Submitted:
01 March 2024
Posted:
04 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
2.1. Analysis of NC/NH2-PEG-rGO Hybrid Membrane
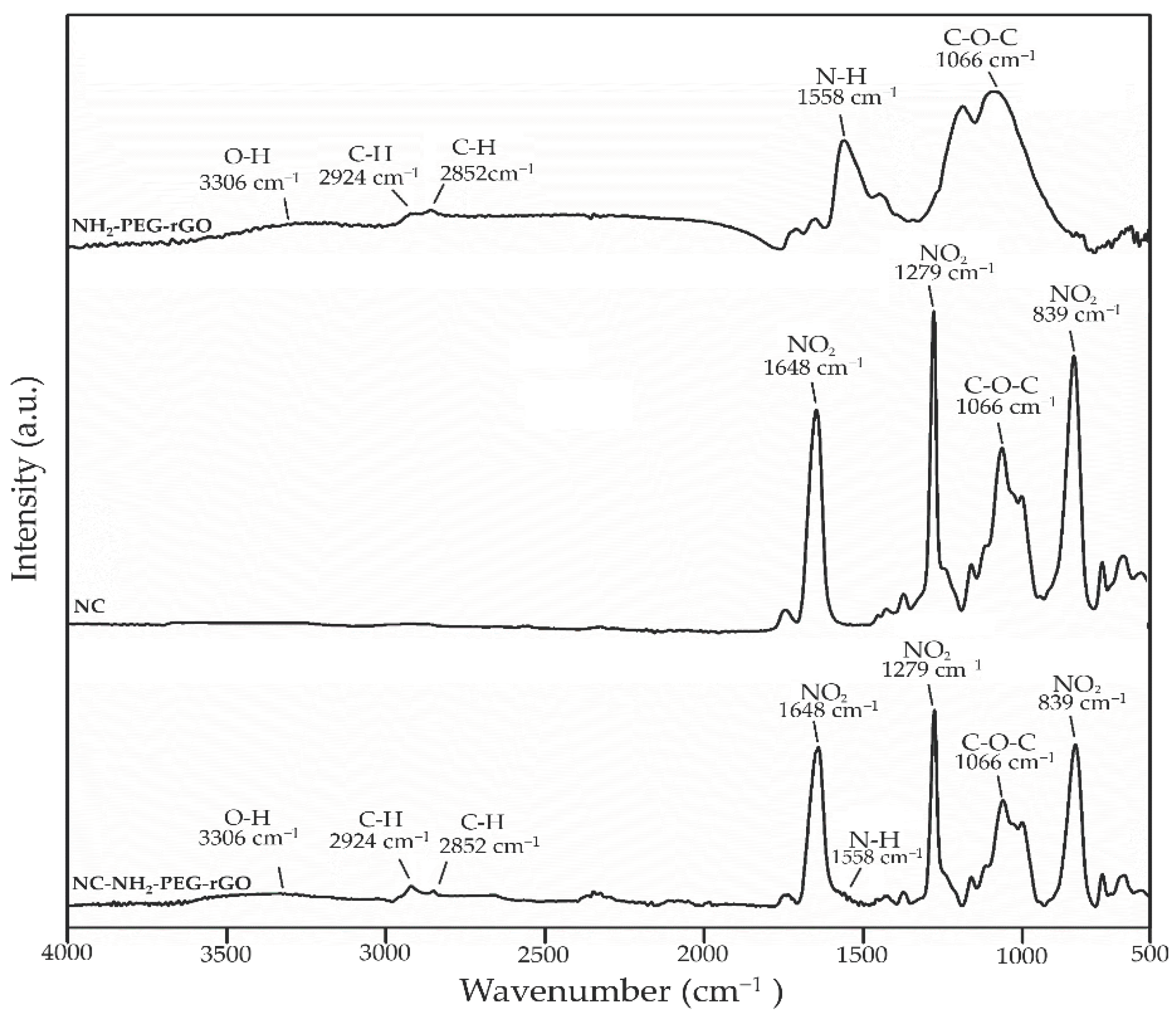
2.1.1. FTIR Analysis
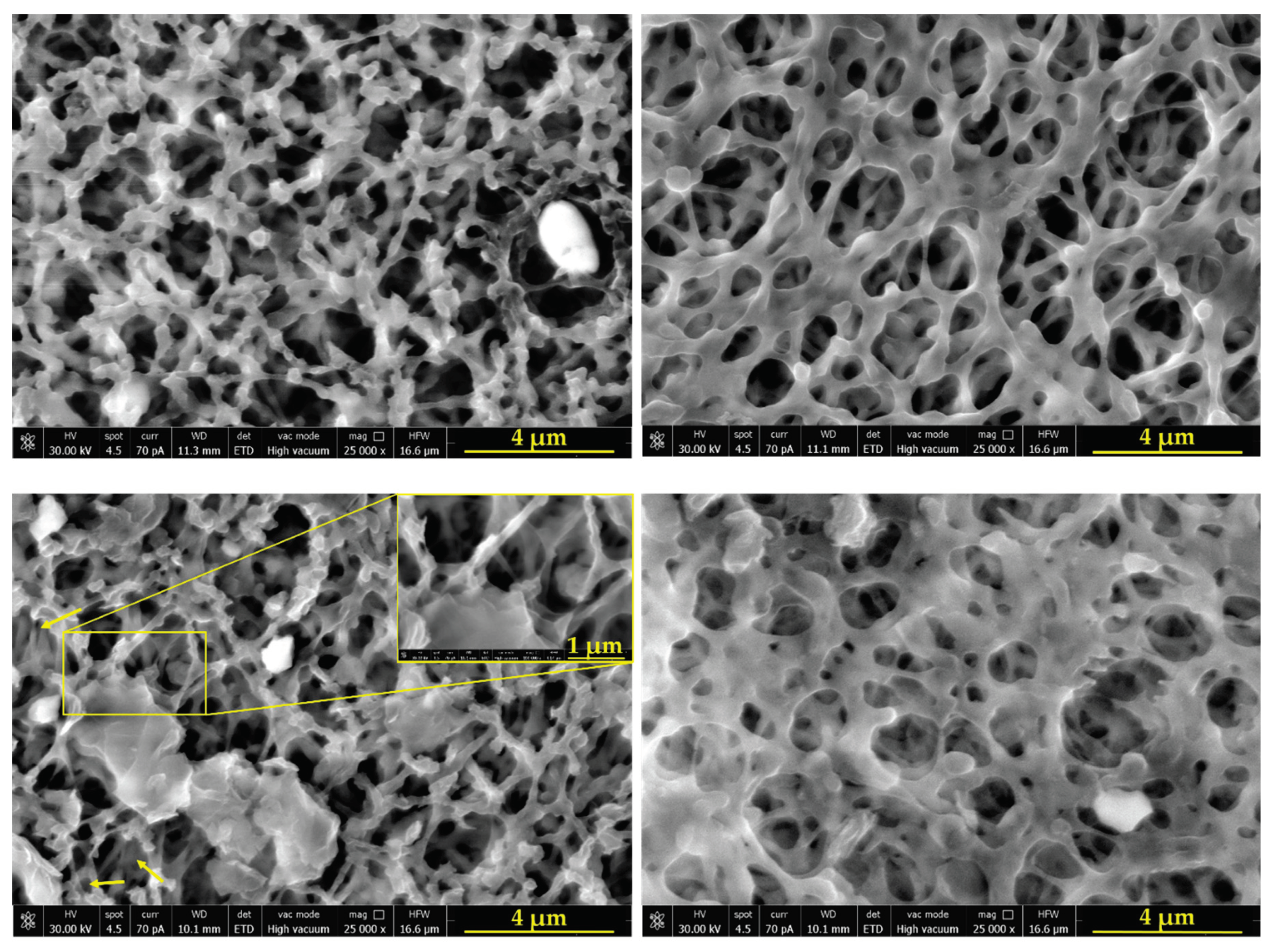
2.1.2. Morphological Characterization
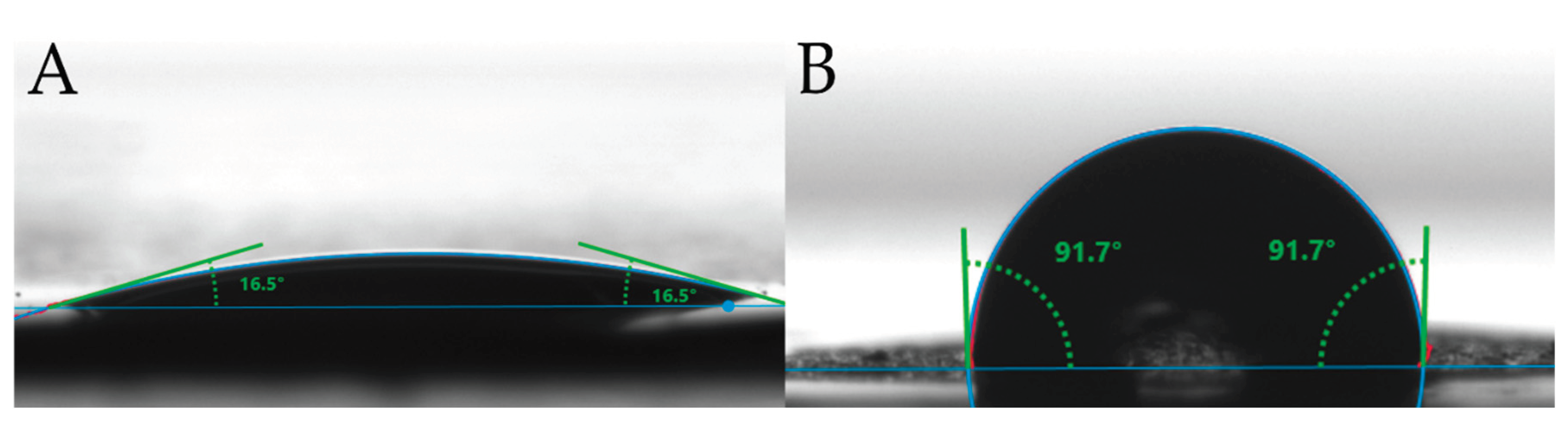
2.1.3. Wettability Characteristics
2.2. Oligo DNA Adsorption, Detection and Extraction Using NC/NH2-PEG-rGO Hybrid Membranes in Complex Media
3. Materials and Methods
3.1. Reagents
3.2. Preparation of Aminated Graphene Dispersion
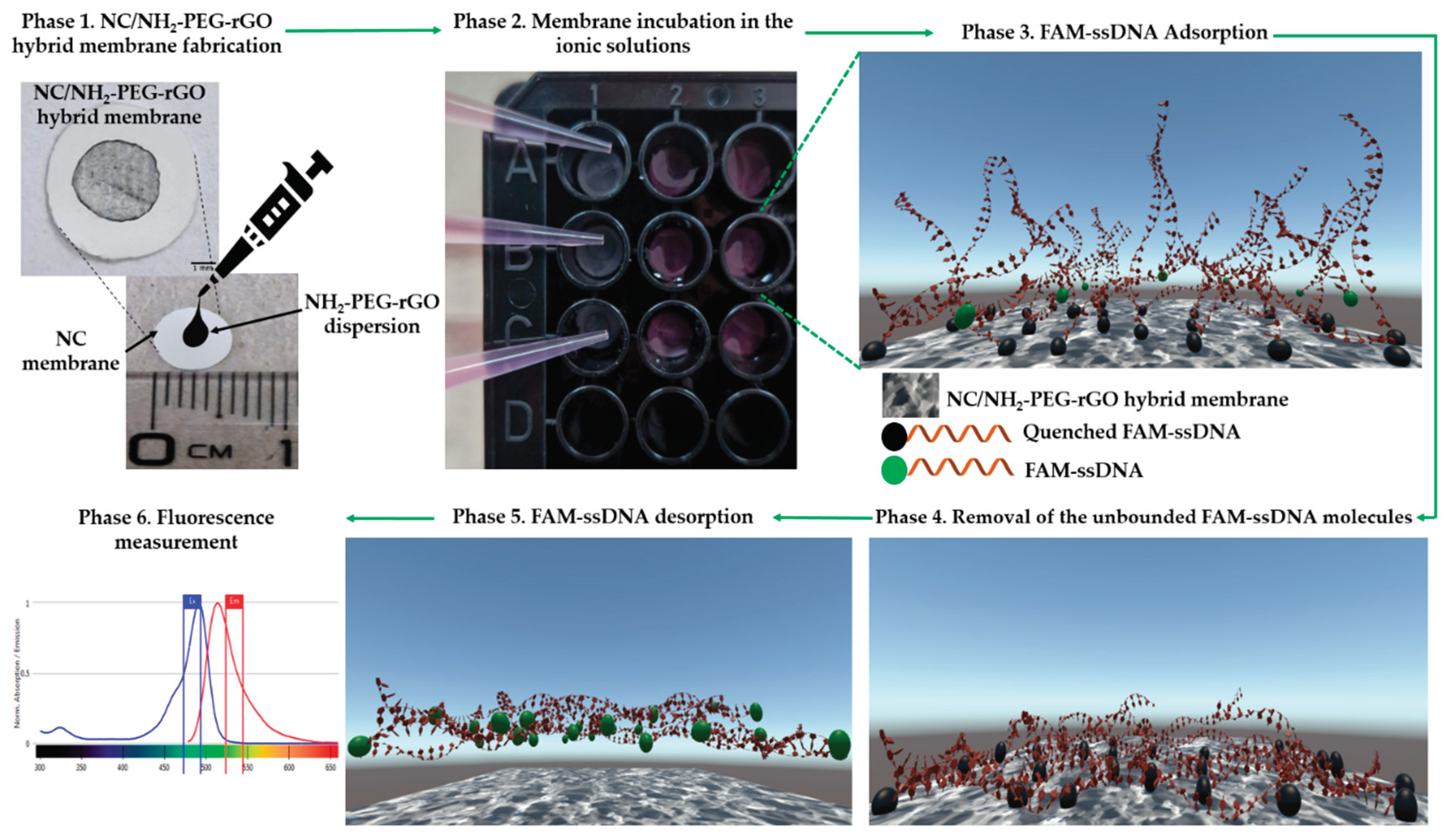
3.3. NC/NH2-PEG-rGO Hybrid Membrane Fabrication
3.4. Adsorption, Extraction, Detection of FAM-Labeled ssDNA in Ionic Solutions from NC/NH2-PEG-rGO Hybrid Membrane
3.5. Spectroflourimeter Assay
3.6. Membrane Characterization
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Lau, H.Y.; Botella, J.R. Advanced DNA-Based Point-of-Care Diagnostic Methods for Plant Diseases Detection. Front. Plant Sci. 2017, 8, 2016. [Google Scholar] [CrossRef]
- Kulkarni, A.; Tanga, S.; Karmakar, A.; Hota, A.; Maji, B. CRISPR-Based Precision Molecular Diagnostics for Disease Detection and Surveillance. ACS Appl. Bio Mater. 2023, 6, 3927–3945. [Google Scholar] [CrossRef]
- Abdel-Latif, A.; Osman, G. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 2017, 13, 1. [Google Scholar] [CrossRef]
- Shetty, P.J. The Evolution of DNA Extraction Methods. Am. J. Biomed. Sci. Res. 2020, 8, 39–45. [Google Scholar] [CrossRef]
- Bramucci, A.R.; Focardi, A.; Rinke, C.; Hugenholtz, P.; Tyson, G.W.; Seymour, J.R.; et al. Microvolume DNA extraction methods for microscale amplicon and metagenomic studies. ISME Commun. 2021, 1, 1–5. [Google Scholar] [CrossRef]
- Huergo, L.F.; Selim, K.A.; Conzentino, M.S.; Gerhardt, E.C.M.; Santos, A.R.S.; Wagner, B.; et al. Magnetic Bead-Based Immunoassay Allows Rapid, Inexpensive, and Quantitative Detection of Human SARS-CoV-2 Antibodies. ACS Sens. 2021, 6, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Muller, T.G.; Khalid, D.; Sonntag-Buck, V.; Heuser, A.M.; Glass, B.; et al. SARS-CoV-2 RNA Extraction Using Magnetic Beads for Rapid Large-Scale Testing by RT-qPCR and RT-LAMP. Viruses 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Mohajeri, N.; Mostafavi, E.; Zarghami, N. The feasibility and usability of DNA-dot bioconjugation to antibody for targeted in vitro cancer cell fluorescence imaging. J. Photochem. Photobiol. B: Biol. 2020, 209, 111944. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Feng, S.; Ji, C.; Shi, M.; He, X.; Xu, F.; et al. Microstructural effects on permeability of Nitrocellulose membranes for biomedical applications. J. Membr. Sci. 2019, 595, 117502. [Google Scholar] [CrossRef]
- Tang, R.; Xie, M.Y.; Li, M.; Cao, L.; Feng, S.; Li, Z.; et al. Nitrocellulose Membrane for Paper-based Biosensor. Appl. Mater. Today 2021, 26, 101305. [Google Scholar] [CrossRef]
- Fan, M.; Li, Y.; Chen, J.; Lin, Y.; Lai, S.; Peng, S.; et al. Plasmonic internal standard-decorated nitrocellulose membranes for duplex detection of circulating tumor biomarkers. Sensors Actuators B: Chem. 2023, 395. [Google Scholar] [CrossRef]
- Das, D.; Namboodiri, S. Selection of a suitable paper membrane for Loop Mediated Isothermal DNA amplification reaction (LAMP) in a point-of-care diagnostic kit – Experimental and CFD analysis. Chem. Eng. Sci. 2020, 229, 116130. [Google Scholar] [CrossRef]
- Toader, G.A.; Nitu, F.R.; Ionita, M. Graphene Oxide/Nitrocellulose Non-Covalent Hybrid as Solid Phase for Oligo-DNA Extraction from Complex Medium. Molecules 2023, 28, 4599. [Google Scholar] [CrossRef]
- Okhay, O.; Tkach, A. A comprehensive review of the use of porous graphene frameworks for various types of rechargeable lithium batteries. J. Energy Storage 2024, 80. [Google Scholar] [CrossRef]
- Khanam, P.N.; AlMaadeed, M.A.; Ouederni, M.; Mayoral, B.; Hamilton, A.; Sun, D. Effect of two types of graphene nanoplatelets on the physico–mechanical properties of linear low–density polyethylene composites. Adv. Manuf. Polym. Compos. Sci. 2016, 2, 67–73. [Google Scholar] [CrossRef]
- Farani, M.R.; Khadiv-Parsi, P.; Riazi, G.H.; Ardestani, M.S.; Rad, H.S. PEGylation of graphene/iron oxide nanocomposite: assessment of release of doxorubicin, magnetically targeted drug delivery and photothermal therapy. Appl. Nanosci. 2020, 10, 1205–1217. [Google Scholar] [CrossRef]
- dos Anjos, E.G.; Moura, N.K.; Antonelli, E.; Baldan, M.R.; Gomes, N.A.; Braga, N.F.; et al. Role of adding carbon nanotubes in the electric and electromagnetic shielding behaviors of three different types of graphene in hybrid nanocomposites. J. Thermoplast. Compos. Mater. 2022, 36, 3209–3235. [Google Scholar] [CrossRef]
- Feicht, P.; Siegel, R.; Thurn, H.; Neubauer, J.W.; Seuss, M.; Szabó, T.; et al. Systematic evaluation of different types of graphene oxide in respect to variations in their in-plane modulus. Carbon 2017, 114, 700–705. [Google Scholar] [CrossRef]
- Wang, J.; Chen, D.; Huang, W.; Yang, N.; Yuan, Q.; Yang, Y. Aptamer-functionalized field-effect transistor biosensors for disease diagnosis and environmental monitoring. Exploration 2023, 3, 20210027. [Google Scholar] [CrossRef] [PubMed]
- Aspermair, P.; Mishyn, V.; Bintinger, J.; Happy, H.; Bagga, K.; Subramanian, P.; et al. Reduced graphene oxide–based field effect transistors for the detection of E7 protein of human papillomavirus in saliva. Anal. Bioanal. Chem. 2020, 413, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Sethi, J.; Suhail, A.; Safarzadeh, M.; Sattar, A.; Wei, Y.; Pan, G. NH2 linker for femtomolar label-free detection with reduced graphene oxide screen-printed electrodes. Carbon 2021, 179, 514–522. [Google Scholar] [CrossRef]
- Krishnan, S.K.; Singh, E.; Singh, P.; Meyyappan, M.; Singh Nalwa, H. A review on graphene-based nanocomposites for electrochemical and fluorescent biosensors. RSC Adv. 2019, 9, 8778–8881. [Google Scholar] [CrossRef] [PubMed]
- Ratre, P.; Nazeer, N.; Kumari, R.; Thareja, S.; Jain, B.; Tiwari, R.; et al. Carbon-Based Fluorescent Nano-Biosensors for the Detection of Cell-Free Circulating MicroRNAs. Biosensors 2023, 13, 226. [Google Scholar] [CrossRef]
- Sargazi, S.; Fatima, I.; Kiani, M.H.; Mohammadzadeh, V.; Arshad, R.; Bilal, M.; et al. Fluorescent-based nanosensors for selective detection of a wide range of biological macromolecules: A comprehensive review. Int. J. Biol. Macromol. 2022, 206, 115–147. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Speranza, G. Functionalization of Carbon Nanomaterials for Biomedical Applications. C J. Carbon Res. 2019, 5, 72. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Z.; Pang, W.; Chen, H.; Yan, G. Graphene oxide based fluorescence super-quencher@QDs composite aptasensor for detection of Ricin B-chain. Sensors Actuators B: Chem. 2019, 301, 127014. [Google Scholar] [CrossRef]
- Metternich, J.T.; Wartmann, J.A.C.; Sistemich, L.; Nißler, R.; Herbertz, S.; Kruss, S. Near-Infrared Fluorescent Biosensors Based on Covalent DNA Anchors. J. Am. Chem. Soc. 2023, 145, 14776–14783. [Google Scholar] [CrossRef]
- Borchers, A.; Pieler, T. Programming Pluripotent Precursor Cells Derived from Xenopus Embryos to Generate Specific Tissues and Organs. Genes 2010, 1, 413–426. [Google Scholar] [CrossRef]
- Becheru, D.F.; Vlăsceanu, G.M.; Banciu, A.; Vasile, E.; Ioniţă, M.; Burns, J.S. Optical Graphene-Based Biosensor for Nucleic Acid Detection; Influence of Graphene Functionalization and Ionic Strength. Int. J. Mol. Sci. 2018, 19, 3230. [Google Scholar] [CrossRef]
- Battisti, A.; Samal, S.K.; Puppi, D. Biosensing Systems Based on Graphene Oxide Fluorescence Quenching Effect. Micromachines 2023, 14, 1522. [Google Scholar] [CrossRef]
- Neema, P.; Tomy, A.M.; Cyriac, J. Chemical sensor platforms based on fluorescence resonance energy transfer (FRET) and 2D materials. TrAC Trends Anal. Chem. 2019, 124, 115797. [Google Scholar] [CrossRef]
- Ueno, Y. Graphene-Based FRET Aptasensors. Anal. Sci. 2020, 37, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.; Ruan, S.; Sun, Y.; Wu, Z.; Xu, J.; Zhang, T.; et al. ATP and ssDNA aptamer-mediated peroxidase-like activity of rGO@PDA@CeO2 nanozyme: Exosomal proteins profiling and detection at physiological pH for colorimetric sensor. Sens. Actuators B: Chem. 2023, 394, 134429. [Google Scholar] [CrossRef]
- Chiticaru, E.A.; Pilan, L.; Ioniţă, M. Electrochemical Detection Platform Based on RGO Functionalized with Diazonium Salt for DNA Hybridization. Biosensors 2022, 12, 39. [Google Scholar] [CrossRef]
- Sedlackova, E.; Bytesnikova, Z.; Birgusova, E.; Svec, P.; Ashrafi, A.M.; Estrela, P.; et al. Label-Free DNA Biosensor Using Modified Reduced Graphene Oxide Platform as a DNA Methylation Assay. Materials 2020, 13, 4936. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Wang, Y.; Du, Z.; Mao, X.; Sun, D.; et al. Studies on binding of single-stranded DNA with reduced graphene oxide–silver nanocomposites. IET Nanobiotechnology 2020, 14, 308–313. [Google Scholar] [CrossRef]
- Mirza-Aghayan, M.; Mohammadi, M.; Addad, A.; Boukherroub, R. Pd nanoparticles supported on reduced graphene oxide as an effective and reusable heterogeneous catalyst for the Mizoroki–Heck coupling reaction. Appl. Organomet. Chem. 2020, 34. [Google Scholar] [CrossRef]
- Zhang, J.; Weng, H.; Miao, X.; Li, Q.; Wang, S.; Xie, H.; et al. Preparation and Characterization of Folate-Targeted Fe 3 O 4 Nanoparticle Codelivering Cisplatin and TFPI-2 Plasmid DNA for Nasopharyngeal Carcinoma Therapy. J. Nanomater. 2017, 2017, 1–10. [Google Scholar] [CrossRef]
- Ding, P.; Su, S.; Song, N.; Tang, S.; Liu, Y.; Shi, L. Influence on thermal conductivity of polyamide-6 covalently-grafted graphene nanocomposites: varied grafting-structures by controllable macromolecular length. RSC Adv. 2014, 4, 18782–18791. [Google Scholar] [CrossRef]
- Tawade, A.K.; Kamble, B.B.; Sharma, K.K.K.; Tayade, S.N. Simultaneous electrochemical investigations of dopamine and uric acid by in situ amino functionalized reduced grahene oxide. SN Appl. Sci. 2020, 2, 1–10. [Google Scholar] [CrossRef]
- Habte, A.T.; Ayele, D.W. Synthesis and Characterization of Reduced Graphene Oxide (rGO) Started from Graphene Oxide (GO) Using the Tour Method with Different Parameters. Adv. Mater. Sci. Eng. 2019, 2019, 1–9. [Google Scholar] [CrossRef]
- Nunes, S.; Ramacciotti, F.; Neves, A.; Angelin, E.M.; Ramos, A.M.; Roldão, É.; et al. A diagnostic tool for assessing the conservation condition of cellulose nitrate and acetate in heritage collections: quantifying the degree of substitution by infrared spectroscopy. Heritage Sci. 2020, 8, 1–14. [Google Scholar] [CrossRef]
- Su, X.; Zhao, Q.; Zhang, D.; Dong, W. Synthesis and membrane performance characterization of self-emulsified waterborne nitrocellulose dispersion modified with castor oil. Appl. Surf. Sci. 2015, 356, 610–614. [Google Scholar] [CrossRef]
- Sun, B.-B.; Yao, B.-H.; He, Y.-Q.; Yang, B. Preparation and Photochromic Performance of Homogeneous Phase Nitrocellulose Membrane Grafting Spirooxazine Moieties. Coatings 2020, 10, 569. [Google Scholar] [CrossRef]
- Chen, J.-J.; Li, Y.; Zheng, X.-M.; He, F.-A.; Lam, K.-H. Enhancement in electroactive crystalline phase and dielectric performance of novel PEG-graphene/PVDF composites. Appl. Surf. Sci. 2018, 448, 320–330. [Google Scholar] [CrossRef]
- Sahu, M.; Reddy, V.R.M.; Kim, B.; Patro, B.; Park, C.; Kim, W.K.; et al. Fabrication of Cu(2)ZnSnS(4) Light Absorber Using a Cost-Effective Mechanochemical Method for Photovoltaic Applications. Materials 2022, 15. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yan, H.; Chen, Z.; Liu, Q.; Feng, Y.; Ding, F.; et al. Amino Functionalization of Reduced Graphene Oxide/Tungsten Disulfide Hybrids and Their Bismaleimide Composites with Enhanced Mechanical Properties. Polymers 2018, 10, 1199. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, F.; Wösten, H.A.B.; Scholtmeijer, K. Creating Surface Properties Using a Palette of Hydrophobins. Materials 2010, 3, 4607–4625. [Google Scholar] [CrossRef] [PubMed]
- Brutin, D.; Starov, V. Recent advances in droplet wetting and evaporation. Chem. Soc. Rev. 2017, 47, 558–585. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Orejon, D.; Song, D.; Zhang, X.; McHale, G.; Takamatsu, H.; et al. Non-wetting of condensation-induced droplets on smooth monolayer suspended graphene with contact angle approaching 180 degrees. Commun. Mater. 2022, 3, 1–9. [Google Scholar] [CrossRef]
- Chi, J.; Ma, B.; Dong, X.; Gao, B.; Elbaz, A.; Liu, H.; et al. A bio-inspired photonic nitrocellulose array for ultrasensitive assays of single nucleic acids. Anal. 2018, 143, 4559–4565. [Google Scholar] [CrossRef] [PubMed]
- Morzy, D.; Rubio-Sánchez, R.; Joshi, H.; Aksimentiev, A.; Di Michele, L.; Keyser, U.F. Cations Regulate Membrane Attachment and Functionality of DNA Nanostructures. J. Am. Chem. Soc. 2021. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Lei, Z.; Zhang, Y. Non-covalent interactions of graphene surface: Mechanisms and applications. Chem 2022, 8, 947–979. [Google Scholar] [CrossRef]
- Jones, S.F.; Joshi, H.; Terry, S.J.; Burns, J.R.; Aksimentiev, A.; Eggert, U.S.; et al. Hydrophobic Interactions between DNA Duplexes and Synthetic and Biological Membranes. J. Am. Chem. Soc. 2021, 143, 8305–8313. [Google Scholar] [CrossRef] [PubMed]
- Matsarskaia, O.; Roosen-Runge, F.; Schreiber, F. Multivalent ions and biomolecules: Attempting a comprehensive perspective. Chemphyschem 2020, 21, 1742–1767. [Google Scholar] [CrossRef] [PubMed]
- MacDermott-Opeskin, H.; McDevitt, C.A.; O’mara, M.L. Comparing Nonbonded Metal Ion Models in the Divalent Cation Binding Protein PsaA. J. Chem. Theory Comput. 2020, 16, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.-X.; Cui, Y.; Zhao, T.; Fu, H.-W.; Koirala, D.; Punnoose, J.A.; Kong, D.-M.; Mao, H. Divalent cations and molecular crowding buffers stabilize G-triplex at physiologically relevant temperatures. Sci. Rep. 2015, 5, 9255–9255. [Google Scholar] [CrossRef]
- Balint, E.; Unk, I. Selective Metal Ion Utilization Contributes to the Transformation of the Activity of Yeast Polymerase eta from DNA Polymerization toward RNA Polymerization. Int. J. Mol. Sci. 2020, 21, 8248. [Google Scholar] [CrossRef]
- Liu, Y.-F.; Ran, S.-Y. Divalent metal ions and intermolecular interactions facilitate DNA network formation. Colloids Surfaces B: Biointerfaces 2020, 194, 111117. [Google Scholar] [CrossRef]
- Nie, X.; Zhang, Z.; Wang, C.-H.; Fan, Y.-S.; Meng, Q.-Y.; You, Y.-Z. Interactions in DNA Condensation: An Important Factor for Improving the Efficacy of Gene Transfection. Bioconjugate Chem. 2018, 30, 284–292. [Google Scholar] [CrossRef]
- Marzo, M.; Liu, D.; Ruiz, A.; Chalmers, R. Identification of multiple binding sites for the THAP domain of the Galileo transposase in the long terminal inverted-repeats. Gene 2013, 525, 84–91. [Google Scholar] [CrossRef]
- Natarajan, V.P.; Zhang, X.; Morono, Y.; Inagaki, F.; Wang, F. A Modified SDS-Based DNA Extraction Method for High Quality Environmental DNA from Seafloor Environments. Front. Microbiol. 2016, 7, 986. [Google Scholar] [CrossRef]
- Martins, N.M.R.; Anbu, S.; Mahmudov, K.T.; Ravishankaran, R.; da Silva, M.F.C.G.; Martins, L.M.D.R.S.; et al. DNA and BSA binding and cytotoxic properties of copper(ii) and iron(iii) complexes with arylhydrazone of ethyl 2-cyanoacetate or formazan ligands. New J. Chem. 2017, 41, 4076–4086. [Google Scholar] [CrossRef]
- Wei, Q.; Dong, J.; Zhao, P.; Li, M.; Cheng, F.; Kong, J.; et al. DNA binding, BSA interaction and SOD activity of two new nickel(II) complexes with glutamine Schiff base ligands. J. Photochem. Photobiol. B: Biol. 2016, 161, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Feizi-Dehnayebi, M.; Dehghanian, E.; Mansouri-Torshizi, H. Probing the biomolecular (DNA/BSA) interaction by new Pd(II) complex via in-depth experimental and computational perspectives: synthesis, characterization, cytotoxicity, and DFT approach. J. Iran. Chem. Soc. 2022, 19, 3155–3175. [Google Scholar] [CrossRef]
- Lima, A.F.; May, G.; Diaz-Colunga, J.; Pedreiro, S.; Paiva, A.; Ferreira, L.; et al. Osmotic modulation of chromatin impacts on efficiency and kinetics of cell fate modulation. Sci. Rep. 2018, 8, 7210. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.R.; Sligar, S.G. Changes in solvation during DNA binding and cleavage are critical to altered specificity of the EcoRI endonuclease. Proc. Natl. Acad. Sci. USA 1998, 95, 2186–2191. [Google Scholar] [CrossRef] [PubMed]
- Blane, A.; Fanucchi, S. Effect of pH on the Structure and DNA Binding of the FOXP2 Forkhead Domain. Biochemistry 2015, 54, 4001–4007. [Google Scholar] [CrossRef]
- Crnolatac, I.; Giestas, L.; Horvat, G.; Parola, A.J.; Piantanida, I. Flavylium Dye as pH-Tunable Fluorescent and CD Probe for Double-Stranded DNA and RNA. Chemosensors 2020, 8, 129. [Google Scholar] [CrossRef]
- Nishiyama, K.; Takezawa, Y.; Shionoya, M. pH-Dependence of the thermal stability of metallo-DNA duplexes containing ligand-type 5-hydroxyuracil nucleobases. Inorganica Chim. Acta 2016, 452, 176–180. [Google Scholar] [CrossRef]
- Budiawan, S.; Purwaningsih, S.; Cahaya, D.I. The study of DNA adduct 8-hydroxy-2′deoxyguanosine (8-OHdG) formation of butylated hydroxyanisole (BHA) and its metabolite ter-butyl hydroquinone (TBHQ) through in vitro reaction with Calf Thymus DNA and 2′deoxyguanosine. IOP Conf. Series Mater. Sci. Eng. 2017, 188, 012010. [Google Scholar] [CrossRef]
- Khrustalev, V.V.; Barkovsky, E.V.; Khrustaleva, T.A. Magnesium and manganese binding sites on proteins have the same predominant motif of secondary structure. J. Theor. Biol. 2016, 395, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Cavallo, L. Frequency and Effect of the Binding of Mg2+, Mn2+, and Co2+Ions on the Guanine Base inWatson-Crick and Reverse Watson-Crick Base Pairs. J. Phys. Chem. B, 2009, 113, 15670–8. [Google Scholar] [CrossRef] [PubMed]
- Akanuma, G.; Kobayashi, A.; Suzuki, S.; Kawamura, F.; Shiwa, Y.; Watanabe, S.; et al. Defect in the Formation of 70S Ribosomes Caused by Lack of Ribosomal Protein L34 Can Be Suppressed by Magnesium. J. Bacteriol. 2014, 196, 3820–3830. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Du, P.; Min, C.; Liao, Y.; Sun, H.; Jiang, Y. Poly(1-amino-5-chloroanthraquinone): Highly Selective and Ultrasensitive Fluorescent Chemosensor For Ferric Ion. J. Fluoresc. 2013, 23, 621–627. [Google Scholar] [CrossRef]
- Selan, O.T.E.; Hawu, H.N.; Poy, L.; Cunha, T.D.; Darmakusuma, D. The substitution of Mg2+ with Mn2+/4+ metal ions in chlorophyll structure isolated from Gliricidia sepium leaves. J. Phys. Conf. Series, 2021, 2017, 012002. [Google Scholar] [CrossRef]
- Fisher, T. (25.02.2024). DNA and RNA Molecular Weights and Conversions. Available online: https://www.thermofisher.com/ro/en/home/references/ambion-tech-support/rna-tools-and-calculators/dna-and-rna-molecular-weights-and-conversions.html.
- Liu, H.; Cao, G. Effectiveness of the Young-Laplace equation at nanoscale. Sci. Rep. 2016, 6, 23936. [Google Scholar] [CrossRef]
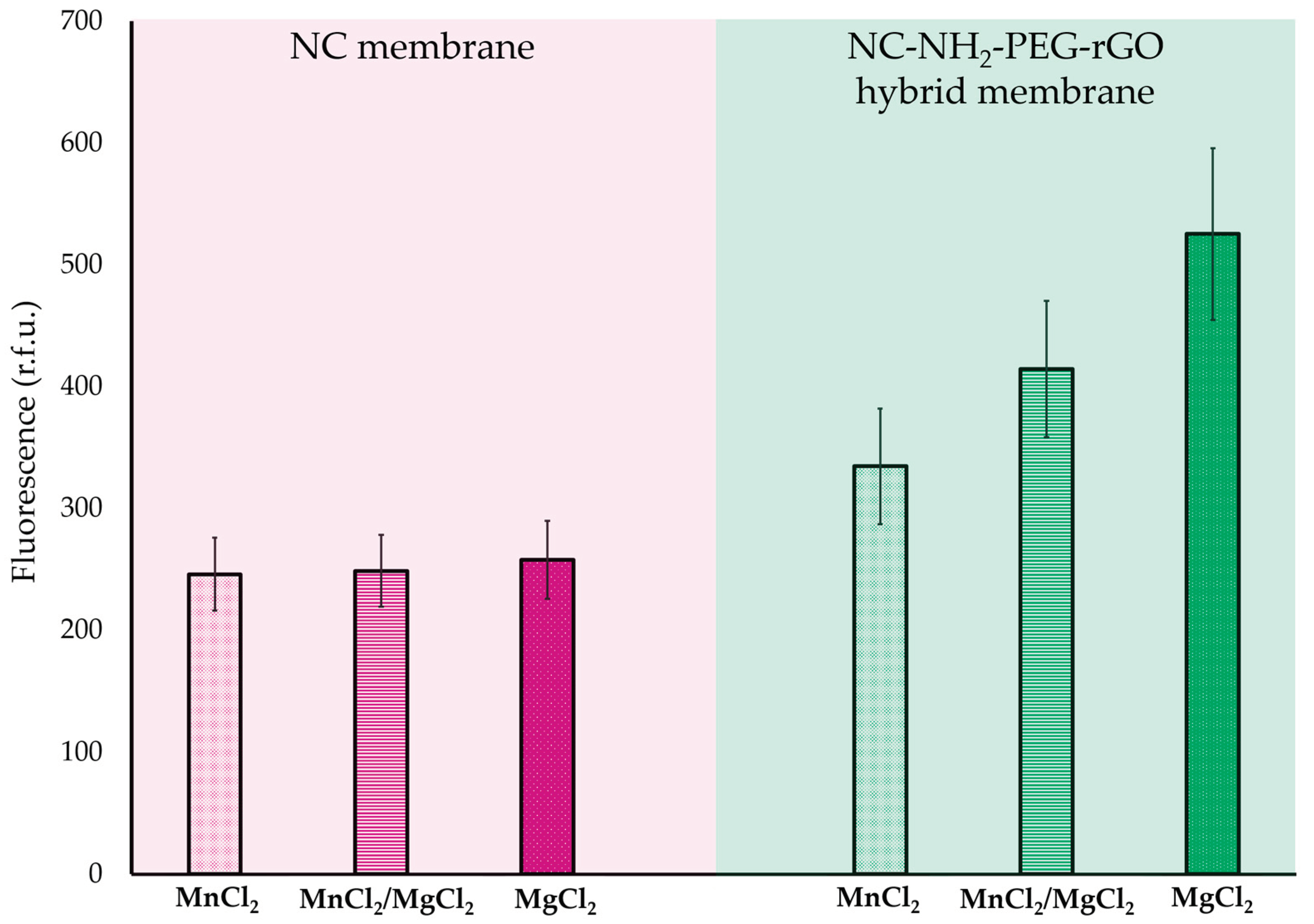
| Ionic particles in complex media | Desorption of ssDNA from the NC Membrane measured in pg, with standard deviation (s.d.) | Desorption of ssDNA from the NC/NH2-PEG-rGO Membrane measured in pg, with standard deviation (s.d.) | ||||
| Time | 60 minutes | |||||
| U/M | pg | s.d. | pg | s.d. | ||
| MnCl2 | 286.41 | ±34.75 | 389.32 | ±54.98 | ||
| MnCl2/MgCl2 | 289.67 | ±34.4 | 482.35 | ±65.23 | ||
| MgCl2 | 300.25 | ±37.1 | 611.12 | ±82.03 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




