godbout, C.; Casas-Selves, M.; Falgueyret, J.P.; Baruah, P.S.; Nicolas, O.; Stocco, R.; Poirier, H.; Martino, G.; Fortin, A.B.; Roulston, A.; Chefson, A.; Dorich, S.; St-Onge, M.; Patel, P.; Pellerin, C.; Ciblat, S.; Pinter, T.; Barabe, F.; El Bakkouri, M.; Parikh, P.; Gervais, C.; Sfeir, A.; Mamane, Y.; Morris, S.J.; Black, W.C.; Sicheri, F.; Gallant, M.2022 Identification of rp-6685, an orally bioavailable compound that inhibits the DNA polymerase activity of poltheta. J Med Chem, 65, 13198-13215. [CrossRef]
- Stockley, M.L.; Ferdinand, A.; Benedetti, G.; Blencowe, P.; Boyd, S.M.; Calder, M.; Charles, M.D.; Edwardes, L.V.; Ekwuru, T.; Finch, H.; Galbiati, A.; Geo, L.; Grande, D.; Grinkevich, V.; Holliday, N.D.; Krajewski, W.W.; MacDonald, E.; Majithiya, J.B.; McCarron, H.; McWhirter, C.L.; Patel, V.; Pedder, C.; Rajendra, E.; Ranzani, M.; Rigoreau, L.J.M.; Robinson, H.M.R.; Schaedler, T.; Sirina, J.; Smith, G.C.M.; Swarbrick, M.E.; Turnbull, A.P.; Willis, S.; Heald, R.A.2022 Discovery, characterization, and structure-based optimization of small-molecule in vitro and in vivo probes for human DNA polymerase theta. J Med Chem, 65, 13879-13891. [CrossRef]
- Chheda, P.R.; Nieto, N.; Kaur, S.; Beck, J.M.; Beck, J.R.; Honzatko, R.; Kerns, R.J.; Nelson, S.W.2022 Promising antimalarials targeting apicoplast DNA polymerase from plasmodium falciparum. Eur J Med Chem, 243, 114751. [CrossRef]
- Rahman, S.; Copeland, W.C.2019 Polg-related disorders and their neurological manifestations. Nat Rev Neurol, 15, 40-52. [CrossRef]
- Wood, R.D.; Doublie, S.2022 Genome protection by DNA polymerase theta. Annu Rev Genet, 56, 207-228. [CrossRef]
- Newman, J.A.; Cooper, C.D.O.; Aitkenhead, H.; Gileadi, O.2015 Structure of the helicase domain of DNA polymerase theta reveals a possible role in the microhomology-mediated end-joining pathway. Structure, 23, 2319-2330. [CrossRef]
- Zahn, K.E.; Averill, A.M.; Aller, P.; Wood, R.D.; Doublie, S.2015 Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat Struct Mol Biol, 22, 304-311. [CrossRef]
- Yousefzadeh, M.J.; Wyatt, D.W.; Takata, K.; Mu, Y.; Hensley, S.C.; Tomida, J.; Bylund, G.O.; Doublie, S.; Johansson, E.; Ramsden, D.A.; McBride, K.M.; Wood, R.D.2014 Mechanism of suppression of chromosomal instability by DNA polymerase polq. PLoS Genet, 10, e1004654. [CrossRef]
- Hogg, M.; Sauer-Eriksson, A.E.; Johansson, E.2012 Promiscuous DNA synthesis by human DNA polymerase theta. Nucleic Acids Res, 40, 2611-2622. [CrossRef]
- Chandramouly, G.; Zhao, J.; McDevitt, S.; Rusanov, T.; Hoang, T.; Borisonnik, N.; Treddinick, T.; Lopezcolorado, F.W.; Kent, T.; Siddique, L.A.; Mallon, J.; Huhn, J.; Shoda, Z.; Kashkina, E.; Brambati, A.; Stark, J.M.; Chen, X.S.; Pomerantz, R.T.2021 Poltheta reverse transcribes RNA and promotes RNA-templated DNA repair. Sci Adv, 7. [CrossRef]
- Chen, C.C.; Feng, W.; Lim, P.X.; Kass, E.M.; Jasin, M.2018 Homology-directed repair and the role of brca1, brca2, and related proteins in genome integrity and cancer. Annu Rev Cancer Biol, 2, 313-336. [CrossRef]
- Ramsden, D.A.; Carvajal-Garcia, J.; Gupta, G.P.2022 Mechanism, cellular functions and cancer roles of polymerase-theta-mediated DNA end joining. Nat Rev Mol Cell Biol, 23, 125-140. [CrossRef]
- Goff, J.P.; Shields, D.S.; Seki, M.; Choi, S.; Epperly, M.W.; Dixon, T.; Wang, H.; Bakkenist, C.J.; Dertinger, S.D.; Torous, D.K.; Wittschieben, J.; Wood, R.D.; Greenberger, J.S.2009 Lack of DNA polymerase theta (polq) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat Res, 172, 165-174. [CrossRef]
- Welcsh, P.L.; King, M.C.2001 Brca1 and brca2 and the genetics of breast and ovarian cancer. Hum Mol Genet, 10, 705-713. [CrossRef]
- Feng, W.; Simpson, D.A.; Carvajal-Garcia, J.; Price, B.A.; Kumar, R.J.; Mose, L.E.; Wood, R.D.; Rashid, N.; Purvis, J.E.; Parker, J.S.; Ramsden, D.A.; Gupta, G.P.2019 Genetic determinants of cellular addiction to DNA polymerase theta. Nat Commun, 10, 4286. [CrossRef]
- Ceccaldi, R.; Liu, J.C.; Amunugama, R.; Hajdu, I.; Primack, B.; Petalcorin, M.I.; O’Connor, K.W.; Konstantinopoulos, P.A.; Elledge, S.J.; Boulton, S.J.; Yusufzai, T.; D’Andrea, A.D.2015 Homologous-recombination-deficient tumours are dependent on poltheta-mediated repair. Nature, 518, 258-262. [CrossRef]
- Caracciolo, D.; Riillo, C.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P.2021 Alternative non-homologous end-joining: Error-prone DNA repair as cancer’s achilles’ heel. Cancers (Basel), 13. [CrossRef]
- Du, Y.; Yamaguchi, H.; Hsu, J.L.; Huang, M.C.2017 Parp inhibitors as precision medicine for cancer treatment. Natl. Sci. Rev., 4, 576–592. [CrossRef]
- Rose, M.; Burgess, J.T.; O’Byrne, K.; Richard, D.J.; Bolderson, E.2020 Parp inhibitors: Clinical relevance, mechanisms of action and tumor resistance. Front Cell Dev Biol, 8, 564601. [CrossRef]
- Li, H.; Liu, Z.Y.; Wu, N.; Chen, Y.C.; Cheng, Q.; Wang, J.2020 Parp inhibitor resistance: The underlying mechanisms and clinical implications. Mol Cancer, 19, 107. [CrossRef]
- Krais, J.J.; Glass, D.J.; Chudoba, I.; Wang, Y.; Feng, W.; Simpson, D.; Patel, P.; Liu, Z.; Neumann-Domer, R.; Betsch, R.G.; Bernhardy, A.J.; Bradbury, A.M.; Conger, J.; Yueh, W.T.; Nacson, J.; Pomerantz, R.T.; Gupta, G.P.; Testa, J.R.; Johnson, N.2023 Genetic separation of brca1 functions reveal mutation-dependent poltheta vulnerabilities. Nat Commun, 14, 7714. [CrossRef]
- Zhou, J.; Gelot, C.; Pantelidou, C.; Li, A.; Yucel, H.; Davis, R.E.; Farkkila, A.; Kochupurakkal, B.; Syed, A.; Shapiro, G.I.; Tainer, J.A.; Blagg, B.S.J.; Ceccaldi, R.; D’Andrea, A.D.2021 A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nat Cancer, 2, 598-610. [CrossRef]
- Pismataro, M.C.; Astolfi, A.; Barreca, M.L.; Pacetti, M.; Schenone, S.; Bandiera, T.; Carbone, A.; Massari, S.2023 Small molecules targeting DNA polymerase theta (poltheta) as promising synthetic lethal agents for precision cancer therapy. J Med Chem, 66, 6498-6522. [CrossRef]
- Rodriguez-Berriguete, G.; Ranzani, M.; Prevo, R.; Puliyadi, R.; Machado, N.; Bolland, H.R.; Millar, V.; Ebner, D.; Boursier, M.; Cerutti, A.; Cicconi, A.; Galbiati, A.; Grande, D.; Grinkevich, V.; Majithiya, J.B.; Piscitello, D.; Rajendra, E.; Stockley, M.L.; Boulton, S.J.; Hammond, E.M.; Heald, R.A.; Smith, G.C.M.; Robinson, H.M.R.; Higgins, G.S.2023 Small-molecule poltheta inhibitors provide safe and effective tumor radiosensitization in preclinical models. Clin Cancer Res, 29, 1631-1642. [CrossRef]
- Gellert, M.; O’Dea, M.H.; Itoh, T.; Tomizawa, J.1976 Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A, 73, 4474-4478. [CrossRef]
- Kalanon, M.; McFadden, G.I.2010 Malaria, plasmodium falciparum and its apicoplast. Biochem Soc Trans, 38, 775-782. [CrossRef]
- Kaur, S.; Nieto, N.S.; McDonald, P.; Beck, J.R.; Honzatko, R.B.; Roy, A.; Nelson, S.W.2022 Discovery of small molecule inhibitors of plasmodium falciparum apicoplast DNA polymerase. J Enzyme Inhib Med Chem, 37, 1320-1326. [CrossRef]
- Sato, S.2011 The apicomplexan plastid and its evolution. Cell Mol Life Sci, 68, 1285-1296. [CrossRef]
- Wilson, R.J.; Denny, P.W.; Preiser, P.R.; Rangachari, K.; Roberts, K.; Roy, A.; Whyte, A.; Strath, M.; Moore, D.J.; Moore, P.W.; Williamson, D.H.1996 Complete gene map of the plastid-like DNA of the malaria parasite plasmodium falciparum. J Mol Biol, 261, 155-172. [CrossRef]
- Seeber, F.; Soldati-Favre, D.2010 Metabolic pathways in the apicoplast of apicomplexa. Int Rev Cell Mol Biol, 281, 161-228. [CrossRef]
- Seow, F.; Sato, S.; Janssen, C.S.; Riehle, M.O.; Mukhopadhyay, A.; Phillips, R.S.; Wilson, R.J.; Barrett, M.P.2005 The plastidic DNA replication enzyme complex of plasmodium falciparum. Mol Biochem Parasitol, 141, 145-153. [CrossRef]
- Milton, M.E.; Choe, J.Y.; Honzatko, R.B.; Nelson, S.W.2016 Crystal structure of the apicoplast DNA polymerase from plasmodium falciparum: The first look at a plastidic a-family DNA polymerase. J Mol Biol, 428, 3920-3934. [CrossRef]
- Pandey, A.R.; Singh, S.P.; Joshi, P.; Srivastav, K.S.; Srivastava, S.; Yadav, K.; Chandra, R.; Bisen, A.C.; Agrawal, S.; Sanap, S.N.; Bhatta, R.S.; Tripathi, R.; Barthwal, M.K.; Sashidhara, K.V.2023 Design, synthesis and evaluation of novel pyrrole-hydroxybutenolide hybrids as promising antiplasmodial and anti-inflammatory agents. Eur J Med Chem, 254, 115340. [CrossRef]
- Lansdon, E.B.; Brendza, K.M.; Hung, M.; Wang, R.; Mukund, S.; Jin, D.; Birkus, G.; Kutty, N.; Liu, X.2010 Crystal structures of HIV-1 reverse transcriptase with etravirine (tmc125) and rilpivirine (TMC278): Implications for drug design. J Med Chem, 53, 4295-4299. [CrossRef]
- Das, K.; Bauman, J.D.; Clark, A.D., Jr.; Frenkel, Y.V.; Lewi, P.J.; Shatkin, A.J.; Hughes, S.H.; Arnold, E.2008 High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci U S A, 105, 1466-1471. [CrossRef]
- Singh, K.; Marchand, B.; Rai, D.K.; Sharma, B.; Michailidis, E.; Ryan, E.M.; Matzek, K.B.; Leslie, M.D.; Hagedorn, A.N.; Li, Z.; Norden, P.R.; Hachiya, A.; Parniak, M.A.; Xu, H.T.; Wainberg, M.A.; Sarafianos, S.G.2012 Biochemical mechanism of HIV-1 resistance to rilpivirine. J Biol Chem, 287, 38110-38123. [CrossRef]
- Centers for disease control and prevention. Antibiotic resistance threats in the united states. U.S. Department of health and human services 2019. Available at https://www.Cdc.Gov/drugresistance/biggest-threats.Html. Accessed on oct. 10, 2023. Available at https://www.cdc.gov/DrugResistance/Biggest-Threats.html.
- Antimicrobial Resistance, C.2022 Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet, 399, 629-655.
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L.J.N.m.2019 Defining and combating antibiotic resistance from one health and global health perspectives. 4, 1432-1442. [CrossRef]
- McEwen, S.A.; Collignon, P.J.J.M.s.2018 Antimicrobial resistance: A one health perspective. 6, 6.2. 10. [CrossRef]
- Chatterjee, A.; Modarai, M.; Naylor, N.R.; Boyd, S.E.; Atun, R.; Barlow, J.; Holmes, A.H.; Johnson, A.; Robotham, J.V.2018 Quantifying drivers of antibiotic resistance in humans: A systematic review. Lancet Infect Dis, 18, e368-e378. [CrossRef]
- Tamma, P.D.; Cosgrove, S.E.2020 Unlikely bedfellows: The partnering of antibiotic stewardship programs and the pharmaceutical industry. Clin Infect Dis, 71, 682-684. [CrossRef]
- Blaskovich, M.A.J.A.I.D. Antibiotic alternatives special issue. ACS Publications: 2021; Vol. 7, pp 2025-2026. [CrossRef]
- O’Neill, J.2016 Tackling drug-resistant infections globally: Final report and recommendations.
- Kornberg, A.; Baker, T.A. DNA replication. W. H. Greeman: 1992; Vol. 3.
- Hejna, J.A.; Moses, R.E. DNA replication. Elsevier Inc.: 2009.
- De Lucia, P.; Cairns, J.1969 Isolation of an e. Coli strain with a mutation affecting DNA polymerase. Nature, 224, 1164-1166. [CrossRef]
- Maul, R.W.; Sanders, L.H.; Lim, J.B.; Benitez, R.; Sutton, M.D.2007 Role of escherichia coli DNA polymerase i in conferring viability upon the dnan159 mutant strain. J Bacteriol, 189, 4688-4695. [CrossRef]
- Garcia-Ortiz, M.V.; Marsin, S.; Arana, M.E.; Gasparutto, D.; Guerois, R.; Kunkel, T.A.; Radicella, J.P.2011 Unexpected role for helicobacter pylori DNA polymerase i as a source of genetic variability. PLoS Genet, 7, e1002152. [CrossRef]
- Rondon, M.R.; Horswill, A.R.; Escalante-Semerena, J.C.1995 DNA polymerase i function is required for the utilization of ethanolamine, 1,2-propanediol, and propionate by salmonella typhimurium lt2. J Bacteriol, 177, 7119-7124. [CrossRef]
- Li, Y.; Korolev, S.; Waksman, G.1998 Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of thermus aquaticus DNA polymerase i: Structural basis for nucleotide incorporation. EMBO J, 17, 7514-7525. [CrossRef]
- Kohlstaedt, L.A.; Wang, J.; Friedman, J.M.; Rice, P.A.; Steitz, T.A.1992 Crystal structure at 3.5 a resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science, 256, 1783-1790. [CrossRef]
- Huang, H.; Chopra, R.; Verdine, G.L.; Harrison, S.C.1998 Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science, 282, 1669-1675. [CrossRef]
- Das, K.; Martinez, S.E.; Bauman, J.D.; Arnold, E.2012 HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat Struct Mol Biol, 19, 253-259. [CrossRef]
- Singh, K.; Flores, J.A.; Kirby, K.A.; Neogi, U.; Sonnerborg, A.; Hachiya, A.; Das, K.; Arnold, E.; McArthur, C.; Parniak, M.; Sarafianos, S.G.2014 Drug resistance in non-B subtype HIV-1: Impact of HIV-1 reverse transcriptase inhibitors. Viruses, 6, 3535-3562. [CrossRef]
- Franklin, M.C.; Wang, J.; Steitz, T.A.2001 Structure of the replicating complex of a pol alpha family DNA polymerase. Cell, 105, 657-667. [CrossRef]
- Johnson, K.A.1993 Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem, 62, 685-713. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




