1. Introduction
The ongoing evolution of global infectious diseases underscores the increasing necessity and criticality of vaccine development. As an emerging vaccine technology, mRNA vaccines, which utilise the intrinsic mechanisms of human cells to produce specific pathogen antigens, provide a new direction for vaccine development. Properly designed mRNA sequences can enhance the targeting and stability of mRNA vaccines, thereby improving their efficacy and durability [
1]. Traditional non-replicating mRNA vaccines typically comprise a 5′ cap structure, a 5′-untranslated region (UTR), an open reading frame (ORF) encoding the target antigen, a 3′-UTR, and a poly(A) tail structure [
2]. UTRs are critical factors that regulate mRNA stability and translation efficiency, playing significant roles in cellular transcription and translation processes [
3]. In mRNA vaccine design, the selection of UTRs directly impacts the expression levels of the vaccine and may also modulate its immunogenicity [
4,
5,
6]. UTRs may be derived from naturally occurring UTRs of highly expressed genes, such as the UTRs from α- and β-globin genes, as demonstrated in the selection made for the SARS-CoV-2 vaccine BNT162b2 [
7]. Alternatively, UTRs from the pathogen itself can be employed, or advantageous UTRs can be identified through systematic evolution and high-throughput screening, facilitated by Artificial Intelligence (AI) tools [
8,
9]. However, considering the variability in UTR efficacy across different cell types, there arises a critical need for the development of alternative UTR sequences optimized for specific application needs and intended cellular targets [
10,
11].
The efficacy of the adaptive immune response to pathogens and vaccines is intricately linked to the activation and functionality of dendritic cells (DCs). Serving as sentinel cells, DCs undertake the critical task of surveilling the body for pathogens and vaccine components. Upon encountering these entities, DCs efficiently process antigens and subsequently migrate to adjacent lymph nodes via the lymphatic system. Within the lymph nodes, DCs orchestrate the presentation of processed antigens to other immune cells, thereby orchestrating the initiation of a targeted and adaptive immune response tailored to the encountered threat [
12]. DCs are a population of specialized antigen-presenting cells (APCs) consisting of different subtypes, among which immature DCs exhibit high migratory capacity, while mature DCs can effectively activate naïve T cells, playing a crucial role in initiating, modulating, and maintaining immune responses [
13]. Following the administration of lipid nanoparticle (LNP)-mRNA vaccines, neutrophils, monocytes, and dendritic cells (DCs) efficiently internalize LNP-mRNA. DCs are primarily responsible for translating mRNA [
14]. Targeting antigen to DCs increases antibody levels [
15]. Dendritic cells in the peripheral blood such as plasmacytoid dendritic cells produce large amounts of type 1 interferon in response to microbial, especially viral, infections and stimulate the corresponding T-cell response [
16].These dendritic cells can be transported to various tissues and organs in the human body through blood circulation [
17]. Therefore, we hypothesized that optimizing the mRNA sequence to enhance its stability and translation efficiency in peripheral blood dendritic cells could improve the efficacy and durability of mRNA vaccines. In this study, we conducted a screening of untranslated regions (UTRs) derived from highly expressed genes in peripheral blood dendritic cells using bioinformatics analysis. We discovered that the expression of reporter genes in peripheral blood dendritic cells was amplified when utilizing both the 5′ UTR and 3′ UTR of TMSB10. Subsequently, we employed LNP encapsulation to deliver the SARS-CoV-2 RBD antigen modified with the UTR of TMSB10 and immunized mice via the intramuscular route. Our findings demonstrated that the modified RBD antigen enhanced antigen-specific humoral and T-cell immunity. This investigation highlights the potential of the original UTR of TMSB10 to enhance the immunogenicity of mRNA vaccines, offering promise for diverse applications in the development of viral mRNA vaccines.
2. Materials and Methods
2.1. Molecular Cloning and mRNA Synthesis
The plasmids with different UTRs (Sequences showed in
Supplementary Table S1) were synthesized by GenScript (Nanjing, China). mRNA was produced using T7 High Yield RNA Transcription kit (Novoprotein, China) on linearized plasmids. Then, Cap 1 was added to the synthesized RNA by using the Cap 1 Capping System (Novoprotein, China) and Pseudo-UTP (Ψ-UTP, APExBIO, USA) was fully substituted for UTP.
2.2. Cell Culture and Transfection
In vitro transient transfection of mRNA was conducted in HEK 293T cells, DC2.4, 16HBE, or RAW264.7 using Lipofectamine® 3000 reagent (Invitrogen, USA) following the manufacturer’s instructions. Briefly, HEK 293T cells and RAW264.7 were cultured in DMEM (Gibco, USA), while DC2.4 and 16HBE were cultured in RPMI 1640, supplemented with 10% FBS (Gibco, USA) and 1% penicillin-streptomycin (Gibco). Cell dissociation was achieved using 0.25% TrypLE (Thermo Fisher Scientific, USA), and cells were seeded in 48-well plates at a density of 50,000 cells per well. After 18 hours, the medium was replaced with 2% FBS-medium, and cells were transfected with mRNA (0.25μg per well) using Lipofectamine 3000 Transfection Reagent. Supernatants were collected 24 hours post-transfection and stored at -80℃.
2.3. Gaussia Luciferase (GLuc) Assay In Vitro
In vitro evaluation of GLuc expression was conducted using the Secrete-Pair™ Gaussia Luciferase Assay Kit. After thawing the collected supernatant on ice, 10 μl of the supernatant was added to a black 96-well plate for measurement. A working solution was prepared according to the kit instructions (100 μl per well), and the chemiluminescence value was measured at 500 ms using a Varioskan Lux (Thermo Scientific, USA).
2.4. Preparation of Lipid-Gluc mRNA Nanoparticles
In brief, SM-102 (AVT, China), DSPC (AVT, China), cholesterol (AVT, China), and DMG-PEG2000 (AVT, China) were dissolved in ethanol at a molar ratio of 50:10:38.5:1.5. The mRNA, solubilized in 50 mM citrate buffer, was mixed with lipid at a N/P ratio of 6:1. The lipid was then mixed with mRNA at a flow rate of 1:3 at room temperature to form vesicles approximately 100 nm in diameter through a T-tube. LNP-encapsulated mRNA samples were dialyzed against PBS (pH 7.4) in dialysis bags (Viskase, USA) for 24 hours and stored at 4°C until use. Encapsulation efficiency was measured using the Quant-iT RiboGreen RNA Assay Kit (Invitrogen, USA) with a Varioskan Lux (Thermo Scientific, USA).
2.5. Gaussia Luciferase (GLuc) Assay In Vivo
LNPs for in vivo imaging were formulated with mRNA encoding Gaussia luciferase (GLuc). The formulated LNPs were administered intramuscularly to mice at a dose of 5 μg of GLuc. Six hours later, tail blood samples were collected from the mice, and the fluorescence value of whole blood was measured using the Secrete-Pair™ Gaussia Luciferase Assay Kit (GeneCopoeia, LF062), following the instructions provided. Following blood collection, coelenterazine was injected intraperitoneally immediately and incubated for 5 minutes. Luciferase expression in different organs was confirmed using an IVIS (PerkinElmer, USA).
2.6. Immunization and Detection of Antigen-Specific Antibodies in Mice
Female BALB/c mice were obtained from the Animal Center of the Third Military Medical University, and the animal experiments were ethically approved by the Laboratory Animal Welfare and Ethics Committee of the Third Military Medical University (Approval No. AMUWE20201373). For vaccinations, groups of 6- to 8-week-old BALB/c mice were immunized on days 0 and 7. The mRNA vaccine and the empty carrier control group were administered via intramuscular injection. Each dose of the mRNA vaccine contained 15 μg of mRNA, approximately 200 μL. The empty LNP (PBS-LNP) used as the control. After 14 days post-immunization, 100 μL of blood was collected from the mouse tail vein, followed by centrifugation at 4°C for 10 minutes at 3000 rpm to isolate the serum. The serum was subsequently stored at -80°C until further analysis. The Mice anti-SARS-CoV-2 (S-RBD) IgG ELISA Kit (FineTesT, China) was employed to measure antibody concentrations. Absorbance at 450 nm was measured, and accurate quantification was performed using a Varioskan Lux (Thermo Scientific, USA).
2.7. Enzyme-Linked Immunospot (ELISPOT) Assays
Cellular immune responses in vaccinated mice were evaluated using IFN-γ and IL-4 pre- coated ELISPOT kits (MabTech, Germany) following the manufacturer’s protocol. Briefly, plates were blocked using RPMI 1640 (Thermo Fisher Scientific, USA) supplemented with 10% FBS and incubated for 30 minutes. Immunized mouse splenocytes were then seeded at 1,000,000 cells per well, along with a peptide pool for SARS-CoV-2 RBD protein [
18] which is a gift from Prof. Shan Guan(2 mg/ml of each peptide). After incubation at 37°C with 5% CO
2 for 36 h, the plates were washed with wash buffer, and biotinylated anti-mouse IFN-γ and IL-4 antibodies were added to each well, followed by a 2-hour incubation at room temperature. Subsequently, AEC substrate solution was added, and after air-drying, the plates were read using an automated ELISPOT reader (AID Classic EliSpot Reader, Germany). The numbers of spot-forming cells (SFC) per 1,000,000 cells were calculated.
2.8. Flow Cytometry Analyses for Mouse Splenocytes
Evaluation of T cell proliferation in immunized mice was conducted using a FACSCalibur flow cytometer (BD Biosciences, USA). Briefly, a total of 1,000,000 mouse splenocytes were stimulated with the SARS-CoV-2 RBD peptide pool (4 mg/ml of each peptide) for 4 hours at 37°C with 5% CO2. Brefeldin A (1 mg/ml, BD Sciences, USA) was then added to the splenocytes and incubated for an additional 4 hours. After two washes with PBS, the splenocytes were permeabilized and stained with fluorescently conjugated antibodies to CD3 (FITC) (BD Pharmingen, USA), CD4 (PerCP-Cyanine5.5) (BD Pharmingen, USA), CD8 (PE-Cyanine7) (BD Pharmingen, USA), CD44 (APC) (BD Pharmingen, USA), CD62L (PE) (BD Pharmingen, USA), and APC/CY7 (BioLegend, USA). Data were analyzed using FlowJo software.
2.9. Data Analysis
All statistical analyses were conducted using GraphPad Prism V8.0.2 software. The student’s t-test was employed for statistical comparisons between groups. A P-value ≤ 0.05 was considered indicative of a significant difference between groups.
3. Results
3.1. Screening and Preliminary Application of the TMSB10-UTR
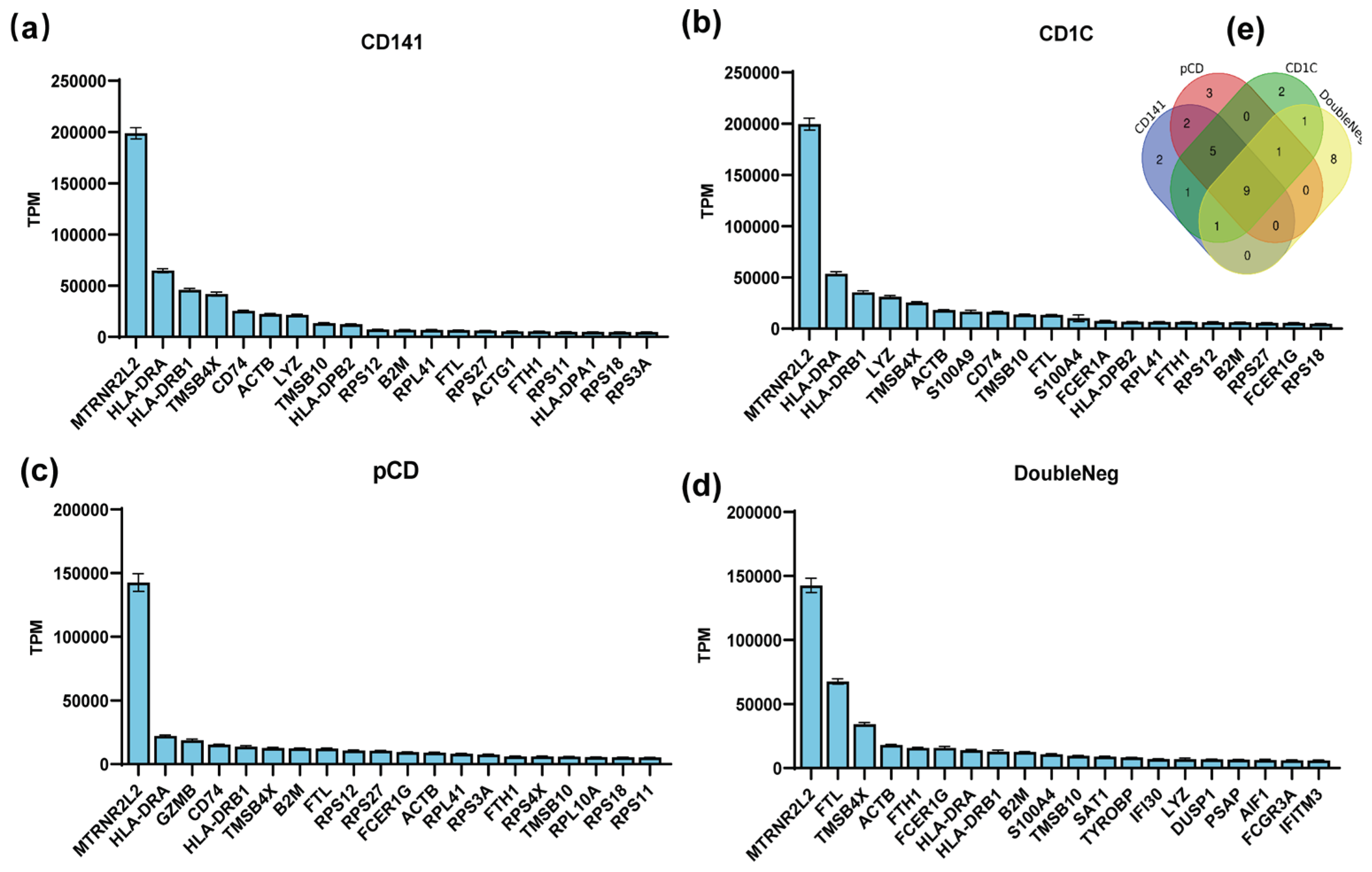
In this study, we conducted an analysis to rank gene abundance in dendritic cells, encompassing both plasmacytoid dendritic cells (pDCs) and myeloid dendritic cells (mDCs) through utilized the human peripheral blood mononuclear cell sequencing dataset GSE94820 from the Gene Expression Omnibus (GEO) database. Top 20 genes with the highest abundance for expression in these cell types were selected (
Figure 1a–d) and among them, nine genes - MTRNR2L2, HLA-DRA, HLA-DRB1, TMSB4X, ACTB, TMSB10, B2M, FTL, and FTH1 - were consistently expressed in all dendritic cells subtypes (
Figure 1e). The high mRNA abundance of these genes implies their significance in antigen-presenting cells, suggesting that their mRNAs have high stability in antigen-presenting cells.
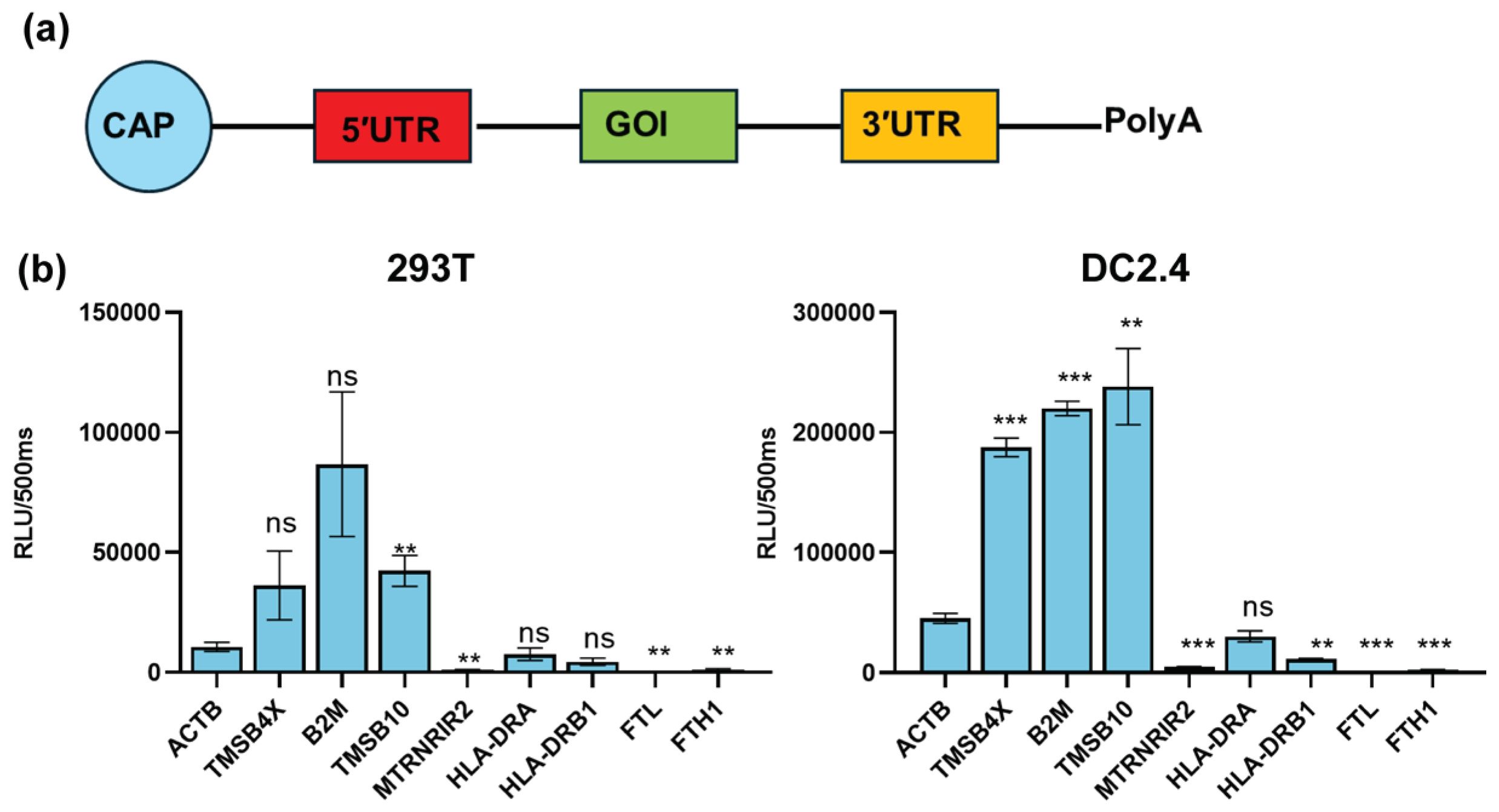
To examine the regulation of target gene expression by UTRs of these nine genes, we inserted the UTRs of these genes into a modified reporter gene Gaussian luciferase (TPA-Gluc) and produced nine mRNAs with Cap 1 and Ploy(A) tail (
Figure 2a). Then the nine mRNAs were transfected into 293T and DC2.4 cells (a model for antigen-presenting cells). After 24 hours we found that the UTRs of TMSB4X, B2M, and TMSB10 exhibited the significant ability to enhance TPA-Gluc expression in antigen-presenting cell types. Notably, the UTR of TMSB10 demonstrated the highest potency in regulating reporter gene expression within DC2.4 cells (
Figure 2b).
3.2. TMSB10-UTR Enhances Target Gene Expression in Antigen-Presenting Cells and IN VIVO
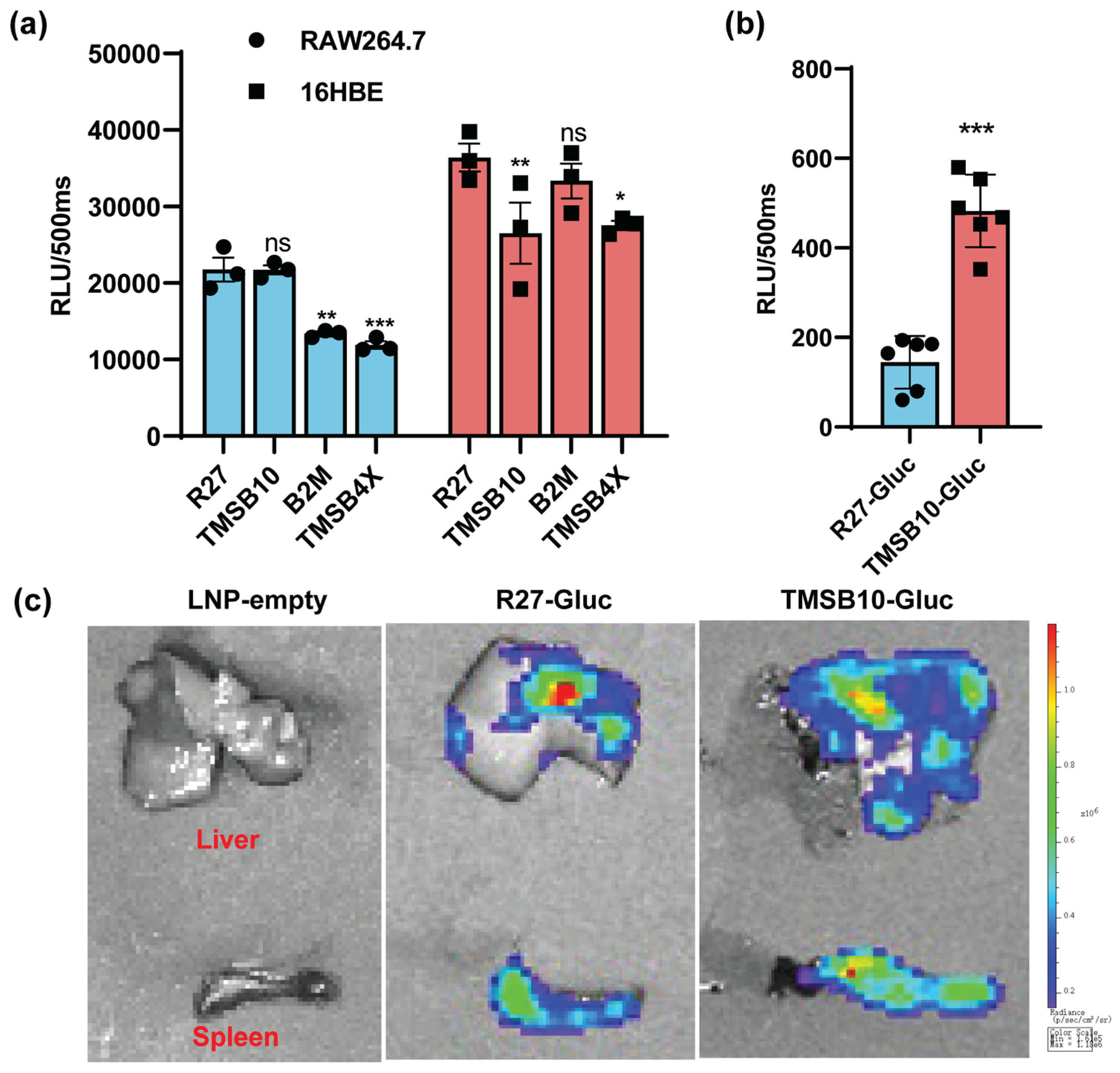
To evaluate the effectiveness of these highly expressed UTRs in comparison to previously reported optimized natural UTR, we utilized the R27-UTR as described by Zeng et al. (2020). Comparative analyses were conducted between R27-UTR and the UTRs of TMSB4X, B2M, and TMSB10 in regulating Gluc gene expression in both antigen-presenting cells and epithelial cells. Our results demonstrated that TMSB10-UTR-Gluc exhibited higher expression levels in mouse macrophage line RAW264.7 cells, while in 16HBE cells, TMSB10-UTR-Gluc expression was lower compared to R27-UTR-Gluc (
Figure 3a). These findings indicate that TMSB10-UTR, while comparable to R27-UTR in expression, possesses a cell-specific advantage over R27-UTR in antigen-delivery cells.
Following this, Gluc-LNP lipid nanoparticles were intramuscularly administered to mice, and bioluminescence values in whole blood were evaluated after 6 hours. Remarkably, the expression of TMSB10-UTR-Gluc exhibited a notable increase compared to R27-UTR-Gluc in the blood (
Figure 3b). Upon dissecting the mice, we observed that TMSB10-UTR exhibited higher expression levels in the spleen and liver compared to R27-UTR-Gluc (
Figure 3c). These findings underscore the substantial promise of TMSB10-UTR in governing predominant target gene expression in animal antigen-presenting cells, offering an effective UTR selection strategy for mRNA vaccine design.
3.3. TMSB10-UTR enhances SARS-CoV-2 mRNA vaccine efficacy
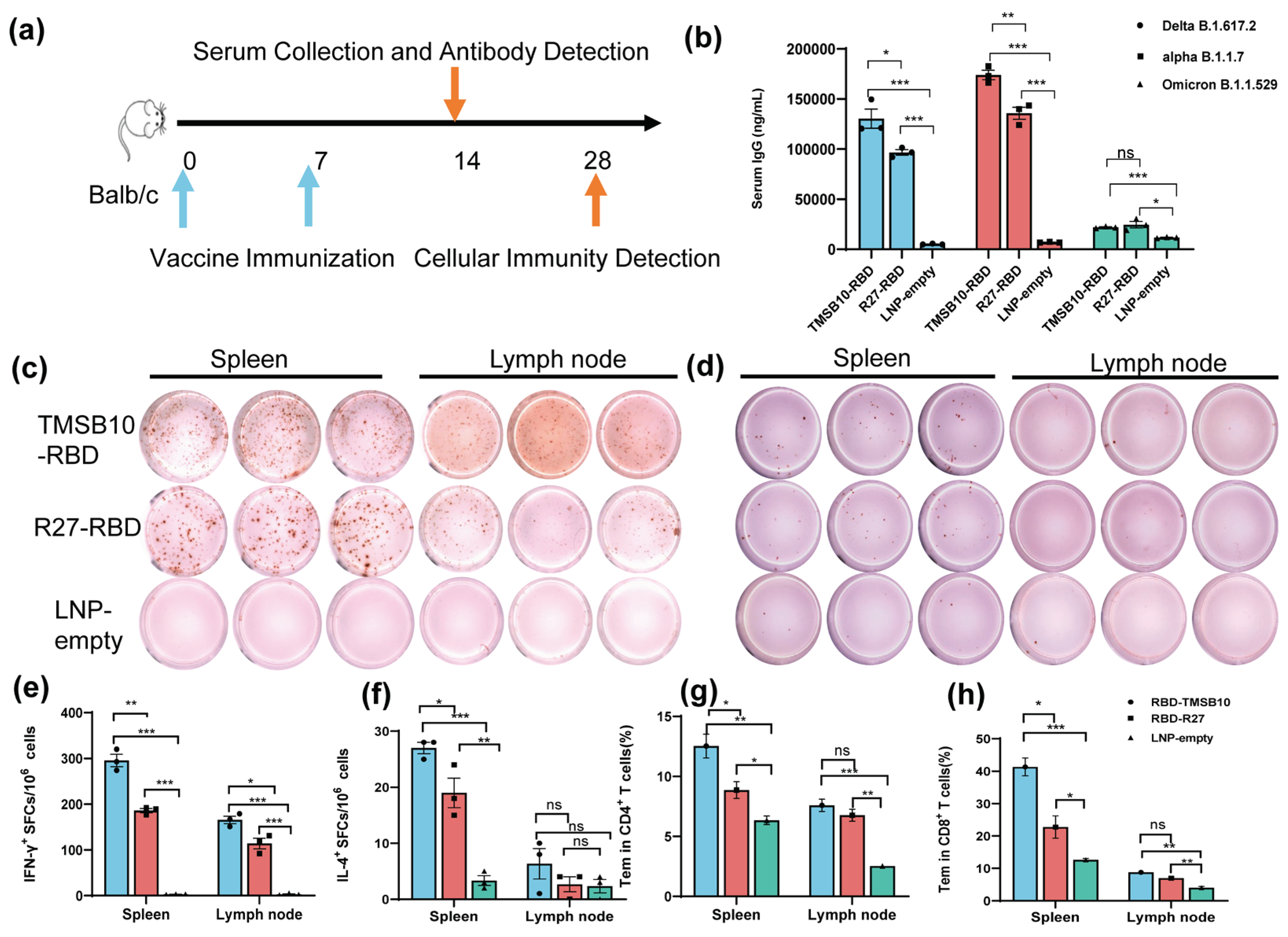
To validate the efficacy of TMSB10-UTR for mRNA vaccine applications, we proceeded to design mRNA sequences incorporating both R27-UTR and TMSB10-UTR. These sequences were developed using the SARS-CoV-2 Delta variant strain RBD (B.1.617.2, GenBank: OK091006.1) as the antigen, along with TPA as the signal peptide. Subsequently, we synthesized the mRNA in vitro, followed by the formulation of mRNA-LNP vaccines. Mice were vaccinated by intramuscular injection on days 0 and 7 (
Figure 4a). On day 14, tail blood samples were collected, corresponding to 1 week after the final immunization, to assess antibody levels (
Figure 4a). Remarkably, the mRNA-LNP vaccine incorporating TMSB10-UTR demonstrated a notable increase in specific immunoglobulin IgG levels against SARS-CoV-2 strains Delta B1.617.2, alpha B.1.1.7 and Omicron B.1.1.529 (
Figure 4b). To compare the cellular immune responses activated level between R27-UTR and TMSB10-UTR, we assessed the production of interferon-γ (IFN-γ) and interleukin-4 (IL-4) in spleen and lymph node using ELISpot assay on the 28th day post-immunization. Splenic lymphocytes and lymph nodes from mice immunized with the TMSB10 UTR exhibited a significantly higher level of IFN-γ and IL-4 secretion compared to those immunized with the R27 UTR (
Figure 4c, d, e, and f). Additionally, the TMSB10 UTR-mRNA vaccine elicited a greater abundance of CD4
+ and CD8
+ T cells in splenocytes compared to the R27-UTR (
Figure 4g and h). These results suggest that the UTR of TMSB10 can indeed enhance the immunogenicity of mRNA vaccines in the organism.
4. Discussion
Dendritic cells are a specialized class of antigen-presenting cells that play a key role in the immune system [
19]. In this paper, we screened the nine genes with the highest mRNA abundance in each dendritic cell subtype (
Figure 1). Among these genes, namely TMSB4X, B2M and TMSB10, which exhibited high expression in dendritic cells, were ultimately selected (
Figure 2). Notably, the UTR of TMSB10 demonstrated comparable efficacy to previously reported optimized R27-UTRs. Moreover, the expression of reporter genes associated with TMSB10 UTR was significantly higher in antigen-presenting cells but notably lower in lung epithelial cells (
Figure 3a), indicating that the UTR of TMSB10 possesses greater specificity for antigen-presenting cells. Subsequent animal experiments revealed that the TMSB10 UTR demonstrated elevated expression levels in the bloodstream. Given that TMSB10 was initially identified as a highly expressed gene in peripheral blood cells, its UTR performance corresponds well with its source (
Figure 3b). Subsequently, we applied it to mRNA vaccine against COVID-19. Experimental results further demonstrated that the UTR of TMSB10 indeed elicits higher humoral immunity (
Figure 4b) and induces stronger T-cell immune responses (
Figure 4c–g). These findings underscore the feasibility of our approach targeting DCs to design UTRs for enhancing their antigen-presenting capabilities. That is not only applicable to SARS-CoV-2, but also to other mRNA vaccines, thus offering broad prospects for application.
However, these natural UTRs, which are highly expressed in antigen-presenting cells, still have room for further improvement. For instance, The TOP motif, a cis-regulatory RNA element, initiates immediately following the m7G cap structure and features a characteristic invariant 5′-cytidine followed by a continuous stretch of 4–15 pyrimidines [
20]. It’s known to negatively regulate mRNA translation [
21]. Upstream Open Reading Frames (uORFs) within the 5′ UTR region also serve to suppress downstream protein expression [
22]. Potential enhancements could include the deletion of the 5′ Terminal OligoPyrimidine (5′ TOP) motif or upstream Open Reading Frames (uORFs) in the 5′UTR. Another strategy involves incorporating aptamers into the 5′UTR sequence to recruit translation-enhancing proteins or cap-binding proteins, thereby boosting the expression levels of downstream genes [
23]. Additionally, strategies could include deleting binding sites in the 5′ and 3′ UTRs for microRNAs that degrade mRNAs or inhibit their translation [
24,
25], or reducing unstructured sequences within the 3′ UTR sequence, among other approaches, all of which have the potential to further enhance protein translation efficiency [
26]. Alternatively, we can employ AI for genetic evolutionary training using our existing library of highly expressed UTRs to further enhance their expression capabilities [
27]. These improvements provide new strategic directions for improving mRNA vaccine efficiency and persistence.
5. Patents
This work has been filed for a Chinese patent under Patent Application No. 202310557980.4
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, X.D. and J.L.; methodology, X.Y. and Y.Z.; software, X.D. and Y.Z.; validation, J.H. and J.L.; formal analysis, X.D. and Y.Z; investigation, X.D.; resources, J.L.; data curation, X.D. and Y.Z; writing—original draft preparation, X.D. and Y.Z; writing—review and editing, X.D., J.H. and J.L; visualization, X.D. and Y.Z.; supervision, J.L.; project administration, J.L.; funding acquisition, X.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Natural Science Foundation Project of Chongqing, grant number cstc2020jcyj-bshX0116.
Institutional Review Board Statement
The animal study protocol was approved by the Laboratory Animal Welfare and Ethics Committee of the Third Military Medical University (AMUWE20201373).
Acknowledgments
We would like to express our gratitude to Prof. Shan Guan for providing the Spike peptides to us.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Metkar, M.; Pepin, C.S.; Moore, M.J. Tailor made: the art of therapeutic mRNA design. Nature reviews. Drug discovery 2024, 23, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.; Li, C.; Xiao, Q.; Zhang, D.; Chen, Y.; Rosenecker, J.; Ding, X.; Guan, S. Recent Advances and Innovations in the Preparation and Purification of In Vitro-Transcribed-mRNA-Based Molecules. Pharmaceutics 2023, 15. [Google Scholar] [CrossRef]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and Challenges in the Delivery of mRNA-based Vaccines. Pharmaceutics 2020, 12. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines — a new era in vaccinology. Nature Reviews Drug Discovery 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Holtkamp, S.; Kreiter, S.; Selmi, A.; Simon, P.; Koslowski, M.; Huber, C.; Türeci, O.; Sahin, U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 2006, 108, 4009–4017. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Sekhon, S.S.; Shin, W.-R.; Ahn, G.; Cho, B.-K.; Ahn, J.-Y.; Kim, Y.-H. Modifications of mRNA vaccine structural elements for improving mRNA stability and translation efficiency. Molecular & Cellular Toxicology 2022, 18, 1–8. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. The New England journal of medicine 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Cenik, C.; Cenik, E.S.; Byeon, G.W.; Grubert, F.; Candille, S.I.; Spacek, D.; Alsallakh, B.; Tilgner, H.; Araya, C.L.; Tang, H.J.G.r. Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. 2015, 25, 1610–1621. [Google Scholar] [CrossRef]
- Zeng, C.; Hou, X.; Yan, J.; Zhang, C.; Li, W.; Zhao, W.; Du, S.; Dong, Y. Leveraging mRNA Sequences and Nanoparticles to Deliver SARS-CoV-2 Antigens In Vivo. Advanced materials (Deerfield Beach, Fla.) 2020, 32, e2004452. [Google Scholar] [CrossRef]
- Sample, P.J.; Wang, B.; Reid, D.W.; Presnyak, V.; McFadyen, I.J.; Morris, D.R.; Seelig, G. Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nature biotechnology 2019, 37, 803–809. [Google Scholar] [CrossRef]
- Orlandini von Niessen, A.G.; Poleganov, M.A.; Rechner, C.; Plaschke, A.; Kranz, L.M.; Fesser, S.; Diken, M.; Löwer, M.; Vallazza, B.; Beissert, T.; et al. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3′ UTRs Identified by Cellular Library Screening. Molecular therapy: the journal of the American Society of Gene Therapy 2019, 27, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nature reviews. Drug discovery 2021, 20, 817–838. [Google Scholar] [CrossRef] [PubMed]
- Macri, C.; Pang, E.S.; Patton, T.; O’Keeffe, M. Dendritic cell subsets. Seminars in cell & developmental biology 2018, 84, 11–21. [Google Scholar] [CrossRef]
- Liang, F.; Lindgren, G.; Lin, A.; Thompson, E.A.; Ols, S.; Röhss, J.; John, S.; Hassett, K.; Yuzhakov, O.; Bahl, K.; et al. Efficient Targeting and Activation of Antigen-Presenting Cells In Vivo after Modified mRNA Vaccine Administration in Rhesus Macaques. Molecular therapy: the journal of the American Society of Gene Therapy 2017, 25, 2635–2647. [Google Scholar] [CrossRef] [PubMed]
- Hinke, D.M.; Andersen, T.K.; Gopalakrishnan, R.P.; Skullerud, L.M.; Werninghaus, I.C.; Grødeland, G.; Fossum, E.; Braathen, R.; Bogen, B. Antigen bivalency of antigen-presenting cell-targeted vaccines increases B cell responses. Cell reports 2022, 39, 110901. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald-Bocarsly, P.J.B. Natural interferon-α producing cells: the plasmacytoid dendritic cells. 2002, 33, S16–S29. [Google Scholar] [CrossRef]
- Cavanagh, L.L.; Von Andrian, U.H. Travellers in many guises: The origins and destinations of dendritic cells. 2002, 80, 448–462. [Google Scholar] [CrossRef]
- Sun, S.; Li, E.; Zhao, G.; Tang, J.; Zuo, Q.; Cai, L.; Xu, C.; Sui, C.; Ou, Y.; Liu, C.; et al. Respiratory mucosal vaccination of peptide-poloxamine-DNA nanoparticles provides complete protection against lethal SARS-CoV-2 challenge. Biomaterials 2023, 292, 121907. [Google Scholar] [CrossRef]
- van Helden, S.F.; van Leeuwen, F.N.; Figdor, C.G. Human and murine model cell lines for dendritic cell biology evaluated. Immunology letters 2008, 117, 191–197. [Google Scholar] [CrossRef]
- Cockman, E.; Anderson, P.; Ivanov, P. TOP mRNPs: Molecular Mechanisms and Principles of Regulation. Biomolecules 2020, 10. [Google Scholar] [CrossRef]
- Yamashita, R.; Suzuki, Y.; Takeuchi, N.; Wakaguri, H.; Ueda, T.; Sugano, S.; Nakai, K. Comprehensive detection of human terminal oligo-pyrimidine (TOP) genes and analysis of their characteristics. Nucleic acids research 2008, 36, 3707–3715. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.P.; Overton, K.W.; Wang, C.L. Tuning gene expression with synthetic upstream open reading frames. Proceedings of the National Academy of Sciences of the United States of America 2013, 110, 11284–11289. [Google Scholar] [CrossRef] [PubMed]
- Tusup, M.; Kundig, T.; Pascolo, S. An eIF4G-recruiting aptamer increases the functionality of in vitro transcribed mRNA. EPH - International Journal of Medical and Health Science 2018, 4. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. Genetics 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Ricci, E.P.; Limousin, T.; Soto-Rifo, R.; Rubilar, P.S.; Decimo, D.; Ohlmann, T. miRNA repression of translation in vitro takes place during 43S ribosomal scanning. Nucleic acids research 2013, 41, 586–598. [Google Scholar] [CrossRef]
- Lai, W.C.; Zhu, M.; Belinite, M.; Ballard, G.; Mathews, D.H.; Ermolenko, D.N. Intrinsically Unstructured Sequences in the mRNA 3′ UTR Reduce the Ability of Poly(A) Tail to Enhance Translation. Journal of molecular biology 2022, 434, 167877. [Google Scholar] [CrossRef]
- McCaffrey, P. Artificial Intelligence for Vaccine Design. Methods in molecular biology (Clifton, N.J.) 2022, 2412, 3–13. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).