Submitted:
28 February 2024
Posted:
28 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introductions
2. Potential Biological Roles of CHI3L1
2.1. Increased Levels of CHI3L1 under the Major Inflammatory Disorders
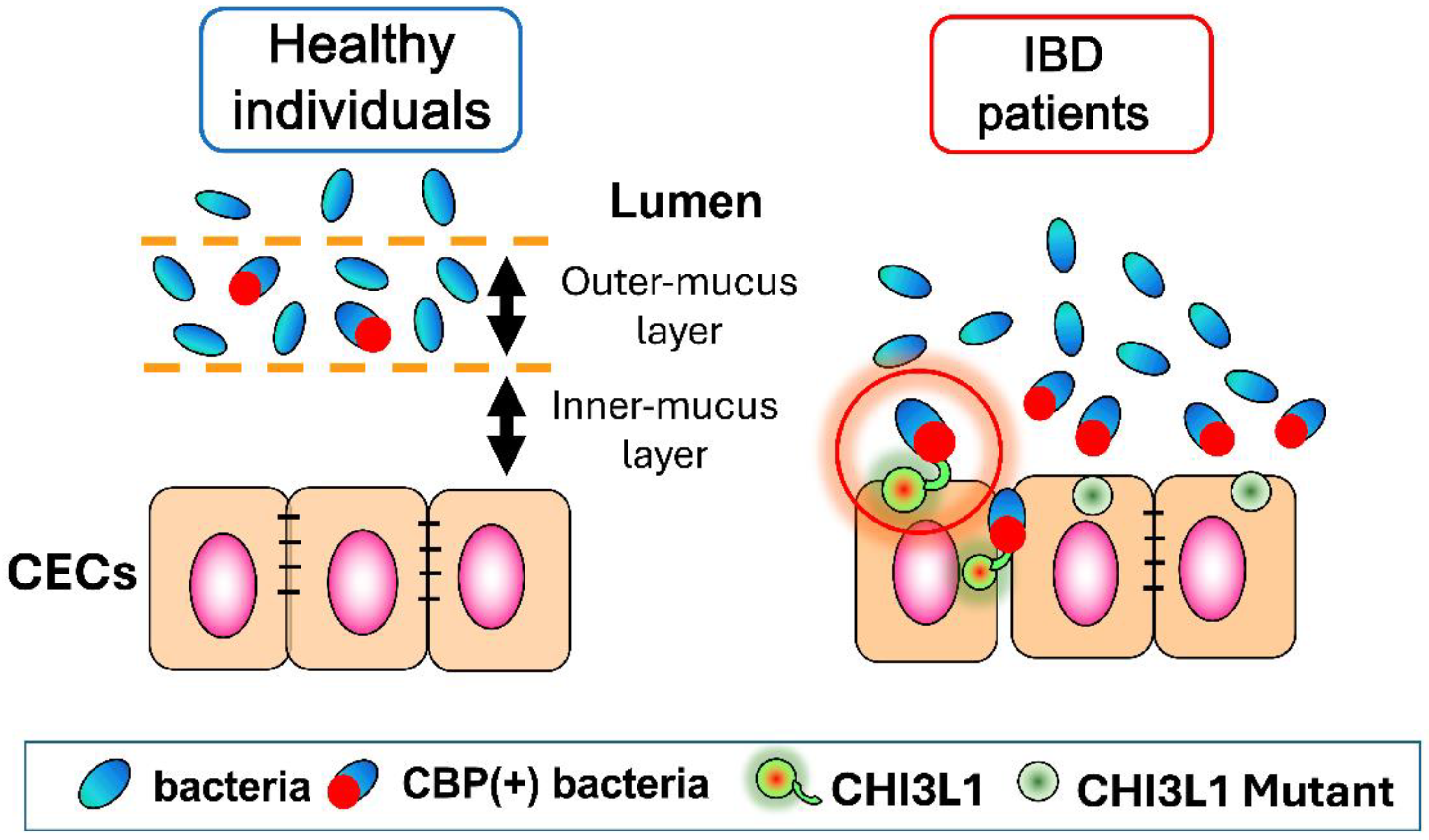
2.1.1. Inflammatory Bowel Disease (IBD)
2.1.2. Multiple Sclerosis (MS)
2.1.3. Alzheimer’s Disease (AD)
2.1.4. Asthma, Chronic Obstructive Pulmonary Disease (COPD)
2.2. CHI3L1 Expression in Various Cell Types
3. CHI3L1-Mediated Host-Microbial Interactions
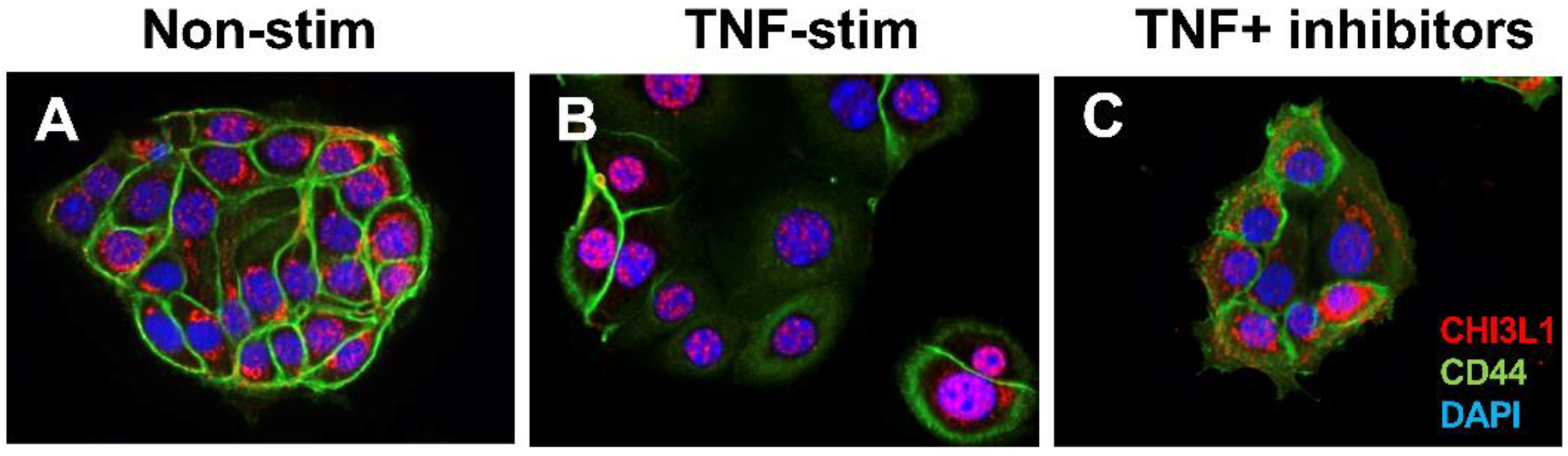
3.1. CHI3L1 as an Enhancer of Bacterial Adhesion and Invasion on Colonic Epithelial Cells
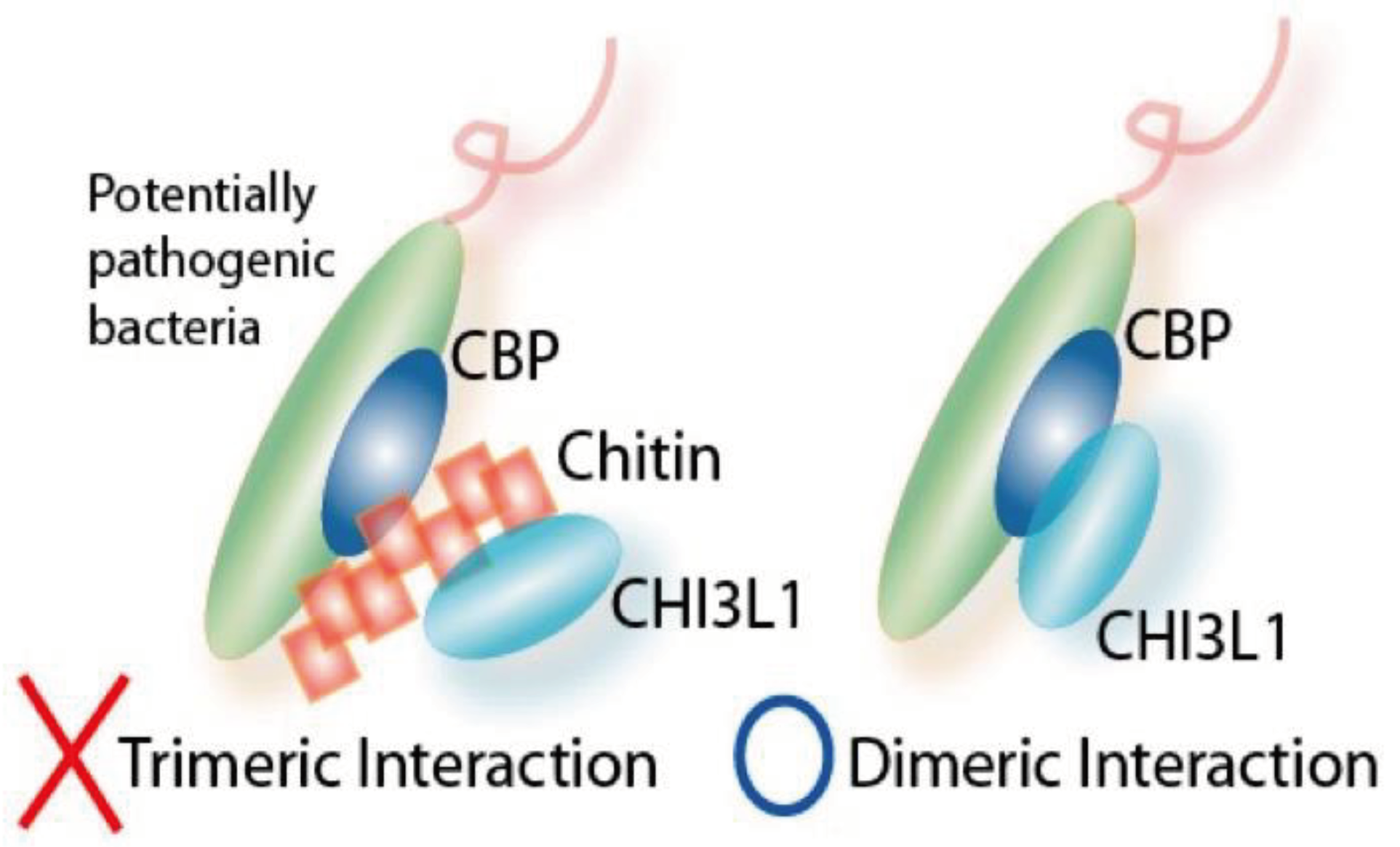
3.2. Interactions between CHI3L1 and Bacterial Chitinase (ChiA) in Escherichia coli
3.3. Potential Role of CHI3L1 as an Inducer of Intestinal Dysbiosis
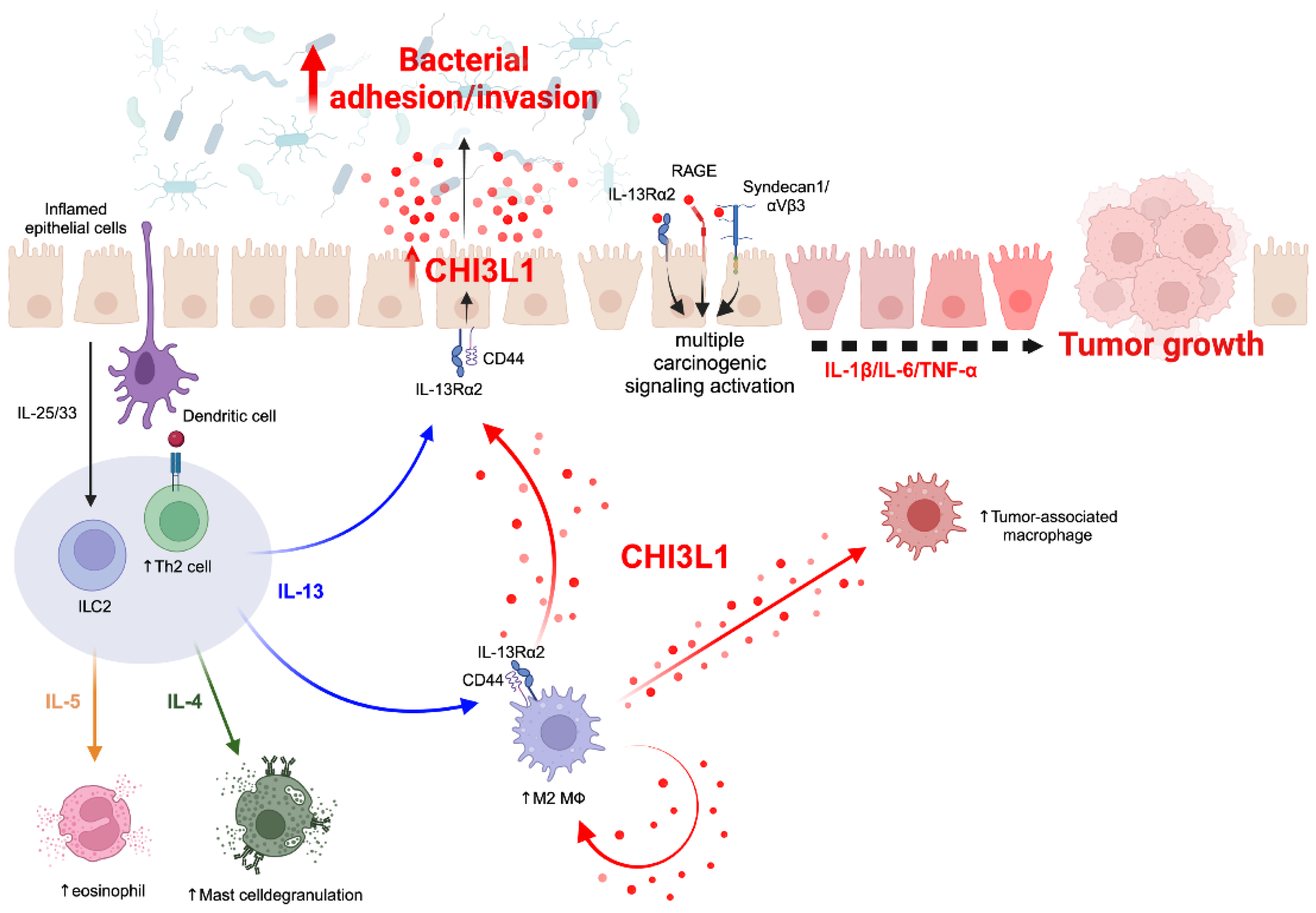
4. Association between CHI3L1 and Chronic Inflammation
4.1. CHI3L1-Associated Chronic Inflammation in Animal Models
4.2. CHI3L1-Associated Chronic Inflammation in Human
5. Therapeutic Potentials of CHI3L1-Blockers/Inhibitors for Various Diseases
5.1. Anti-CHI3L1 Antibody
5.2. Methylxanthine Derivatives Including Caffeine as a Chitinase Inhibitor
5.3. Chitin Microparticles and Chito-Oligosaccharides
6. Conclusions
Acknowledgments
Grant Supports
References
- Podolsky, D.K. Inflammatory bowel disease (1). N. Engl. J. Med. 1991, 325, 928–37. [Google Scholar] [CrossRef]
- Podolsky, D.K. Inflammatory bowel disease (2). N. Engl. J. Med. 1991, 325, 1008–1016. [Google Scholar] [CrossRef]
- Podolsky, D.K. Inflammatory bowel disease. N. Engl. J. Med. 2002, 347, 417–429. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current understanding of dysbiosis in disease in human and animal models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Ke, X.; You, K.; Pichaud, M.; Haiser, H.J.; Graham, D.B.; Viamakis, H.; Porter, J.A.; Xavier, R.J. Gut bacterial metabolites modulate endoplasmic reticulum stress. Genome Biol. 2021, 22, 292. [Google Scholar] [CrossRef]
- You, K.; Wang, L.; Chou, C.H.; Liu, K.; Nakata, T.; Jaiswal, A.; Yao, J.; Lefkovith, A.; Omar, A.; Perrigoue, J.G.; et al. QRICH1 dictates the outcome of ER stress through transcriptional control of proteostasis. Science 2021, 371, eabb6896. [Google Scholar] [CrossRef]
- Mizoguchi, E. Chitinase 3-like-1 exacerbates intestinal inflammation by enhancing bacterial adhesion and invasion in colonic epithelial cells. Gastroenterology 2006, 130, 398–411. [Google Scholar] [CrossRef]
- Kawada, M.; Chen, C.C.; Arihiro, A.; Nagatani, K.; Watanabe, T.; Mizoguchi, E. Chitinase 3-like-1 enhances bacterial adhesion to colonic epithelial cells through the interaction with bacterial chitin-binding protein. Lab Invest 2008, 88, 883–895. [Google Scholar] [CrossRef]
- Low, D; Tran, H.T.; Lee, I.A.; Dreux, N.; Kamba, A.; Reinecker, H.C.; Darfeuille-Michaud, A.; Barnich, N.; Mizoguchi, E. Chitin-binding domains of Escherichia coli ChA mediate interactions with intestinal epithelial cells in mice with colitis. Gastroenterology 2013, 145, 602-612. [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active enZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acid Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Lee, C.G.; Dela Cruz, C.S.; Ma, B.; Ahangari, F.; Zhou, Y.; Halaban, R.; Sznol, M.; Elias, J.A. Chitinase-like proteins in lung injury, repair, and metastasis. Proc. Am. Thora. Soc. 2012, 9, 57–61. [Google Scholar] [CrossRef]
- Houston, D.R.; Recklies, A.D.; Krupa, J.C.; van Aalten, D.M. Structure and ligand-induced conformational change of the 39-kDa glycoprotein from human articular chondrocytes. J. Biol. Chem. 2003, 278, 30206–30212. [Google Scholar] [CrossRef]
- Kawada, M.; Hachiya, Y.; Arihiro, A.; Mizoguchi, E. Role of mammalian chitinases in inflammatory conditions. Keio J. Med. 2007, 56, 21–27. [Google Scholar] [CrossRef]
- Hamid, R.; Khan, M.A.; Ahmad, M.; Ahmad, M.M.; Abdin, M.Z; Musarrat. J.; Javed, S. Chitinases: an update. J. Pharm. Bioallied Sci. 2013, 5, 21-29. [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Zhu, Z.; Zheng, T.; Homer, R.J.; Kim, Y.K.; Chen, N.Y.; Cohn, L.; Hamid, Q.; Elias, J.A. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 2004, 304, 1678–1682. [Google Scholar] [CrossRef]
- Chupp, G.I.; Lee, C.G.; Jarjour, N.; Shim, Y.M.; Holm, C.T.; He, S.; Dziura, J.D.; Reed, J.; Coyle, A.J.; Kiener, P.; et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N. Eng. J. Med. 2007, 357, 2016–2027. [Google Scholar] [CrossRef]
- Blazevic, N.; Rogic, D.; Pelajic, S.; Miler, M.; Glancic, G.; Ratkajec, V.; Vrkolina, N.; Bakula, D.; Hrabar, D.; Pavic T. YKL-40 as a biomarker in various inflammatory diseases: A review. Biochem. Med. (Zagreb). 2024, 34: 010502. [CrossRef]
- Zhao, T.; Su, Z.; Li, Y.; Zhang, X.; You, Q. Chitinase-3 like-protein-1 function and its role in diseases. Signal Transduct. Target Ther. 2020, 5, 201. [Google Scholar] [CrossRef]
- Koutroubakis, I.E.; Petinaki, E.; Dimoulios, P.; Vardas, E.; Roussomoustakaki, M.; Maniatis, A.N.; Kouroumalis, E.A. Increased serum levels of YKL-40 in patients with inflammatory bowel disease. Int. J. Colorectal Dis. 2003, 18, 254–259. [Google Scholar] [CrossRef]
- Vind, I,; Johansen, J.S.; Price, P.A.; Munkholm, P. Serum YKL-40, a potential new marker of disease activity in patients with inflammatory bowel disease. Scand. J. Gastroenterol. 2003, 38, 599-605. [CrossRef]
- Punzi, L.; Podswiadek, M.; D’Inca, R.; Zaninotto, M.; Bernardi, D.; Pienani, M.; Sturniolo, G.C. Serum human cartilage glycoprotein 39 as a marker of arthritis associated with inflammatory bowel disease. Ann. Rheum. Dis. 2003, 62, 1224–1226. [Google Scholar] [CrossRef]
- Erzin, Y.; Uzun, H.; Karatas. A.; Celik, A.F. Serum YJK-40 as a marker of disease activity and structure formation in patients with Crohn’s disease. J. Gastroenterol. Hepatol. 2008, 23: e357-362. [CrossRef]
- Pieczarkowski, S.; Kowalska-Deptuch, K.; Kwinta, P.; Wedrychowicz, A.; Tomaski, P.; Stochel-Gaudyn, A.; Fyderek, K. Serum concentration of fibrosis markers in children with inflammatory bowel disease 2020, 60, 61-74. [CrossRef]
- Douadi, C.; Vazeille, E.; Chambon, C.; Hebraud, M.; Fargeas, M.; Dodel, M.; Coban, D.; Pereira, B.; Birer, A.; Sauvanet, P.; et al. Anti-TNF agents restrict adherent-invasive Escherichia coli replication within macrophages through modulation of chitinase 3-like 1 in patients with Crohn’s disease. J. Crohns Colitis 2022, 16, 1140–1150. [Google Scholar] [CrossRef]
- Deutschmann, C.; Roggenbuck, D.; Schierack, P. The loss of tolerance to CHI3L1-A putative role in inflammatory bowel disease? Clin. Immunol. 2019, 199, 12–17. [Google Scholar] [CrossRef]
- Akesson, J.; Hojjati, S.; Hellberg, S.; Reffetseder, J.; Khademi, M.; Rynkowski, R.; Kockum, I.; Altafini, C.; Lubovac-Pilav, Z.; Mellergard, J.; et al. Proteomics reveal biomarkers for diagnosis, disease activity and long-term disability outcomes in multiple sclerosis. Nat. Commun. 2023, 14, 6903. [Google Scholar] [CrossRef]
- Talaat, F.; Abdelatty, S.; Ragaie, C.; Dahshan, A. Chitinase-3-like 1-protein in CFS: a novel biomarker for progression in patients with multiple sclerosis. Neurol. Sci. 2023, 44, 3243–3252. [Google Scholar] [CrossRef]
- Ahmad, I.; Wergeland, S.; Overland, E.; Bϕ, L. An association of chitinase-3 like-protein-1 with neural deterioration in multiple sclerosis. A.S.N. Neuro. 2023, 15, 17590914231198980. [Google Scholar]
- Lamancova, P.; Urban, P.; Maslankova, J.; Rabajdova, M.; Marekova, M. Correlation of selected serum protein levels with the degree of disability and NEDA-3 status in multiple sclerosis phenotypes. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 3933–3941. [Google Scholar]
- Donder, A.; Ozdemir, H.H. Serum YKL-40 levels in patients with multiple sclerosis. Arq. Neuropsiquiatr. 2021, 79, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Magliozzi, R.; Pezzini, F.; Pucci, M.; Rossi, S.; Facchiano, F.; Marastoni, D.; Montagnana, M.; Lippi, G.; Reynolds, R.; Calabrese, M. Changes in cerebrospinal fluid balance of TNF and TNF receptors in Naïve multiple sclerosis patients: Early involvement in compartmentalized intrathecal inflammation. Cells 2021, 10, 1712. [Google Scholar] [CrossRef] [PubMed]
- Cubas-Nunez, L.; Gil-Perotin, S.; Castillo-Villalba, J.; Lopez, V.; Tarazona, L.S.; Gasque-Rubio, R.; Carratala-Bosca, S.; Alcala-Vicente, C.; Perez-Miralles, F.; Lassmann, H.; et al. Potential role of CHI3L1+ astrocytes in progression in MS. Neurol. Neuroimmunol. Neuroinflamm. 2021, 8, e972. [Google Scholar] [CrossRef] [PubMed]
- Mahler, M.R.; Sφndergaard, H.B.; Buheit, S.; von Essen, M.R.; Christensen, J.R.; Enevold, C.; Sellebjerg, F. Multiplex assessment of cerebrospinal fluid biomarkers in multiple sclerosis. Mult. Scle.r. Relat. Disord. 2020, 45, 102391. [Google Scholar] [CrossRef] [PubMed]
- Picon, C.; Tejeda-Velarde, A.; Fernandez-Velasco, J.I.; Comabella, M.; Alvarez-Lafuente, R.; Quintane, E.; de la Maza, S.S.; Monreal E, Villarrubia, N.; Alvarez-Carmeno, J.C.; et al. Identification of the immunological changes appearing in the CSF during the early immunofluorescence process occurring in multiple sclerosis. Front. Immunol. 2021, 12, 685139. [CrossRef]
- Neumann, A.; Ohlei, O.; Kucukali, F.; Bos, I.; Timsina, J.; Vos, S.; Prokopenko, D.; Tijms, B.M.; Andreasson, U.; Blennow, K.; et al. Multivariate GWAS of Alzheimer’s disease CSF biomarker profiles implies GRIN2D in synaptic functioning. Genome. Med. 2023, 15, 79. [Google Scholar] [CrossRef]
- Connolly, K.; Lehoux, M.; O’Rourke, R.; Assetta, B.; Erdemir, G.A.; Elias, J.A.; Lee, C.G.; Huang, Y.W. Potential role of chitinase-3-like protein (CHI3L1/YKL-40) in neurodegeneration and Alzheimer’s disease. Alzheimers Dement. 2023, 19, 9–24. [Google Scholar] [CrossRef]
- Sanfilippo, C.; Castrogiovanni, P.; Imbesi, R.; Nunnari, G.; Rosa, M.D. Postsynaptic damage and microglial activation in AD patients could be linked CXCR4/CXCL12 expression levels. Brain Res. 2020, 1749, 147127. [Google Scholar] [CrossRef]
- Moreno-Rodriguez, M.; Perez, S.E.; Nadeem, M.; Malek-Ahmadi, M.; Mufson, E.J. Frontal cortex chitinase and pentraxin neuroinflammatory alterations during the progression of Alzheimer’s disease. J. Neuroinflammation 2020, 17, 58. [Google Scholar] [CrossRef]
- Wang, L.; Gao, T.; Cai, T.; Li, K.; Zheng, P.; Liu, J. Alzheimer’s Disease Neuroimaging initiative. Cerebrospinal fluid levels of YKL-40 in prodromal Alzheimer’s disease. Neurosci. Lett. 2020, 715, 134658. [CrossRef]
- Lleo, A., Alcolea, D.; Martinez-Lage, P.; Scheitens, P.; Parnetti, L.; Poirier, J.; Simonse, A.H.; Verbeek, M.M.; Rosa-Neto, P.; Slot, R.E.R.; et al. Longitudinal cerebrospinal fluid biomarker trajectories along the Alzheimer’s disease continuum in the BIOMARKAPD study. Alzheimers Dement. 2019, 15, 742-753. [CrossRef]
- Dhiman, K.; Blennow, K.; Zetterberg, H.; Martins, R.N.; Gupta, V.B. Cerebrospinal fluid biomarkers for understanding multiple aspect of Alzheimer’s disease pathogenesis. Cell Mol. Life Sci. 2019, 76, 1833–1863. [Google Scholar] [CrossRef] [PubMed]
- Nordengen, K.; Kirseborn, R.E.; Henjum, K.; Selnes, P.; Gisladottir, B.; Wettergreen, M.; Torsetnes, S.B.; Grontvedt, G.R.; Waterloo, K.K.; Aarsland, D.; et al. Glial activation and inflammation along the Alzheimer’s disease continuum. J. Neuroinflammation 2019, 16, 46. [Google Scholar] [CrossRef]
- Pan, R.; Zhu, X.; Zhou, Y.; Ding, L.; Cul, Y. Diagnostic value of YKL-40 for patients with asthma: A meta-analysis. Allergy Asthma Proc. 2021; 42, e167. [CrossRef]
- Paplinska-Goryca, M.; Misiukiewicz-Stepien, P.; Proboszcz, M.; Nejman-Gryz, P.; Gorska, K.; Krenke, R. The expression of TSLP, IL-33, and IL-17A in monocyte derived dendritic cells from asthma and COPD patients are related to epithelial-macrophage interactions. Cells 2020, 9: 1944. [CrossRef]
- Hubner, K.; Karwelat, D.; Pietsch, E.; Beinborn, I.; Winterberg, S.; Bedenberder, K.; Benedikter, B.; Schmeck, B.; Vollmeister, E. NF-κB-mediated inhibition of microRNA-149-5p regulates Chitinase-3-like 1 expression in human airway epithelial cells. Cell Signal. 2020, 67, 109498. [Google Scholar] [CrossRef] [PubMed]
- Knihtila, H.; Kotaniemi-Syrjanen, A.; Pelkonen, A.S.; Savinko, T.; Malmberg, L.P.; Makela, M.J. Serum chitinase-like protein YKL-40 is linked to small airway function in children with asthmatic symptoms. Pediatr. Allergy Immunol. 2019, 30, 803–809. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, X.; Liu, Y.; Zhang, L.; Wang, J.; Hansbro, P.M.; Wang, L.; Wang, G.; Hsu, A.C.Y. Chitinase-like protein YKL-40 correlates with inflammatory phenotypes, anti-asthma responsiveness and future exacerbations. Respir. Res. 2019, 20, 95. [Google Scholar] [CrossRef] [PubMed]
- Yoosuf, N.; Maciejewski, M.; Ziemek, D.; Jelinsky, S.A.; Folkersen, L.; Muller. M.; Sahistrom, P.; Vivar, N,; Catrina, A.; Berg, L.; et al. Early prediction of clinical response to anti-TNF treatment using multi-omics and machine learning in rheumatoid arthritis. Rheumatology (Oxford). 2022, 61, 1680-1689. [CrossRef]
- Parlak, E.; Laloglu, E. Analysis of Chitinase-3-like protein 1, IL-1-alpha, and IL-6 as novelinflammatory biomarker for COVID-19. J. Interferon Cytokine Res. 2022, 42, 536–541. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, R.; Sciorati, C.; Lore, N.I.; Capobianco, A.; Tresoldi, C.; Cirillo, D.M.: Ciceri, F.; Rovere-Querini, P.; Manfredi, A.A. Chitinase- 3-like protein-1 at hospital admission predicts COVID-19 outcome: a prospective cohort study. Sci Rep 2022, 12, 7606. [CrossRef]
- Kimura, Y.; Nakai, Y.; Shin, J.; Hara, M.; Takeda, Y.; Kubo, S.; Jeremiah, S.S.; Ino, Y.; Akiyama, T.; Moriyama, K.; et al. Identification of serum prognostic biomarkers of severe COVID-19 using a quantitative proteomic approach. Sci Rep 2021, 11, 20638. [Google Scholar] [CrossRef]
- Hammoudeh, S.M.; Hammoudeh, A.M.; Bhamidimarri, P.M.; Al Safar, H.; Mahboub, B.; Kunstner, A.; Busch, H.; Halwani, R.; Hamid, Q.; Rahmani, M.; et al. Systems immunology analysis reveals the contribution of pulmonary and extrapulmonary tissues to the immunopathogenesis of severe COVID-19 patients. Front. Immunol. 2021, 12, 595150. [Google Scholar] [CrossRef] [PubMed]
- Kang, Q.; Xu, J.; Luo, H.; Tan, N.; Chen, H.; Cheng, R.; Pan, J.; Han, Y.; Liu, D.; Xi, H.; et al. Direct antiviral agent treatment leads to rapid and significant fibrosis regression after HCV eradication. J Viral Hepat 2021, 28, 1284–1292. [Google Scholar] [CrossRef]
- Harrison, S.A.; Ratziu, V.; Boursier, J.; Francque, S.; Bedossa, P.; Majd, Z.; Cordonnier, G.; Sudrik, F.B.; Darteil, R.; Liebe, R.; et al. A blood-based biomarker panel (NIS4) for non-invasive diagnosis of non-alcoholic steatohepatitis and liver fibrosis: a prospective derivation and global validation study. Lancet Gastroenterol. Hepatol. 2020, 5, 970–985. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhong, M.; Wang, W.; Li, Y.H. Chi3l1 regulates APAP-induced liver injury by promoting macrophage infiltration. Eur. Rev. Med. Pharmacol. Sci. 2019; 23, 4996-5003. [CrossRef]
- Teratani, Y. Chitinase 3-like-1 expression is upregulated under inflammatory conditions in human oral epithelial cells. Kurume Med. J. 2023, 68, 221–228. [Google Scholar] [CrossRef]
- Duruk, G., Laloglu, E. Relationship between dental caries and YKL-40 levels in saliva. J. Clin. Pediatr. Dent. 2022, 46, 137-142. [CrossRef]
- Laucyte-Cibulskiene, A.; Ward, L.J.; Ebert, T.; Tosti, G.; Tucci, C.; Hernandez, L.; Kautzky-Willer, A.; Herrenro, M.T.; Kautky-Willer, A.; Herrero, M.T.; et al. Role of GDF-15, YKL-40 and MMP9 in patients with end-stage kidney disease: focus on sex-specific associations with vascular outcomes and all-cause mortality. Biol. Sex Differ. 2021, 12, 50. [Google Scholar] [CrossRef]
- Puthumana, J.; Thiessen-Philbrook, H.; Xu, L.; Coca, S.G.; Garg, A.X.; Himmelfarb, J.; Bhatraju, P.K.; Ikizler, T.A.; Siew, E.D.; Ware, L.B.; et al. Biomarkers of inflammation and repair in kidney disease progression. J. Clin. Invest. 2021, 131, e139927. [Google Scholar] [CrossRef]
- Schrauben, S.J.; Shou, H.; Zhang, X.; Anderson.; A.H.; Bonventre, J.V.; Chen, J.; Coca, S.; Furth, S.L.; Greenberg, J.H.; Gutierrez, O.M.; et al. Association of multiple plasma biomarker concentrations with progression of prevalent diabetic kidney disease: Findings from the chronic renal insufficiency cohort (CRIC) study. J. Am. Soc. Nephhrol. 2021, 32, 115-126. [CrossRef]
- Greenberg, J.H.; Abraham, A.G.; Xu, Y.; Schelling, J.R.; Feiman, H.I.; Sabbisetti, W.S.; Gonzalez, M.C.; Coca, S.; Schrauben, S.J.; Waikar, S.S.; et al. Plasma biomarkers of tubular injury and inflammation are associated with CKD progression in children. J Am. Soc. Nephrol. 2020, 31, 1067–1077. [Google Scholar] [CrossRef]
- Malhotra, R.; Katz. R.; Jotwani, V.; Ambrosius, W.; Raphael, K.L.; Haley, W.; Rastogi, A.; Cheung, A.K.; Freedman, B.I.; Punzi, H.; et al. Urine markers of kidney tubule cell injury and kidney function decline in SPRINT trial participants with CKD. Clin. J. Am. Soc. Nephrol. 2020, 15, 349–358. [CrossRef]
- Wallentin, L.; Eriksson, N.; Oiszowka, M.; Grammer, T.B.; Hagstrom, E.; Held, C.; Keleber, M.E.; Koenig, W.; Marz, W.; Stewart, R.A.H.; et al. Plasma proteins associated with cardiovascular death in patients with chronic coronary heart disease: A retrospective study. PLoS Med. 2021, 18, e1003513. [Google Scholar] [CrossRef]
- Folkersen, L.; Gustafsson, S.; Wang, Q.; Hansen, D.H.; Hedman, A.K.; Schork, A.; Page, K.; Zhernakova, D.V.; Wu, Y.; Peters, J.; et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30, 931 individuals. Nat. Metab. 2020, 2, 1135–1148. [Google Scholar] [CrossRef]
- Arain, F.; Abraityte, A.; Bogdanova, M.; Solberg, O.G.; Micheisen, A.E.; Lekva, T.; Aakhus, S.; Holm, S.; Halvorsen, B.; Finsen, A.V.; et al. YKL-40 (Chitinase-3-like protein 1) serum levels in aortic stenosis. Circ. Heart Fail. 2020, 13, e006643. [Google Scholar] [CrossRef]
- Kim, E.G.; Kim, M.N.; Hong, J.Y.; Lee, J.W.; Kim, S.Y.; Kim, K.W.; Lee, C.G.; Elias, J.A.; Song, T.W.; Sohn, M.Y. Chitinase 3-like 1 contributes to food allergy via M2 macrophage polarization. Allergy Asthma Immunol. Res. 2020, 12, 1012–1028. [Google Scholar] [CrossRef]
- Sianipar, I.R.; Sestramita, S.; Pradnjaparamita, T.; Yunir, E.; Harbuwono. D S.; Soewondo, P.; Tahapary, D.L. The role of intestinal-fatty acid binding proteins and chitinase-3-like protein 1 across the spectrum of dysglycemia. Diabetes Metab. Syndr. 2022, 16, 102366. [CrossRef]
- Omidian, M.; Mahmoudi, M.; Javanbakht, M.H.; Eshraghian, M.R.; Abshirini, M.; Daneshzad, E.; Hasani, H.; Alvandi, E.; Djalai, M. Effects of vitamin D supplementation on circulatory YKL-40 and MCP-1 biomarkers associated with vascular diabeteic complications; A randomized, placebo-controlled, double-blind clinical trial. Diabetes Metab. Syndr. 2019, 13, 2873–2877. [Google Scholar] [CrossRef] [PubMed]
- Yuksel, I.T.; Cetin, B.A.; Koroglu, N.; Mathyk, B.A.; Erdem, B. Inflammatory marker YKL-40 levels in intrahepatic cholestasis of pregnancy. Gynecol. Endocrinol. 2019, 35, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Bulanik, M.; Sagsoz, N.; Sayan, C.D.; Yeral, M.I.; Kisa, U. Comparison of serum Ykl-40 and ischemia modified albumin levels between pregnant women with hyperemesis gravidarum and normal pregnant women. Med. Arch. 2019, 73, 97–100. [Google Scholar] [CrossRef]
- Sobkowiak, P.; Narozna, B.; Wojsyk-Banaszak, I.; Breborowicz, A.; Szczepankiewicz, A. Expression of proteins associated with airway fibrosis differs between children with allergic asthma and allergic rhinitis. Int. Immunopathol. Pharmacol. 2021, 35, 2058738421990493. [Google Scholar] [CrossRef] [PubMed]
- Permain, J.; Appleton, L.; Ho, S.S.C.; Coffey, M.; Ooi, C.Y.; Keenan, J.I.; Day, A.S. Children with cystic fibrosis have elevated levels of fecal chitinase-3-like-1. J. Pediatr. Gastroenterol. Nutr. 2022, 75, 48–51. [Google Scholar] [CrossRef] [PubMed]
- Topcu, D.B.; Tugcu, G.; Er, B.; Polat, S.E.; Hizal, M.; Yalcin, E.E.; Ersoz, D.D.; Coplu, L.; Ozcelik, U.; Kiper, N.; et al. Increased plasma YKL-40 level and chitotriosidase activity in cystic fibrosis patients. Inflammation 2022, 45, 627–638. [Google Scholar] [CrossRef]
- Buisson, A.; Vazeille, E.; Minet-Quinard, R.; Goutte, M.; Bouvier, D.; Goutorbe, F.; Pereira, B.; Barnich, Bommelaer, G. Faecal chitinase 3-like 1 is a reliable marker as accurate as faecal calprotectin in detecting endoscopic activity in adult patients with inflammatory bowel disease. Aliment. Pharmacol. Ther. 2016, 43, 1069-1079. [CrossRef]
- Aomatsu, T.; Imaeda, H.; Matumoto, K.; Kimura, E.; Yoden, A.; Tamai, H.; Fujiyama, Y.; Mizoguchi, E.; Andoh, A. Faecal chitinase 3-like -1: a novel biomarker of disease activity in paediatric inflammatory bowel disease. Aliment. Pharmacol. Ther. 2011, 34, 941–948. [Google Scholar] [CrossRef]
- Low, D.; Subramaniam, R.; Lin, L.; Aomatsu, T.; Mizoguchi, A.; Ng, A.; DeGruttola, A.K.; Lee, C.G.; Elias, J.A.; Andoh, A.; et al. Chitinase 3-like 1 induces survival and proliferation of intestinal epithelial cells during chronic inflammation and colitis-associated cancer by regulating S100A9. Oncotarget 2015, 6, 36535–36550. [Google Scholar] [CrossRef]
- Comabella, M.; Fernandez, M.; Martin, R.; Rivera-Vallve, S.; Borras, E.; Chiva, C.; Julia, E.; Rovira, A.; Canto, E.; Alvarez-Cermeno, J.C.; et al. Celebrospinal fluid chitinase 3-like 1 levels are assocaited with conversion to multiple sclerosis. Brain 2010; 133, 1082-1093. [CrossRef]
- Bhardwaj, R.; Yester, J.; Singh, S.K.; Biswas, D.D.; Surace, M.J.; Waters, M.R.; Hauser, K.F.; Yao, Z.; Boyce, B.F.; Kordula, T. RelB/p50 complexes regulate cytokine-induced YKL-40 expression. J. Immunol. 2015, 194, 2862–2870. [Google Scholar] [CrossRef]
- Burman, J.; Raininko, R.; Blennow, K.; Zetterberg, H.; Axeisson, M.; Malmestrom, C. YKL-40 is a CSF biomarker of intrathecal inflammation in secondary progressive multiple sclerosis. J. Neuroimmunol. 2016, 292, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Dichev, V.; Kazakova, M.; Sarafian, V. YKL-40 and neuron-specific enolase in neurodegeneration and neuroinflammation. Rev. Neurosci. 2020, 31, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Lee, I.A.; Low, D.; Kamba, A.; Mizoguchi, A.; Shi, H.N.; Lee, C.G.; Elias, J.A.; Mizoguchi, E. Chitinase 3-like 1 synergistically activates IL-6 mediated STAT3 phosphorylation in intestinal epithelial cells in murine models of infectious colitis. Inflamm. Bowel Dis. 2014, 20, 835–846. [Google Scholar] [CrossRef]
- Eurich, K.; Segawa, M.; Toei-Shimizu, S.; Mizoguchi, E. Potential role of chitinase 3-like 1 in inflammation-associated carcinogenic changes of epithelial cells. World J. Gastroenterol. 2009, 15, 5249–5259. [Google Scholar] [CrossRef]
- Russo, C.; Valle, M.S.; Casabona, A.; Malaguarnera, L. Chitinase Signature in the Plasticity of Neurodegenerative Diseases. Int J Mol Sci. 2023, 24, 6301. [Google Scholar] [CrossRef]
- Temmerman, J.; Engelborghs, S.; Bjerke, M.; D’haeseleer, M. Cerebrospinal fluid inflammatory biomarkers for disease progression in Alzheimer’s disease and multiple sclerosis: a systematic review. Front Immunol. 2023, 14, 1162340. [Google Scholar] [CrossRef]
- Dage, J.L.; Eloyan, A.; Thangarajah, M.; Hammers, D.B.; Fagan, A.M.; Gray, J.D.; Schindler, S.E.; Snoddy, C.; Nudelman, K.N.H.; Faber, K.M.; et al. Cerebrospinal fluid biomarkers in the Longitudinal Early-onset Alzheimer’s Disease Study. Alzheimers Dement. 2023, 19, S115–S125. [Google Scholar] [CrossRef]
- Pelkmans, W.; Shekari, M.; Brugulat-Serrat, A.; Sánchez-Benavides, G.; Minguillón, C.; Fauria, K.; Molinuevo, J.L.; Grau-Rivera, O.; González Escalante, A.; Kollmorgen, G.; et al. Astrocyte biomarkers GFAP and YKL-40 mediate early Alzheimer’s disease progression. Alzheimers Dement. 2024, 20, 483–493. [Google Scholar] [CrossRef]
- Ferrari-Souza, J.P.; Ferreira, P.C.L.; Bellaver, B.; Tissot, C.; Wang, Y.T.; Leffa, D.T.; Brum, W.S.; Benedet, A.L.; Ashton, N.J.; De Bastiani, M.A.; et al. Astrocyte biomarker signatures of amyloid-β and tau pathologies in Alzheimer’s disease. Mol. Psychiatry. 2022, 27, 4781–4789. [Google Scholar] [CrossRef]
- Lananna, B.V.; McKee, C.A.; King, M.W.; Del-Aguila, J.L.; Dimitry, J.M.; Farias, F.H.G.; Nadarajah, C.J.; Xiong, D.D.; Guo, C.; Cammack, A.J.; et al. Chi3l1/YKL-40 is controlled by the astrocyte circadian clock and regulates neuroinflammation and Alzheimer’s disease pathogenesis. Sci. Transl. Med. 2020, 12, eaax3519. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Dobricic, V.; Ohlei, O.; Bos, I.; Vos, S.J.B.; Prokopenko, D.; Tijms, B.M.; Andreasson, U.; Blennow, K.; Vandenberghe, R.; et al. TMEM106B and CPOX are genetic determinants of cerebrospinal fluid Alzheimer’s disease biomarker levels. Alzheimers Dement. 2021, 17, 1628–1640. [Google Scholar] [CrossRef]
- Kim, M.A.; Shin, Y.S.; Pham le, D.; Park, H.S. Adult asthma biomarkers. Curr. Opin. Allergy Clim. Immunol. 2014;14.49-54. [CrossRef]
- Ober, C.; Tan, Z.; Sun, Y.; Possick, J.D.; Pan, L.; Nicolae, R.; Radford, S.; Parry, R.R.; Heinzmann, A.; Deichmann, K.A.; et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N. Engl. J. Med. 2008, 358, 1682–91. [Google Scholar] [CrossRef]
- Gomez, J.L.; Crisafi, G.M.; Holm, C.T.; Meyers, D.A.; Hawkins, G.A.; Bleecker, E.R.; Jarjour, N.; Severe Asthma Research Program (SARP) Investigators.; Cohn, L.; Chupp, G.L. Genetic variation in chitinase 3-like 1 (CHI3L1) contributes to asthma severity and airway expression of YKL-40. J. Allergy Clin. Immunol. 2015, 136, 51-58. [CrossRef]
- James, A.J.; Reinius, L.E.; Verhoek, M.; Gomes, A.; Kupczyk, M.; Hammar, U.; Ono, J.; Ohta, S.; Izuhara, K.; Bel, E.; et al. Increased YKL-40 and Chitotriosidase in Asthma and Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit Care Med. 2016, 193, 131–42. [Google Scholar] [CrossRef]
- Ahangari, F.; Sood, A.; Ma, B.; Takyar, S.; Schuyler, M.; Qualls, C.; Dela Cruz, C.S.; Chupp, G.L.; Lee, C.G.; Elias, J.A. Chitinase 3-like-1 regulates both visceral fat accumulation and asthma-like Th2 inflammation. Am. J. Respir. Cri.t Care Med. 2015, 191, 746–57. [Google Scholar] [CrossRef]
- Johansen, J.S.; Johansen, H.S.; Price, P.A. A new biochemical marker for joint injury. Analysis of YKL-40 in serum and synovial fluid. Br. J. Rheumatol. 1993, 32, 949-955. [CrossRef]
- Johansen, J.S.; Achultz, N.A.; Jensen, B.V. Plasma YKL-40: a potential new cancer biomarker? Future Oncol. 2009, 5, 1065–1082. [Google Scholar] [CrossRef]
- Subramaniam, R.; Mizoguchi, A.; Mizoguchi, E. Mechanistic roles of epithelial and immune cell signaling during the development of colitis-associated cancer. Cancer Res. Front 2016, 2, 1–21. [Google Scholar] [CrossRef]
- Kawada, M.; Seno, H.; Kanda, K.; Nakanishi, Y.; Akitake, R.; Komekado, H.; Kawada, K.; Sakai, Y.; Mizoguchi, E.; Chiba, T. Chitinase 3-like 1 promotes macrophage recruitment and angiogenesis in colorectal cancer. Oncogene 2021, 31, 3111–3123. [Google Scholar] [CrossRef]
- Ray, A.L.; Castillo, E.F.; Morris, K.T.; Nofchissey, R.A.; Weston, L.L.; Samedi, V.G.; et al. Blockade of MK2 is protective in inflammation-associated colorectal cancer development. Int. J. Cancer 2016, 138, 770–775. [Google Scholar] [CrossRef]
- Zhao, T.; Zeng, J.; Xu, Y.; Su, Z.; Chong, Y.; Ling, T.; Xu, H.; Shi, H.; Zhu, M.; Mo, Q.; et al. Chitinase-3 like-protein-1 promotes glioma progression via the NF-κB signaling pathway and tumor microenvironment reprogramming. Theranostics 2022, 12, 6989–7008. [Google Scholar] [CrossRef]
- Guetta-Terrier, C.; Karambizi, D.; Akosman, B.; Zepecki, J.P.; Chen, J.S.; Kamle, S.; Fajardo, J.E.; Fiser, A.; Singh, R.; Toms, S.A.; et al. Chi3l1 is a modulator of glioma stem cell states and a therapeutic target in glioblastoma. Cancer Res. 2023, 83, 1984–1999. [Google Scholar] [CrossRef] [PubMed]
- Sleisenger and Fordtran’s Gastrointestinal and Liver, Disease.; Pathophysiology/ diagnosis/management, edited by Mark, Feldman.; et al., Elsevier 2020.
- Barnich, N.; Carvalho, F. A.; Glasser, A. L.; Darcha, C.; Jantscheff, P.; Allez, M.; Peeters, H.; Bommelaer, G.; Desreumaux, P.; Colombel, J. F.; et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 2007, 117, 1566-74. [CrossRef]
- Tran, H.T.; Barnich, N.; Mizoguchi, E.; et al. Potential role of chitinases and chitin-binding proteins in host-microbial interactions during the development of intestinal inflammation. Histol. Histopathol. 2011, 26, 1453–64. [Google Scholar]
- Chaudhuri, S.; Bruno,J.C.; Alonzo, F. 3rd.; Xayarath, B.; Cianciotto, N.P.; Freitag, N.E. Contribution of chitinases to Listeria monocytogenes pathogenesis. Appl. Enviro. Microbiol. 2010, 76, 7302–7305. [CrossRef]
- Tanaka, H.; Akutsu, H.; Yabuta, I.; Hara, M.; Sugimoto, H.; Ikegami, T.; Watanabe, T.; Fujiwara, T. A novel chitin-binding mode of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12 revealed by solid-state NMR. FEBS Lett. 2018, 592, 3173–3182. [Google Scholar] [CrossRef]
- Fusetti, F.; Pijning, T.; Kalk, K.H.; Bos, E.; Dijkstra, B.W. Crystal structure and carbohydrate-binding properties of the human cartilage glycoprotein-39. J. Bio. Chem. 2003, 278, 37753–37760. [Google Scholar] [CrossRef]
- Moran, A. P.; Gupta, A.; Joshi, L. Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut 2011, 10: 1412-25. [CrossRef]
- Park, D.; Arabyan, N.; Williams, C.C.; Song, T.; Mitra, A.; Weimer, B.C.; Maverakis, E.; Lebrilla, C.B. Salmonella typhimurium enzymatically landscapes the host Intestinal epithelial cell (IEC) surface glycome to increase invasion. Mo.l Cell Proteo. 2016, 15, 3653–3664. [Google Scholar] [CrossRef]
- Johansson Malin, E.V. Mucus layers in inflammatory bowel disease. Inflamm. Bowel Dis. 2014, 20, 2124–31. [CrossRef]
- Hansson Gunnar, C.; and Malin Ev, J. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut microbes 2010, 1, 51-54. [CrossRef]
- Bohr, S.; Patel, S.J.; Vask, R.; Shen, K.; Golberg, A.; Berthiaume, F.; Yarmush, M.L. The Role of CHI3L1 (Chitinase-3-Like-1) in the Pathogenesis of Infections in Burns in a Mouse Model. PLoS ONE 2015, 10, e0140440. [Google Scholar] [CrossRef]
- Siwczak, F.; Cseresnyes, Z.; Hassan, M.I.A.; Aina, K.O.; Caristedt, S.; Sigmund, A.; Groger, M.; Surewaard, B.G.J.; Werz, O.; Figge, M.T. et al. Human macrophage polarization determines bacterial persistence of Staphylococcus aureus in a liver-on-chip-based infection model. Biomaterials 2022, 287, 121632. [CrossRef]
- Bonet-Rossinyol, Q.; Camprubi-Font, C.; Lopez-Siles, M.; Martinez-Medina, M. Identification of differences in gene expression implicated in the adherent-invasive Escherichia coli phenotype during in vitro infection of intestinal epithelial cells. Front. Cell. Infect. Microbiol. 2023, 13, 1228159. [Google Scholar] [CrossRef]
- Bringer, M.A.; Billard, E.; Glasser, A.L.; Colombel, J.F.; Darfeuille-Michaud, A. Replication of Crohn’s disease-associated AIEC within macrophages is dependent on TNF-α secretion. Lab. Invest. 2012, 92, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Kinugasa, T.; Tsunoda, T.; Mizoguchi, E.; Okada, T.; Sudo, T.; Kawahara, A.; Akiba, J.; Akagi, Y. Chitinase 3-like 1, carcinoembryonic antigen-related cell adhesion molecule 6, and ectopic claudin-2 in the carcinogenic process of ulcerative colitis. Anticancer Res. 2022, 42, 4119–4127. [Google Scholar] [CrossRef]
- Nell, S.; Suerbaum, S.; Josenhans, C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat. Rev. Microbiol. 2010, 8, 564–577. [Google Scholar] [CrossRef]
- Santana, P.T.; Rosas, S.L.B.; Ribeiro, B.E.; Marinho, Y.; de Souza, H.S.P. Dysbiosis in Inflammatory Bowel Disease: Pathogenic Role and Potential Therapeutic Targets. Int. J. Mol. Sci. 2022, 23, 3464. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cui, W.; Li, X. Interaction between commensal bacteria, immune response and the intestinal barrier in inflammatory bowel disease. Front. Immunol. 2021, 12, 761981. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Long, X.-B.; Cao, P.-P.; Wang, N.; Liu, Y.; Cui, Y.-H.; Huang, S.-K.; Liu, Z. Clara cell 10-kD protein suppresses Chitinase 3-like 1 expression associated with eosinophilic chronic rhinosinusitis. Am. J. Respir. Cri.t Care Med. 2010, 181, 908–916. [Google Scholar] [CrossRef]
- Zeng, X.; Cheung, S.K.K.; Shi, M.; Or, P.M.Y.; Li, Z.; Liu, J.Y.H.; Ho, W.L.H.; Liu, T.; Lu, K.; Rudd, J.A.; et al. Astrocyte-specific knockout of YKL-40/Chi3l1 reduces Aβ burden and restores memory functions in 5xFAD mice. J. Neuroinflamm. 2023, 20, 290. [Google Scholar] [CrossRef]
- Choi, J.Y.; Yeo, I.J.; Kim, K.C.; Choi, W.R.; Jung, J.-K.; Han, S.-B.; Hong, J.T. K284-6111 Prevents the amyloid beta-induced neuroinflammation and impairment of recognition memory through inhibition of NF-κB-mediated CHI3L1 expression. J. Neuroinflamm. 2018, 15, 224. [Google Scholar] [CrossRef]
- Anwar, M.M.; Fathi, M.H. Early Approaches of YKL-40 as a Biomarker and Therapeutic Target for Parkinson’s Disease. Neurodegener. Dis. Manag. 2023, 13, 85–99. [Google Scholar] [CrossRef]
- Birmpili, D.; Charmarke Askar, I.; Pham-Van, L.D.; Kuntzel, T.; Spenlé, C.; Riou, A.; Bagnard, D. Toward a combination of biomarkers for molecular characterization of multiple sclerosis. Int. J. Mol. Sci. 2022, 23, 14000. [Google Scholar] [CrossRef]
- Kwak, E.J.; Hong, J.Y.; Kim, M.N.; Kim, S.Y.; Kim, S.H.; Park, C.O.; Kim, K.W.; Lee, C.G.; Elias, J.A.; Jee, H.M.; et al. Chitinase 3-like 1 drives allergic skin inflammation via Th2 immunity and M2 macrophage activation. Clin. Exp. Allergy 2019, 49, 1464–1474. [Google Scholar] [CrossRef]
- Lee, C.G.; Hartl, D.; Lee, G.R.; Koller, B.; Matsuura, H.; Da Silva, C.A.; Sohn, M.H.; Cohn, L.; Homer, R.J.; Kozhich, A.A.; et al. Role of breast regression protein 39 (BRP-39)/Chitinase 3-like-1 in Th2 and IL-13-Induced tissue responses and apoptosis. J. Exp. Med. 2009, 206, 1149–1166. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, H.; Hartl, D.; Kang, M.-J.; Dela Cruz, C.S.; Koller, B.; Chupp, G.L.; Homer, R.J.; Zhou, Y.; Cho, W.-K.; Elias, J.A.; et al. Role of breast regression protein-39 in the pathogenesis of cigarette smoke-induced inflammation and emphysema. Am. J. Respir. Cell Mol. Biol. 2011, 44, 777–786. [Google Scholar] [CrossRef]
- Zhou, Y.; He, C.H.; Herzog, E.L.; Peng, X.; Lee, C.-M.; Nguyen, T.H.; Gulati, M.; Gochuico, B.R.; Gahl, W.A.; Slade, M.L.; et al. Chitinase 3-like-1 and its receptors in Hermansky-Pudlak Syndrome-associated lung disease. J. Clin. Invest. 2015, 125, 3178–3192. [Google Scholar] [CrossRef]
- Jung, Y.Y.; Kim, K.C.; Park, M.H.; Seo, Y.; Park, H.; Park, M.H.; Chang, J.; Hwang, D.Y.; Han, S.B.; Kim, S.; et al. Atherosclerosis is exacerbated by Chitinase-3-like-1 in amyloid precursor protein transgenic mice. Theranostics 2018, 8, 749–766. [Google Scholar] [CrossRef]
- Tsantilas, P.; Lao, S.; Wu, Z.; Eberhard, A.; Winski, G.; Vaerst, M.; Nanda, V.; Wang, Y.; Kojima, Y.; Ye, J.; et al. Chitinase 3 like 1 Is a Regulator of Smooth Muscle Cell Physiology and atherosclerotic lesion stability. Cardiovasc. Res. 2021, 117, 2767–2780. [Google Scholar] [CrossRef]
- Kim, M.; Chang, J.Y.; Lee, D.W.; Kim, Y.R.; Son, D.J.; Yun, J.; Jung, Y.S.; Lee, D.H.; Han, S.; Hong, J.T. Chitinase 3 like 1 deficiency ameliorates lipopolysaccharide-induced acute liver injury by inhibition of M2 macrophage polarization. Mol. Immunol. 2023, 156, 98–110. [Google Scholar] [CrossRef]
- Lee, D.H.; Han, J.H.; Lee, Y.S.; Jung, Y.S.; Roh, Y.S.; Yun, J.S.; Han, S.B.; Hong, J.T. Chitinase-3-like-1 deficiency attenuates ethanol-induced liver injury by inhibition of sterol regulatory element binding protein 1-dependent triglyceride synthesis. Metabolism 2019, 95, 46–56. [Google Scholar] [CrossRef]
- Kim, A.D.; Kui, L.; Kaufmann, B.; Kim, S.E.; Leszczynska, A.; Feldstein, A.E. Myeloid-specific deletion of Chitinase-3-like 1 protein ameliorates murine diet-induced steatohepatitis progression. J. Mol. Med. 2023, 101, 813–828. [Google Scholar] [CrossRef]
- Pizano-Martínez, O.; Yañez-Sánchez, I.; Alatorre-Carranza, P.; Miranda-Díaz, A.; Ortiz-Lazareno, P.C.; García-Iglesias, T.; Daneri-Navarro, A.; Vázquez-Del Mercado, M.; Fafutis-Morris, M.; Delgado-Rizo, V. YKL-40 expression in CD14+ liver cells in acute and chronic injury. World J. Gastroenterol. 2011, 17, 3830–3835. [Google Scholar] [CrossRef] [PubMed]
- Di Rosa, M.; Szychlinska, M.A.; Tibullo, D.; Malaguarnera, L.; Musumeci, G. Expression of CHI3L1 and CHIT1 in osteoarthritic rat cartilage model. A Morphological Study. Eur J. Histochem. 2014, 58, 2423. [Google Scholar] [CrossRef] [PubMed]
- Verheijden, G.F.; Rijnders, A.W.; Bos, E.; Coenen-de Roo, C.J.; van Staveren, C.J.; Miltenburg, A.M.; Meijerink, J.H.; Elewaut, D.; de Keyser, F.; Veys, E.; et al. Human cartilage glycoprotein-39 as a candidate autoantigen in rheumatoid arthritis. Arthritis Rheum. 1997, 40, 1115–1125. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Park, H.-J.; Lim, S.; Koo, J.-H.; Lee, H.-G.; Choi, J.O.; Oh, J.H.; Ha, S.-J.; Kang, M.-J.; Lee, C.-M.; et al. Regulation of Chitinase-3-like-1 in T cell elicits Th1 and cytotoxic responses to inhibit lung metastasis. Nat. Commun. 2018, 9, 503. [Google Scholar] [CrossRef]
- Gomez, J.L.; Crisafi, G.M.; Holm, C.T.; Meyers, D.A.; Hawkins, G.A.; Bleecker, E.R.; Jarjour, N.; Cohn, L.; Chupp, G.L. Genetic Variation in Chitinase 3-like 1 (CHI3L1) contributes to asthma severity and airway expression of YKL-40. Journal of Allergy and Clin. Immunol. 2015, 136, 51-58.e10. [CrossRef]
- Salomon, J.; Matusiak, Ł.; Nowicka-Suszko, D.; Szepietowski, J.C. Chitinase-3-Like Protein 1 (YKL-40) reflects the severity of symptoms in atopic dermatitis. Journal of Immunol. Res. 2017, 2017, e5746031. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, Y.; Peng, Q.; Shu, X.; Wang, G.; Wu, X. Serum YKL-40 Level Is Associated with severity of interstitial lung disease and poor prognosis in dermatomyositis with anti-MDA5 antibody. Clin. Rheumatol. 2019, 38, 1655–1663. [Google Scholar] [CrossRef]
- Hamilton, D.; Lehman, H. Asthma phenotypes as a guide for current and future biologic therapies. Clinic. Rev. Allerg. Immunol. 2020, 59, 160–174. [Google Scholar] [CrossRef]
- Kamba, A.; Lee, I.-A.; Mizoguchi, E. Potential Association between TLR4 and Chitinase 3-like 1 (CHI3L1/YKL-40) signaling on colonic epithelial cells in inflammatory bowel disease and colitis-associated cancer. Curr. Mol. Med. 2013, 13, 1110–1121. [Google Scholar] [CrossRef]
- Huang, J.; Gu, Z.; Xu, Y.; Jiang, L.; Zhu, W.; Wang, W. CHI3L1 (Chitinase 3 Like 1) upregulation ss associated with macrophage signatures in esophageal cancer. Bioengineered 12, 7882–7892. [CrossRef]
- Chen, A.; Jiang, Y.; Li, Z.; Wu, L.; Santiago, U.; Zou, H.; Cai, C.; Sharma, V.; Guan, Y.; McCarl, L.H.; et al. Chitinase-3-like 1 protein complexes modulate macrophage-mediated iImmune suppression in glioblastoma. J. Clin. Invest. 2021, 131. [Google Scholar]
- Zhao, T.; Zeng, J.; Xu, Y.; Su, Z.; Chong, Y.; Ling, T.; Xu, H.; Shi, H.; Zhu, M.; Mo, Q.; et al. Chitinase-3 like-protein-1 promotes glioma progression via the NF-κB signaling pathway and tumor microenvironment reprogramming. Theranostics 2022, 12, 6989–7008. [Google Scholar] [CrossRef] [PubMed]
- Cendrowicz, E.; Sas, Z.; Bremer, E.; Rygiel, T.P. The role of macrophages in cancer development and therapy. Cancers (Basel) 2021, 13, 1946. [Google Scholar] [CrossRef]
- Taifour, T.; Attalla, S.S.; Zuo, D.; Gu, Y.; Sanguin-Gendreau, V.; Proud, H.; Solymoss, E.; Bui, T.; Kuasne, H.; Papavasiliou, V.; et al. The tumor-derived cytokine Chi3l1 induces neutrophil extracellular traps that promote T cell exclusion in triple-negative breast cancer Immunity 2023, 56, 2755-2772.e8. [CrossRef]
- Jeon, S.H.; Lee, Y.S.; Yeo, I.J.; Lee, H.P.; Yoon, J.; Son, D.J.; Han, S.-B.; Hong, J.T. Inhibition of Chitinase-3-like-1 by K284-6111 reduces atopic skin inflammation via repressing lactoferrin. Immune. Netw. 2021, 21, e22. [Google Scholar] [CrossRef]
- Yang, P.-S.; Yu, M.-H.; Hou, Y.-C.; Chang, C.-P.; Lin, S.-C.; Kuo, I.-Y.; Su, P.-C.; Cheng, H.-C.; Su, W.-C.; Shan, Y.-S.; et al. Targeting protumor factor Chitinase-3-like-1 secreted by Rab37 vesicles for cancer immunotherapy. Theranostics 2022, 12, 340–361. [Google Scholar] [CrossRef]
- Yu, J.E.; Yeo, I.J.; Son, D.J.; Yun, J.; Han, S.-B.; Hong, J.T. Anti-Chi3L1 antibody suppresses lung tumor growth and metastasis through inhibition of M2 polarization. Mol. Oncol. 2022, 16, 2214–2234. [Google Scholar] [CrossRef]
- Ma, B.; Akosman, B.; Kamle, S.; Lee, C.-M.; He, C.H.; Koo, J.S.; Lee, C.G.; Elias, J.A. CHI3L1 Regulates PD-L1 and Anti–CHI3L1–PD-1 antibody elicits synergistic antitumor responses. J Clin. Invest. 131, e137750. [CrossRef]
- Ma, B.; Kamle, S.; Akosman, B.; Khan, H.; Lee, C.-M.; Lee, C.G.; Elias, J.A. CHI3L1 enhances melanoma lung metastasis via regulation of T cell co-stimulators and CTLA-4/B7 axis. Front. Immunol. 2022, 13, 1056397. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Nebhan, C.A.; Moslehi, J.J.; Balko, J.M. Immune-Checkpoint Inhibitors: Long-term implications of toxicity. Nat. Rev. Clin. Oncol. 2022, 19, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Som, A.; Mandaliya, R.; Alsaadi, D.; Farshidpour, M.; Charabaty, A.; Malhotra, N.; Mattar, M.C. Immune checkpoint inhibitor-induced colitis: A Comprehensive Review. World J. Clin. Cases 2019, 7, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.A.; Kamba, A.; Low, D.; Mizoguchi, E. Novel methylxanthine derivative-mediated anti-inflammatory effects in inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Rao, F.V.; Andersen, O.A.; Vora, K.A.; Demartino, J.A.; van Aalten, D.M. Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes. Chem. Biol. 2005, 12, 973–980. [Google Scholar] [CrossRef]
- Lee, I.A.; Low, D.; Kamba, A.; Llado, V.; Mizoguchi, E. Oral caffeine administration ameliorates acute colitis by suppressing chitinase 3-like 1 expression in intestinal epithelial cells. J. Gastroenterol. 2014, 49, 1206–1216. [Google Scholar] [CrossRef]
- Mizoguchi, E.; Sadanaga, T.; Okada, T.; Minagawa, T.; Akiba, J. Does caffeine have a double-edged sword role in inflammation and carcinogenesis in the colon? Intest. Res. 2023, 21, 306–317. [Google Scholar] [CrossRef]
- Debono, M.; Gordee, R.S. Antibiotics that inhibit fungal cell wall development. Annu. Rev. Microbiol. 1994, 48, 471–497. [Google Scholar] [CrossRef]
- Hakala, B.E.; White, C.; Recklies, A.D. Human cartilage gp-39, a major secretory product of articular chondrocytes and synovial cells, is a mammalian member of a chitinase protein family. J. Biol. Chem. 1993, 268, 25803–25810. [Google Scholar] [CrossRef]
- Lee, C.G.; Da, Silva, C.A.; Lee, J.Y.; Hartl, D.; Elias, J. A. Chitin regulation of immune responses: an old molecule with new roles. Curr. Opin. Immunol. 2008, 20, 1-6. [CrossRef]
- Lee, C.G.; Da, Silva, C.A.; Dela, Cruz, C.S.; Ahangari, F.; Ma, B.; Kang, M. J.; He, C. H.; Takyar, S.; Elias, J. A. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu. Rev. Physiol. 2011, 73, 479-450. [CrossRef]
- Nagatani, K.; Wang, S.; Llado, V.; Lau, C.W.; Li, Z.; Mizoguchi, A.; Nagler, C.R.; Shibata, Y.; Reinecker, H.C.; Mora, J.R.; et al. Chitin microparticles for the control of intestinal Inflammation. Inflamm. Bowel Dis. 2012, 18, 1698–1710. [Google Scholar] [CrossRef] [PubMed]
- Letzel, M.; Synstad, B.; Eijsink, V.G.H.; Peter-Katalinic, J.; Peter, M.G. Libraries of chitin-oligosaccharides of mixed acetylation patterns and their interactions with chitinases. Adv. Chitin Sci. 2000, 4, 545–552. [Google Scholar]
- Cederkvist, F.H.; Parmer, M.P.; Varum, K.M.; Eijsink, V.G.H.; Sorlie, M. Inhibition of a family 18 chitinase by chitooligosaccharides. Carbohydr. Polym. 2008, 74, 41–49. [Google Scholar] [CrossRef]
| Disease | Samples | General Overview | Ref. |
|---|---|---|---|
| IBD | Serum, CECs, Stool, Mφ, Peripheral MDMs, |
|
[23,24,25,26] |
| MS | CSF, Serum, Tissue |
|
[27,28,29,30,31,32,33,34,35] |
| AD | CSF, Tissue |
|
[36,37,38,39,40,41,42,43] |
| Asthma, COPD | Serum, BECs, |
|
[44,45,46,47,48] |
|
|
|
[49] |
| COVID-19 | Serum, Tissue, |
|
[50,51,52,53] |
| Liver Diseases | Serum, Tissue |
|
[54,55,56] |
| Oral Disease | Tissue, Saliva |
|
[57,58] |
| Kidney Disease | Serum, Urine |
|
[59,60,61,62,63] |
| Heart Disease | Serum, |
|
[64,65,66] |
| Food Allergy | Serum |
|
[67] |
| Diabetes Mellitus | Serum |
|
[68,69] |
| Pregnancy | Serum, Urine |
|
[70,71] |
| Rhinitis | Serum |
|
[72] |
| Cystic Fibrosis | Stool, Serum, |
|
[73,74] |
| Disease | Model | Features | Ref. |
|---|---|---|---|
| Eosinophilic Chronic Rhinosinusitis | CC10 WT/KO mice with OVA sensitization |
|
[121] |
| Alzheimer’s disease | 5xFAD mice model Aβ1–42-induced AD mice model |
|
[122,123] |
| Parkinson’s disease | LPS-induced PD rats model |
|
[124] |
| Multiple sclerosis | EAE-PLP mice model |
|
[125] |
| Atopic dermatitis | Filaggrin mutated mice with OVA sensitization |
|
[126] |
| Asthma | BRP-39 WT/KO mice with OVA sensitization |
|
[127] |
| COPD | BRP-39 WT/KO mice with cigarette smoke |
|
[128] |
| Hermansky-Pudlak syndrome | Hps1 mutation mice with Bleomycin |
|
[129] |
| Atherosclerosis | ApoE-/- mice with high-fat diet |
|
[130,131] |
| Liver sepsis | LPS-induced mice model |
|
[132] |
| Alcoholic liver injury | CHI3L1 WT/KO mice with the Lieber-DeCarli ethanol liquid diet |
|
[133] |
| NASH | CHI3L1 WT/KO mice (CreLyz) with choline-deficient high-fat diet |
|
[134] |
| Chronic liver injury | CCl4 i.p. injection rats model |
|
[135] |
| Obesity | CHI3L1 WT/KO mice with HFD |
|
[95] |
| CKD | Ischemia/reperfusion injury mice model with microaneurysm clip |
|
[60] |
| Osteoarthritis | Osteoarthritis rats model with ACLT |
|
[136] |
| Rheumatic arthritis | RA mice model with HC gp-39 (CHI3L1) i.p. injection |
|
[137] |
| IBD | CHI3L1 WT/KO with AOM/DSS CHI3L1 WT/KO with S.typhimurium/AIEC inoculation |
|
[77,82] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





