1. Introduction
Loop-mediated amplification (LAMP) assays are highly specific (due to the large number of primer pairs), generate much more amplicon DNA than conventional PCR, and can detect the amplification product by colorimetry or fluorescence. Fluorescent indicators (dyes) can be included in a LAMP reaction, which, as expected, provides greater sensitivity than colorimetric detection [
1]. LAMP allows specific amplification of target nucleic acids with greater efficiency than typical PCR [
2] and most importantly, under isothermal conditions [
3,
4]. The simplicity of this screening method allows for fast and straightforward DNA analysis. Second, the nature of isothermal amplification massively reduces the complexity of the instrument that must be designed and built without the need for sequential thermocycling stages and the associated expensive and specialized thermocycling equipment, most often restricted to a dedicated laboratory [
5].
Current methods based on DNA analysis, which determine the presence of species-specific biological materials, are fast and have excellent sensitivity [
6]. This study aimed to develop an
in-house method that exploits a novel amplification technique called loop-mediated amplification (LAMP), particularly using the device SaLux19 (Max-Planck Innovation prototype, Germany), allowing fluorescent
real-time control of amplification and focusing on DNA identification from
Sus scrofa biological fluid (venous blood) from forensically sized samples (~2 μl or less, diluted 100x, 1000x, at their appropriate concentration units within the range from ng to pg of DNA [
7]).
The sample for this study was selected according to the following criteria:
Pork and its derivates are religiously forbidden for some groups, such as Jews and Muslims [
8,
9]. Moreover, the consumption of pig tissue-related products can trigger the development of allergies in sensitive individuals [
9,
10].
2. Materials and Methods
2.1. SaLux19 Device
Principle of real-time SaLux19 point of care (POC)–based testing
This pocket-size device has dimensions 14,5 cm x 9,5cm x 4cm (L x W x H) and is equipped with a portable Xlayer power charger QC 3.0 (
Figure 1). By nature, the SaLux19 device can detect specific DNA or cDNA genetic codes. This POC modality allows the determination of both, as previously reverse-transcribed mRNA–based pathogens or DNA pathogens and all biological tissue containing DNA specific to the corresponding species and individuals. The specificity of detection is based on the match in the sequence of a piece of DNA with synthetic “baits” (primers) that bind to the target to be analysed. The SaLux19 biosensor is manufactured for working temperature being set to 64°C. The principle exploits the presence of DNA polymerase with strand-displacement activity, newly formed stem-loop structures, and a new self-primed 3′-end creation [
5]. A fluorescent dye with the primer binds to any newly formed double helix. The binding of the fluorescent dye causes a shift in the fluorescent colour of the dye. This shift in fluorescence is obtained by a light source (light-emitting diode, LED), a set of optical filters with cut-off -510nm, and the photodetectors. The method utilized by SaLux19 is known as loop-mediated isothermal amplification (LAMP). During a real-time LAMP reaction, the amount of shifted fluorescence increases exponentially with time until saturation is reached due to the limited reagent. In this sense, a chain reaction is initiated by the presence of a few molecules of the agent to be detected, and the increase in fluorescence correlates with the input amount of DNA and corresponds to the Ct value acquired by loop amplification. The biosensor display is a part of the microdevice architecture. The maximum possible observation time allowed by the SaLux19 biosensor to detect only a few molecules of the specific substance to be analysed is 40 minutes.
2.2 DNA Extraction and Quantitation
DNA was extracted using a column-based Quick-DNA Miniprep Kit (cat. no. D3024, ZymoResearch, USA). The nDNA amount was quantified using a standard curve constructed by amplifying known amounts of the human DNA control (cat. no. 431266, Thermo Fisher Scientific, USA).
Absolute quantification of the nDNA of Sus scrofa
DNA extracted from nonprecipitating Sus scrofa blood was quantified using iQ SybrGreen Supermix qPCR (cat. no. 1708880, Bio-Rad, USA) targeting the TATA-binding protein (TBP) gene, described as a 2-allele single gene [
11,
12]. TBP genes from different species were aligned in ClustalO (
https://www.ebi.ac.uk/) and primers were designed in PrimerQuestTM (Integrated DNA Technologies tool, USA) in their most homologous sequences. Each 20 µL reaction consisted of 1x Supermix, 10 μM forward: tgctgttgacaagttggtt; reverse primers: tattctgtcctgcaatactg; and 2 µL of unknown DNA. TaqMan human control DNA was used as a standard and ranged from 0.001–10 ng/µL. Samples were run in duplicate on a CFX Connect Real-Time PCR System (Bio-Rad, USA) at 95 °C for 3 min and then 40 cycles of 95 °C for 15 sec and 58 °C for 45 sec. The threshold cycle (Ct) was automatically determined by CFX Manager Dx software v. 3.1 (Bio-Rad, USA).
Relative quantification and determination of the mtDNA:nDNA ratio
Relative quantification of mtDNA was performed using the same iQ SYBR Green Supermix (cat.no. 1708880, Bio-Rad, USA). The changes in the relative quantity of mtDNA with respect to nDNA were determined as the ratio of the number of copies of the mitochondrial 16S gene to that of the TBP gene (nDNA) in the same tube. The adapted mathematical formula by Livak [
13]
2x2
ΔCt (Ct(n)- Ct(mt)) was used for analysis. PCR was carried out in duplicate for each DNA sample. The PCR primers used in the present study were published in the study of Boessenkool et al. [
14], and the PCR temperature profile was the same as that used for absolute quantification.
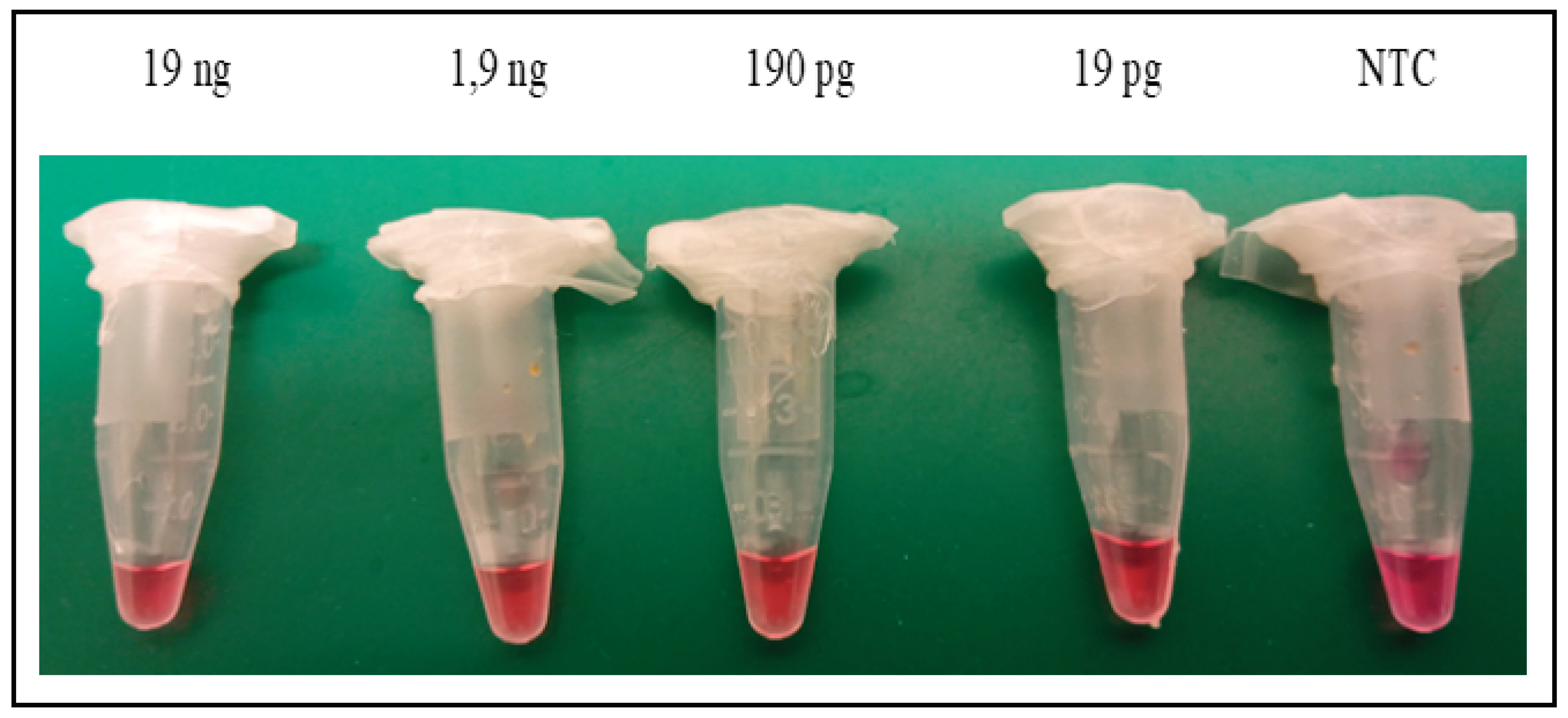
Two dilutions of Sus scrofa DNA were used for the LAMP assay experiments: 100x (at a concentration of ≈127 pg/μL of nDNA with a ratio of mtDNA/nDNA ≈ 8.75 and 1000x diluted DNA (at a concentration of ≈13 pg/μL of nDNA).
As a quick alternative to standard nucleic acid extraction procedures, direct lysis was performed using a DipStick DNA Extraction Kit [
15].
2.3 Primers
1) LAMP primers targeting the mitochondrial cytochrome c oxidase subunit I (COI) gene of Sus scrofa (accession number MN124249.1) were obtained from Jawla et al. 2021 [
16] and synthesized by Eurofins Genomics Europe Shared Services GmbH (Germany). The size of the LAMP product is forensically friendly (226 bp).
2) LAMP primers targeting the human Y-amelogenin gene (AMEL-Y) (accession number NG_008011.2) were used for monitoring male DNA amplification using TaqMan™ Control Genomic DNA (cat. no. 4312660, Thermo Fisher Scientific, USA). The primers used were acquired from the published article of Nogami et al. 2008 [
17] and synthesized by Eurofins Genomics Europe Shared Services GmbH (Germany). The length of the amplification product (202 bp) is sufficient for the detection of AMEL-Y in body fluids, as demonstrated in another study [
18].
2.4 LAMP Assay
Preliminary tests for the identification of Sus scrofa DNA were performed exactly according to the New England Biolabs (NEB, U.K.) web protocols with visual control of colour pH changes [
19] (
Figure 2a) or the typical LAMP protocol [
20], where the intercalating dye SYBRgreen Gel Stain (cat.no. S7563, Thermo Fisher Scientific, USA), at a dilution of 1:10, was loaded into the inner part of the tube lid (
Figure 2b). Reactions were carried out in 0.5 ml reaction microtubes within a thermoblock (Eppendorf, Germany).
Own
real-time LAMP assay optimization was performed on both a real-time PCR system (CFX Connect PCR system, Bio-Rad, USA) and the SaLux19 device (MPI-SERATEC, Germany). The protocol used was identical to the protocol used on the NEB website [
19] and included the same concentration of primer mixture and the addition of 10 000x diluted SYBR Green Gel Stain (cat. no. S7563, Thermo Fisher Scientific, USA) or recommended fluorescent LAMP dye (cat. no. B1700S, NEB, U.K.). LAMP was performed according to the written instructions on the NEB website [
19,
20,
21] (
Figure 3).
2.5. Assay Validation
Here, we explored a method based on isothermal amplification to rapidly identify DNA originating from Sus scrofa on the SaLux19 device. Assay validation reactions were prepared in a final reaction volume of 25 μl with nuclease-free water and incubated at 64 °C for 40 minutes on CFX Connect PCR system (Bio-Rad, USA) or SALUX19 (MPI-SERATEC, Germany). Nerbe-Plus 0,1-ml microtubes (cat. no. 04-032-0350 Nerbe plus GmbH & Co. KG, Germany) have proven useful for both real-time devices.
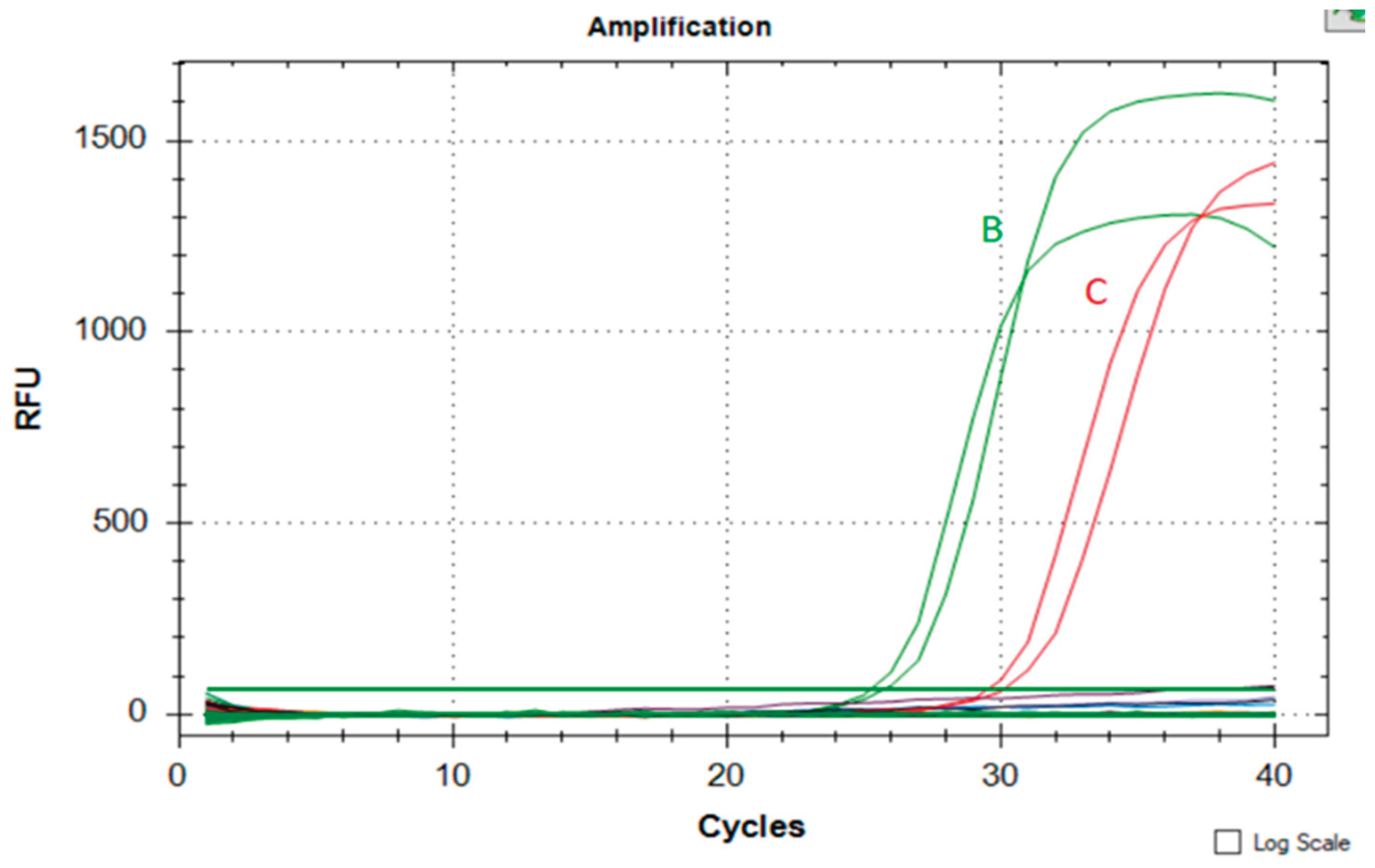
DNA samples from both eukaryotic and prokaryotic species, e.g.,
Panthera tigris, Panthera leo, Panthera pardus, Caracal serval, and
Bos taurus "Salers bull", the latter mentioned as a representative of phylogenetically related
Artiodactyla, further
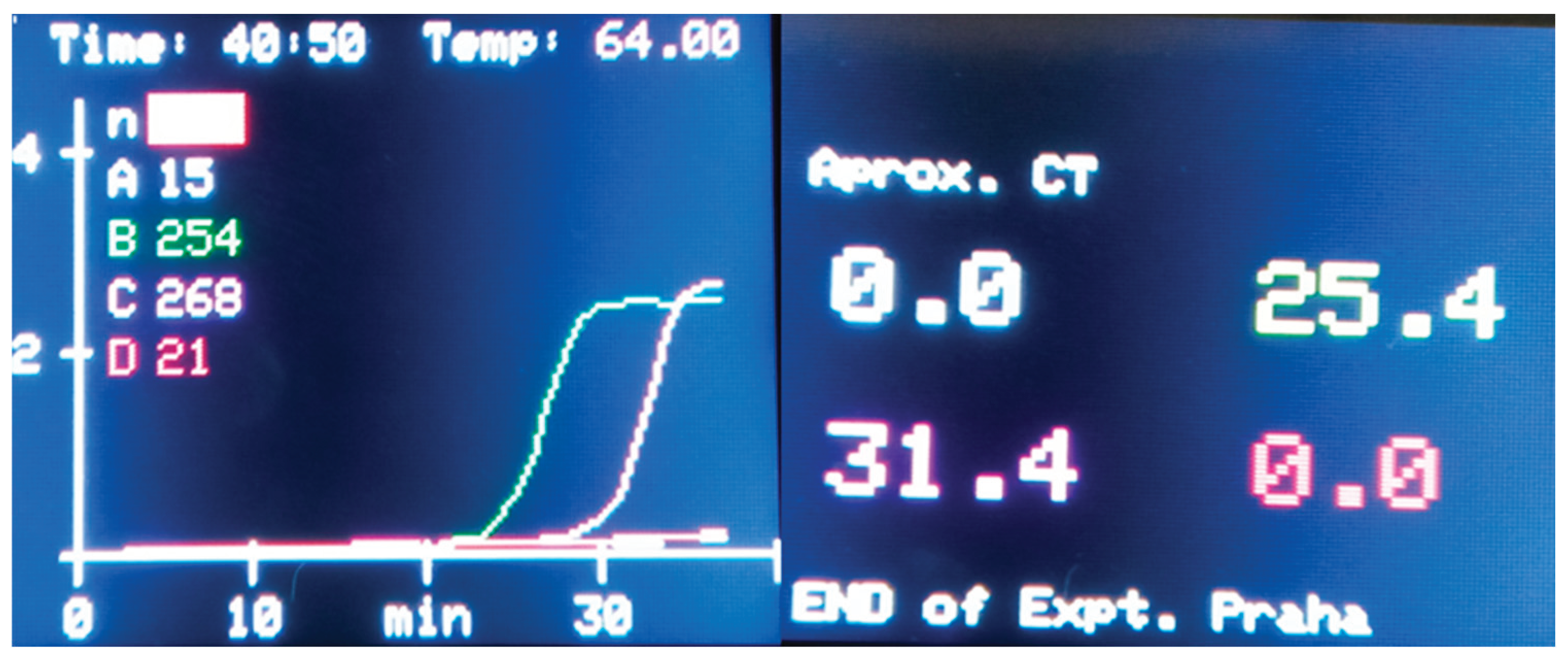
Homo sapiens and the ZymoBIOMICS® Microbial Community Standard (ZymoResearch, USA), were cross-tested for assay specificity. A specificity test revealed no amplification signal for any of the examined species. When SYBR Green Gel Stain was replaced with a fluorescent dye for LAMP assays, there was a significant increase in Y-axis-based relative fluorescent units (RFUs), as shown in
Figure 3 and
Figure 4.
Positive control for amplification
Human male TaqMan™ Control Genomic DNA (cat. no. 431266; Thermo Fisher Scientific, USA) was used as a positive control for amplification. We prepared a dilution series exactly according to the Thermo Fisher application notes [
22]. Once de-frosted, the standard was diluted into the needed aliquots, and stored at –20 °C, to prevent repeated freeze-thaw. When assessing sensitivity, successful amplification of the human AMEL-Y target was achieved by repeated use of at least 10 copies of this DNA control (data not shown).
Test arrangement
The standardized SaLux19 run aiming for species identification should include both LAMP diagnostic reagents with primer cocktails in all positions and an examined sample in one device position, next to a suitable target that serves as a control for a successful LAMP reaction (positive control). For this purpose, human male DNA focused on the Y-amelogenin gene was selected. Finally, one device position is occupied with no template control (NTC), allowing the room´s purity to be checked.
3. Results
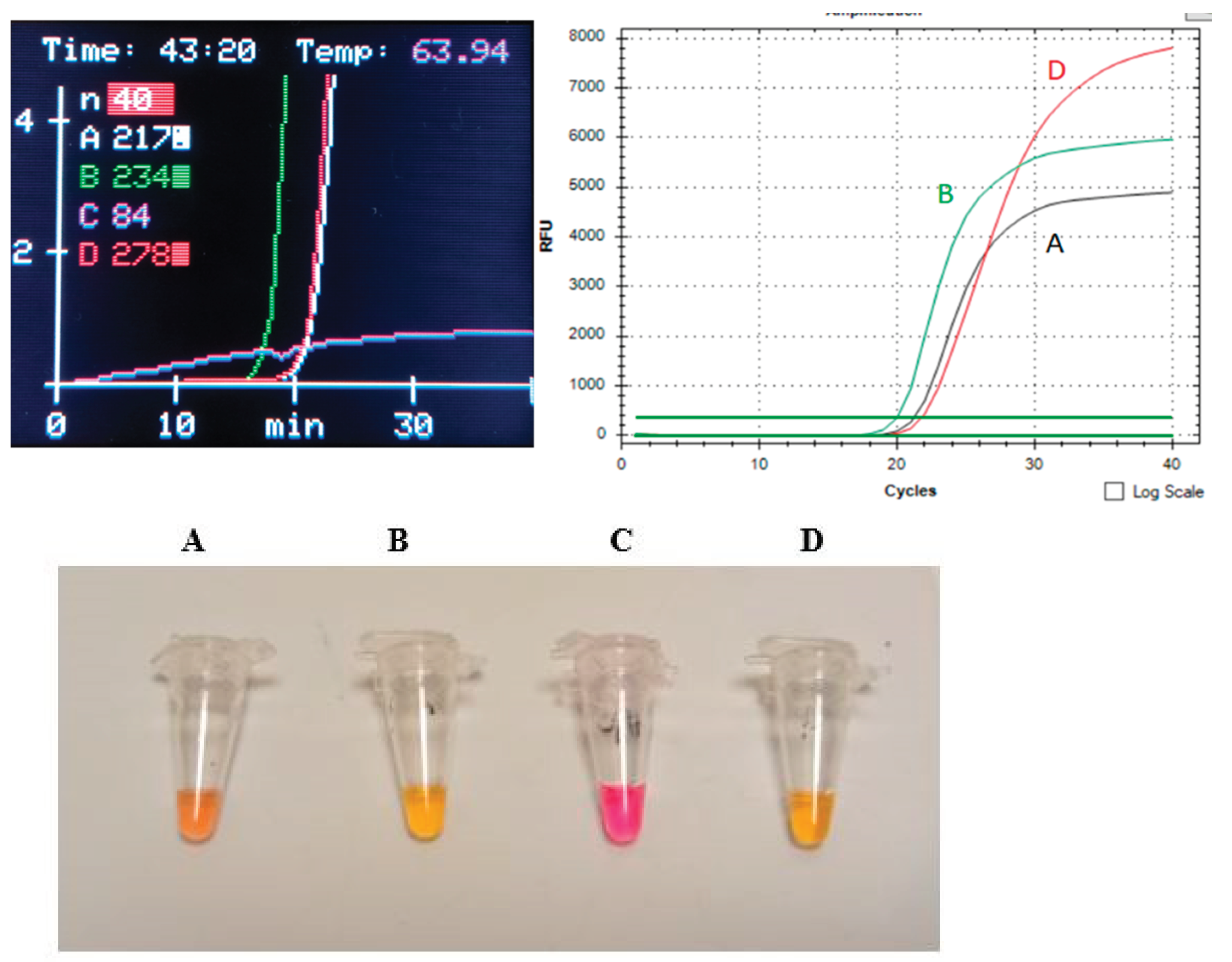
The final, standardized run on
SaLux19 included 10 μl of S
us scrofa whole nonprecipitating blood lysed in direct DipStick extraction buffer (position
A),
Sus scrofa DNA at a concentration of 127 pg/μl nDNA as a positive control (position
B), human male DNA at 30 copies of the AMEL-Y gene for checking out of own amplification (position
D), and no template control (NTC, position
C). In addition to the possibility of
real-time run monitoring, the addition of fluorescent dye directly to the colorimetric master-mixes allows us to assess the results with the naked eye, as shown in
Figure 4.
Figure 4.
The standard LAMP assay run on SALUX19 Sus scrofa cells. An analogous test with the same concentrations of Sus scrofa DNA was performed using a CFX Connect real-time PCR system (Bio-Rad, USA).
Figure 4.
The standard LAMP assay run on SALUX19 Sus scrofa cells. An analogous test with the same concentrations of Sus scrofa DNA was performed using a CFX Connect real-time PCR system (Bio-Rad, USA).
4. Discussion
Here, we report the design, optimization, preliminary tests, and validation of a rapid and portable analysis system for identifying DNA from body fluids of Sus scrofa origin on the SaLux19 device. The SaLux19 LAMP-based point of care (POC) testing includes fluorescent LAMP assay reagents, requires only this pocket-size equipment, and gives results in a short time. The data presented in this report demonstrated that the LAMP analysis system is sensitive enough. Tests of the specificity of the LAMP assay were successfully performed on a real-time the CFX Connect PCR System (Bio-Rad, USA), allowing the testing of more than 4 samples. The developed LAMP assay efficiently excluded 8 other prokaryotes (Pseudomonas aeruginosa, Escherichia coli, Salmonella enterica, Lactobacillus fermentum, Enterococcus faecalis, Staphylococcus aureus, Listeria monocytogenes, and Bacillus subtilis) and 8 eukaryotic species (members of Big cats and Bovidae mentioned in the assay validation chapter, Homo sapiens and two yeasts: Saccharomyces cerevisiae and Cryptococcus neoformans).
In addition to the apparent advantages of monitoring one's reaction in real-time, another benefit of our system is the double evaluation: when the system enables both fluorescent and colorimetric detection and reading based on the color change of the test tubes. Two steps facilitated the dual evaluation: both the direct addition of the fluorescent dye to the colorimetric master-mix and the transfer of reaction mixtures from 0.5 ml thermoblock tubes to thin-walled 0.1 ml tubes, which permits more efficient heat transfer and start-up in quantitative PCR instruments or SaLux19. Moreover, the closed wells eliminated cross-contamination to ensure accuracy, and a less time–consuming procedure was achieved when blood was directly lysed with sodium-hydroxide –based DipStick buffer.
The need for the rapid detection of biological analytes at their
points of care (POC) led to the development of portable instruments enabling either nucleic acid amplification tests or antigen detection. LAMP assays, when coupled with user-friendly instruments. can deliver the results in tens of minutes (SMART-LAMP [
23], DNAiTECH Gen2 [
24], GENIE II (OptiGene Ltd., UK) [
25]. The SaLux19 instrument offers the following advantages: besides the ability to monitor the progress of the run on the built-in display, there is another benefit of portability and the option of recharging via a USB adapter or even a small power bank.
It is appropriate to discuss the suitable sizes of amplified targets for processed meat products here. Potential degradation of DNA during food processing can adversely impact the amplification of longer amplicons. It has been reported that small amplicons with fragments less than 150 bp are recommended due to their increased sensitivity (LOD) in qPCR assays compared to longer fragments exceeding 200 bp [
26]. On the other hand, several studies have been published in which the LAMP assay was shown to have the same detection capability as qPCR [
27,
28,
29,
30].
How the presented research pipeline, where whole nonprecipitating blood was examined, would work on heat-processed meat products remains to be elucidated. In the meantime, this methodology represents a rational, simple, robust, and low-cost approach for identifying Sus scrofa body fluids.
Acknowledgments
We thank the Max Plank Society, the Max-Planck-Innovation GmbH, and the Max-Planck Foundation for their support during the initial development of the LAMP-detection equipment used in this study.
Conflicts of Interest
W.S. is a co-founder of the SaLux U.G., which produces the SaLux19 device.
References
- Scott, A.C.T.; Layne, T.R.; Landers, J.P. Low-Cost Isothermal Amplification Microdevice for Rapid Colorimetric Detection Applied to Body Fluid Identification and Y-Screening 2021.
- Notomi, Tsugunori, et al. “Loop-mediated isothermal amplification of DNA.” Nucleic acids research 28.12 (2000): e63-e63. [CrossRef]
- Khan, M.; Wang, R.; Li, B.; Liu, P.; Weng, Q.; Chen, Q. Comparative Evaluation of the LAMP Assay and PCR-Based Assays for the Rapid Detection of Alternaria Solani. Front. Microbiol. 2018, 9, 2089. [Google Scholar] [CrossRef] [PubMed]
- Kashir, J.; Yaqinuddin, A. Loop Mediated Isothermal Amplification (LAMP) Assays as a Rapid Diagnostic for COVID-19. Med. Hypotheses 2020, 141, 109786. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Lin, C.-W.; Chuang, H.-S. LAMP-Based Point-of-Care Biosensors for Rapid Pathogen Detection. Biosensors 2022, 12, 1068. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, Y.; Kananizadeh, P.; Ohadian Moghadam, S.; Alizadeh, A.; Pourmand, M.R.; Mohammadi, N.; Afshar, D.; Ranjbar, R. The Sensitivity and Specificity of Loop-Mediated Isothermal Amplification and PCR Methods in Detection of Foodborne Microorganisms: A Systematic Review and Meta-Analysis. Iran. J. Public Health 2021. [Google Scholar] [CrossRef] [PubMed]
- Deliveyne, N.; Austin, J.J.; Cassey, P. Developing Loop Mediated Isothermal Amplification (LAMP) Assays for Rapid, Presumptive DNA Detection of an Invasive Reptile (Boa Constrictor). Wildl. Res. 2023. [Google Scholar] [CrossRef]
- Girish, P.S.; Barbuddhe, S.B.; Kumari, A.; Rawool, D.B.; Karabasanavar, N.S.; Muthukumar, M.; Vaithiyanathan, S. Rapid Detection of Pork Using Alkaline Lysis- Loop Mediated Isothermal Amplification (AL-LAMP) Technique. Food Control 2020, 110, 107015. [Google Scholar] [CrossRef]
- Thangsunan, P.; Temisak, S.; Morris, P.; Rios-Solis, L.; Suree, N. Combination of Loop-Mediated Isothermal Amplification and AuNP-Oligoprobe Colourimetric Assay for Pork Authentication in Processed Meat Products. Food Anal. Methods 2021, 14, 568–580. [Google Scholar] [CrossRef]
- Aida, A.A.; Che Man, Y.B.; Wong, C.M.V.L.; Raha, A.R.; Son, R. Analysis of Raw Meats and Fats of Pigs Using Polymerase Chain Reaction for Halal Authentication. Meat Sci. 2005, 69, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Shimada, M.; Nakadai, T.; Tamura, T.-A. TATA-Binding Protein-like Protein (TLP/TRF2/TLF) Negatively Regulates Cell Cycle Progression and Is Required for the Stress-Mediated G(2) Checkpoint. Mol. Cell. Biol. 2003, 23, 4107–4120. [Google Scholar] [CrossRef] [PubMed]
- Duttke, S.H.C.; Doolittle, R.F.; Wang, Y.-L.; Kadonaga, J.T. TRF2 and the Evolution of the Bilateria. Genes Dev. 2014, 28, 2071–2076. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Boessenkool, S.; Epp, L.S.; Haile, J.; Bellemain, E.; Edwards, M.; Coissac, E.; Willerslev, E.; Brochmann, C. Blocking Human Contaminant DNA during PCR Allows Amplification of Rare Mammal Species from Sedimentary Ancient DNA. Mol. Ecol. 2012, 21, 1806–1815. [Google Scholar] [CrossRef] [PubMed]
- DipStick DNA Extraction Kit Available online: https://bento.bio/product/dipstick-dna-extraction-kit/.
- Jawla, J.; Kumar, R.R.; Mendiratta, S.K.; Agarwal, R.K.; Singh, P.; Saxena, V.; Kumari, S.; Boby, N.; Kumar, D.; Rana, P. On-Site Paper-Based Loop-Mediated Isothermal Amplification Coupled Lateral Flow Assay for Pig Tissue Identification Targeting Mitochondrial CO I Gene. J. Food Compos. Anal. 2021, 102, 104036. [Google Scholar] [CrossRef]
- Nogami, H.; Tsutsumi, H.; Komuro, T.; Mukoyama, R. Rapid and Simple Sex Determination Method from Dental Pulp by Loop-Mediated Isothermal Amplification. Forensic Sci. Int. Genet. 2008, 2, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Burgos, G.; Barrionuevo-Pérez, K.; Restrepo, T.; Tejera, E.; De Waard, J.H.; Garzón-Salazar, A.; Gusmão, L. High-Performance LAMP-Based Method for Human Sex Identification Using Y Chromosome-Specific Genetic Markers. Forensic Sci. Int. Genet. Suppl. Ser. 2022, 8, 225–227. [Google Scholar] [CrossRef]
- Colorimetric LAMP Master Assay Available online: https://www.neb.com/en/protocols/2016/08/15/warmstart-colorimetric-lamp-2x-master-mix-typical-lamp-protocol-m1800.
- Typical LAMP Protocol Available online: https://www.neb.com/en/protocols/2014/11/21/typical-lamp-protocol-m0275.
- WarmStart LAMP Kit Available online: https://www.neb.com/en/protocols/2016/08/15/warmstart-lamp-kit-dna-rna-protocol-e1700.
- ThermoFisher Application Notes: Creating Standard Curves with Genomic DNA or Plasmid DNA Templates Available online:https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2FApplication-Notes%2Fcms_042486.pdf.
- García-Bernalt Diego, Juan, et al. "SMART-LAMP: A smartphone-operated handheld device for real-time colorimetric point-of-care diagnosis of infectious diseases via loop-mediated isothermal amplification." Biosensors 12.6 (2022): 424.
- Quinteiro, Javier, et al. "In-the-Field Authentication of Grapevine (Vitis vinifera L.) cv: Albariño Using Chlorotype Discrimination and a Single SNP Interrogation by LAMP." Australian Journal of Grape and Wine Research 2023 (2023).
- Frickmann, Hagen, et al. "Loop-mediated isothermal amplification-based detection of typhoid fever on an automated Genie II Mk2 system–A case-control-based approach." Acta tropica 190 (2019): 293-295. [CrossRef]
- Wang, Y.; Teo, E.; Lin, K.J.; Wu, Y.; Chan, J.S.H.; Tan, L.K. Quantification of Pork, Chicken, Beef, and Sheep Contents in Meat Products Using Duplex Real-Time PCR. Foods 2023, 12, 2971. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cao, W.; Zhang, Y.; Chang, Y.; Huang, H.; Wei, T.; Wu, J.; Ye, L.; Shi, L. Identification for Meat Adulteration (Pork, Beef, Sheep and Duck) in Foodstuff by Microfluidic Chip-Based Real-Time Fluorescent LAMP. J. Food Compos. Anal. 2023, 119, 105223. [Google Scholar] [CrossRef]
- Wang, H.; Meng, X.; Yao, L.; Wu, Q.; Yao, B.; Chen, Z.; Xu, J.; Chen, W. Accurate Molecular Identification of Different Meat Adulterations without Carryover Contaminations on a Microarray Chip PCR-Directed Microfluidic Lateral Flow Strip Device. Food Chem. Mol. Sci. 2023, 7, 100180. [Google Scholar] [CrossRef] [PubMed]
- Sakalar, E.; Kaynak, A. Practical Molecular Detection Method of Beef and Pork in Meat and Meat Products by Intercalating Dye Based Duplex Real-Time Polymerase Chain Reaction. Int. J. Food Prop. 2016, 19, 31–40. [Google Scholar] [CrossRef]
- Kanchanaphum, P. ; Maneenin Sugunya; Chaiyana Woranittha Analysis of Pork Meat Using Loop Mediated Isothermal Amplification (LAMP) to Confirm Halal Status. Int. J. Biosci. IJB 2014, 62–68. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).