2. Materials and Methods
2.1. BioGRID
The quantitative SAINT analysis was used to identify SARS-CoV-2 viral-host proximity interactions in human or model system cells [
11,
12,
13,
14,
15,
16,
17] and those with a Bayesian FDR =< 0.01 were high confidence. Scores are the sum of peptide counts from four mass spec runs with a higher score indicating a higher degree of connectivity between proteins.
STRING [
152,
153] (
https://string-db.org/) is a database of known and predicted PPIs. The curated interactions are direct (physical) and indirect (functional) associations. The interactions came from different sources (genomic context, high-throughput experiments, co-expression, previous knowledge, etc.) which are channeled into seven independent channels. In this paper, we established the PPI network according to the Version: 11.5 of the STRING database. We constructed PPI networks by mapping proteins to the STRING database with a confidence score of >0.9 (highest confidence) with the information from all seven sources.
Protein enrichment is to some extent based on prior knowledge, and the statistical enrichment of the annotated features may not be an intrinsic property of the input. We have used a selected set of protein by BioGRID as functional seeds. Using Cytoscape software, we visualized and analyzed PPI networks, which offer diverse plugins for multiple analyses. Cytoscape represents PPI networks as graphs with nodes illustrating proteins and edges depicting associated interactions.
2.2. CYTOSCAPE and Network Topology Analysis
Cytoscape [
154,
155] through Network Analyzer was used to analyze the topological parameters of networks. We examined network architecture for topological parameters such as clustering coefficient, centralization, density, network diameter, and so on. Our analysis included undirected edges for every network. We termed the number of connected neighbors of a node in a network as the degree of a node. P(k) is used to describe the distribution of node degrees, which counts the number of nodes with degree k where k=0, 1, 2, … We calculated the power law of distribution of node degrees, which is one of the most crucial network topological characteristics. The coefficient R-Squared value (R
2), also known as the coefficient of determination, gives the proportion of variability in the dataset. We also examined other network parameters, including the distribution of various topological features. We did calculation of Hub and Bottleneck nodes based on relevant topological parameters. By examining the PPI network, we found the top 7 hub nodes. These nodes had significantly higher degree values than the others and were primarily in two central modules that were closely connected and compact.
CentiScaPe - Centralities for undirected, directed and weighted networks. Centiscape [
156] computes specific centrality parameters describing the network topology. These parameters facilitate users in locating the most important nodes within a complex network. The computation of the plugin produces both numerical and graphical results, facilitating the identification of key nodes even in extensive networks. Integrating network topological quantification with other numerical node attributes can cause relevant node identification and functional classification, as well as the topological location of proteins in their specific cellular compartments.
2.3. Evaluation of the HUB-and-Spoke Model
Many properties of a scale-free network depend on the value of the degree exponent of the power-law, γ [
157]. Therefore, it is interesting to establish how the network properties vary with γ. The estimation of the expected maximum degree (also known as the natural cut-off) for a scale-free network, which represents the expected size of the largest hub, is based on the following formula [
158]:
where Kmax and Kmin are the expected maximum and minimum degree of a node, respectively. N is the system size, in terms of the number of nodes. Based on Eq. 1, when γ<2 (as in our case) the link acquisition rate of the largest hub is faster than the growth of the network in terms of the number of nodes it contains. In this scenario, the high-degree nodes are attractive. Here the dynamic is of the “winner takes all” type. This leads to a hub-and-spoke network of topology, where all nodes are within a short distance of each other. Our interactome has a gamma value of 1.81, which favors at least one large topological module (metabolic module). A topological module represents an area of the network densely packed with nodes and links wherever nodes have a larger tendency to be connected to the nodes of the same area instead of the nodes placed outside the zone itself.
2.4. Cluster Analysis
For the cluster analysis, we have used the K-Means Clustering method [
159]. K-Means Clustering is an Unsupervised Learning algorithm (centroid-based clustering algorithm) used by STRING to group the protein dataset into different functional clusters. Centroid-based algorithms are efficient, effective, simple and sensitive to initial conditions and outliers. This makes it useful in handling networks. Here, for K, which defines the number of pre-defined clusters, we have used the value of 10 after various manual attempts to search the most reliable clusters in terms of compactness, metabolic functionality, and p-value.
2.5. GO and KEGG Pathway Analyses
To better research and show the biological function of proteins, we performed GO analysis, which included biological process (BP), cellular component (CC) and molecular function (MF). When the P value was below 0.05, we considered the results had statistical significance.
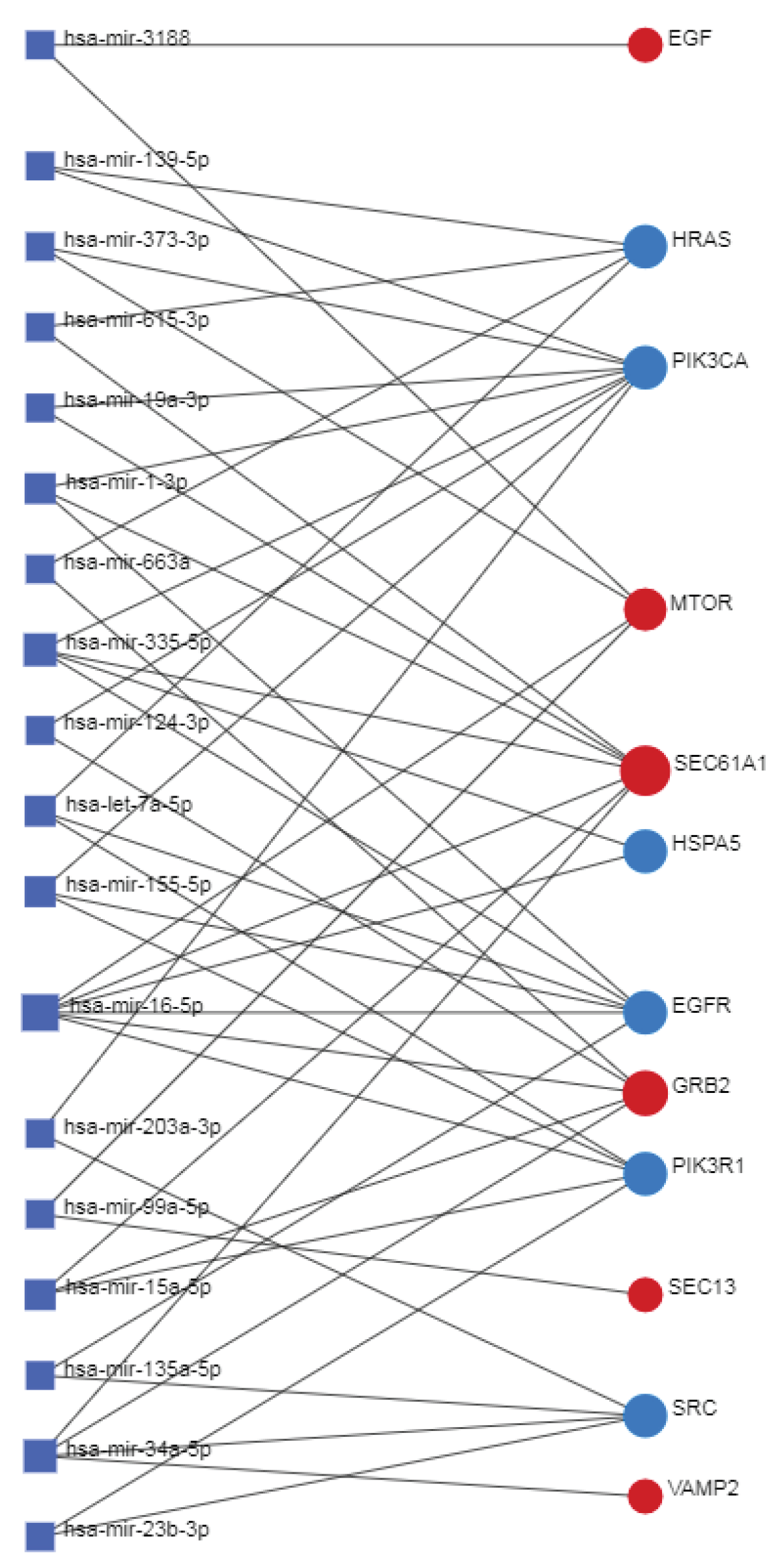
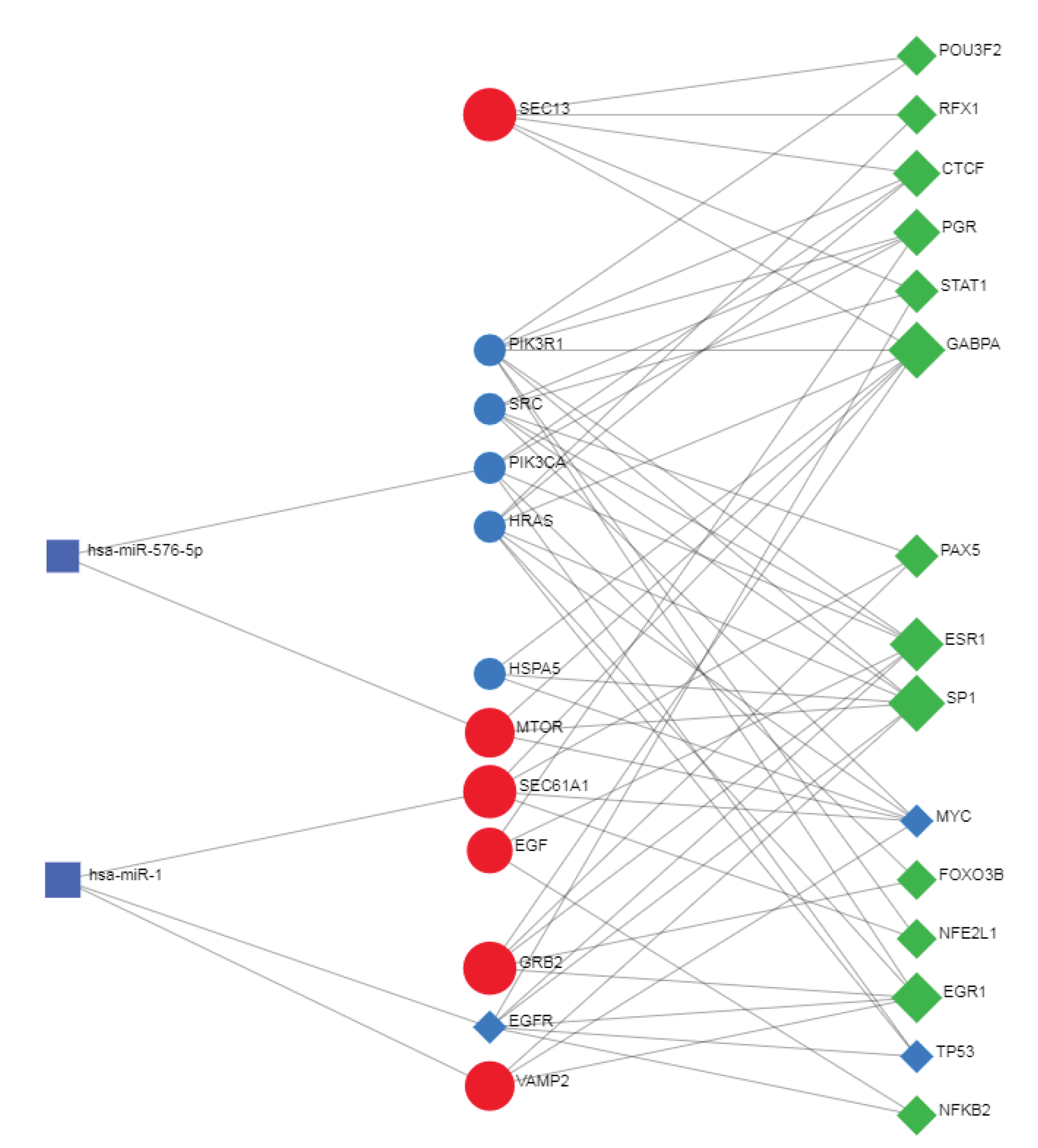
2.6. Network Analyst -- Comprehensive Gene Expression Profiling via Network Visual Analytics: TFs and miRNAs
The Network Analyst [
160,
161] interprets gene lists in a network. It enables the analysis of results present in the network via a powerful online network visualization framework. In protein-protein network analyses, the system also involves the existing relationships between genes, proteins, miRNAs, and human transcription factors, creating a co-regulatory network that is very useful for understanding the mutual relationships between these biological actors.
Databases: Gene-miRNA interactions - miRTarBase v8.0 Comprehensive experimentally validated miRNA-gene interaction data collected from miRTarBase.
TF-gene-interactions - ENCODE Transcription factor and gene target data derived from the ENCODE ChIP-seq data. The BETA Minus algorithm is used to selecting only peak intensity signals <500 and predicted regulatory potential scores <1 from the ENCODE ChIP-seq data for TF-gene-interactions.
Signaling - SIGNOR 2.0. The data is based on data from the SIGnaling Network Open Resource.
RegNetwork: Regulatory Network Repository of Transcription Factor and microRNA Mediated Gene Regulations. RegNetwork is a data repository of five-type transcriptional and posttranscriptional regulatory relationships for human and mouse:
tF → TF
TF → gene
tF → miRNA
miRNA → TF
miRNA → gene
This repository integrates curated regulations and the potential regulations inferred based on the transcription factor binding sites. Transcription factor (TF) and microRNA (miRNA) function at the transcriptional and posttranscriptional levels. It will be valuable for studying gene regulatory systems by integrating the prior knowledge of the transcriptional regulations between TF and target genes, and the posttranscriptional regulations between miRNA and targets. The conservation knowledge of the transcription factor binding site (TFBS) can also be implemented to couple the potential regulatory relationships between regulators and their targets. From RegNetwork, we can query and identify the combinatorial and synergic regulatory relationships among TFs, miRNAs, and genes [
162].
2.7. Protein Intrinsic Disorder and Secondary Structure Prediction
We have used two servers on line, Jpred 4 and IUPred2A. Jpred is a web server that takes protein sequences, and from these predicts the location of secondary structures using a neural network called Jnet. They show the prediction as a graph. IUPred2A [
163,
164] is a combined web interface that allows to identify disordered protein regions using IUPred2 and disordered binding regions using ANCHOR2. IUPred2A can identify disordered protein regions by analyzing their sequence, regardless of whether they are stable. Upon visually inspecting the graphic outputs of both predictive systems, we quickly identified disordered segments in most of the examined proteins, whether viral or human. These results were not displayed because they required a large space.
2.8. SARS2-HUMAN Proteome Interaction Database (SHPID)
We have collected in a single database all the files made available online by BioGRID, containing all the curated physical interactions of the 31 SARS-CoV-2 proteins gained through experiments in human cellular systems with viral baits, followed by purification and characterization with mass spectrometry. These Data are available as a zip file containing multiple zip-files (32 zip-files) each comprising Interactions and Post-Translational Modifications for each single SARS-CoV-2 protein for 33,823 interactions (as June 2023). The database therefore contains the set of all possible real interactions existing between the SARS-CoV-2 proteome with all the proteins of the human proteome. We highlight that not all interactions are real, but some could derive from artifacts of the method, such as non-biological interactions, only because of the random encounter between proteins in the system used. An encounter that would never have happened in the reality of an infection. However, the interactions derive from BioGRID where all, even those with the lowest score, have a significant statistic with an F.D.R. =< 0.01. This allows us to identify as many significant comparisons as possible while maintaining a low false positive rate, i.e., the probability of a false positive is less than 1%, so only 338 interactions among all are truly null. This database is the comprehensive repository of all interactions acknowledged biologically possible between the virus and its human host. The database also contains interactions between individual viral proteins, where known. As part of database search actions, you can ask who interacts with whom, with queries that use single human or viral proteins. The search can include multiple sets of proteins.
2.9. Highlighting the Nodes of a STRING Network Involved in the Same Biological Process (GO)
STRING makes visible all the nodes involved in the same biological process evidenced through its mapped databases onto the proteins (GO, KEGG, REACTOME, and so on) by activating the process itself with a click of the cursor on the process line. Activation means that all nodes involved in the same metabolic process stain similarly. Nodes involved in multiple processes are colored multiple times. This tool is very useful when one wants to analyze the involvement of multiple nodes in many metabolic processes visually, distinguishing the effect of different processes between nodes and identifying which nodes represent the crossing points. If individual nodes do not show any coloration under the effect of clicking, this identifies certain components of a path, or group, that a specific activated process does not influence. The relationships that determine the coloring of the nodes depend on the knowledge base that STRING organizes for a specific network by extracting data and information from the scientific literature in PubMed.
2.10. Comparison between GO Pairs in Enriched Networks
In modeled networks, STRING uses two parameters to analytically define the enriched biological terms. Strength is the measure of how large an enrichment is, expressed as Log10 [Log10 (observed/expected)], while False Discovery Rate (fdr) is the measure of the statistical significance of an enrichment given as a p-value after the Beniamini-procedure Hochberq. The higher the Strength value, the greater the biological effect due to genetic enrichment, indirectly indicating increased gene expression, while the smaller the p value, the greater the certainty that event will occur. Since STRING characterizes biological functions as pairs in which strength and fdr often show very different numerical values from each other, we use the product P [P = strength x -log10 p-value] to get a quantitative evaluation. This product will be greater when “Strength” has a very high value and p has a slight value [the most favorable situation for evaluating an effect positively is that represented by the extremes of their numerical values, very high and slight, respectively]. This facilitates comparisons and evaluation of pairs. Two pairs, one characterized by S = 0.35 and fdr = 1.0e-11, and another characterized by S = 1.9 and fdr = 1.0e-6, could lead one to think that the first is statistically more significant. If we analyze the P value, we have 3.85 and 11.4, respectively. This tells us that the increase in gene expression in the second case is functionally prevalent. The higher the value of the product, the more reliable the result of one pair will be over another. We consider that strength = 1 means a 10-fold genetic enrichment. However, it is important to remember that all fdr values reported by STRING in its biological functionality characterizations (GO, KEGG, etc.) are always significant and never greater than 0.05.
3. Results
3.1. Source of the Data
Fundamental experimental data supporting the role of SARS-CoV-2 in human infection are accumulating. BioGRID, one of the most important biomedical interaction repository, compiled comprehensive datasets of all physical interactions between the proteins of SARS-CoV-2 and the human proteome through the BioGRID COVID-19 Coronavirus Curation Project [
8,
10]. Curators selected interaction data caming from purification processes where researchers used physical methods such as Affinity Capture-MS and Proximity Label-MS. Interactions and their molecular interactors were classified into various levels of significance. With the protein ORF7b (P0DTD8 - NS7B_SARS2, UniProt), BioGRID classified 1,708 unique curated physical interactors [
11,
12,
13,
14,
15,
16,
17], involved in 2,753 interactions (accessed in July 2023). They are unique in being non-redundant and having high confidence interactions at high throughput, associated with high score values of statistical filtering, as determined by using SAINT (Significance Analysis of INTeractome) express version 3.6.0 [
11,
12,
13,
14,
15,
16,
17].
3.2. The Representation of ORF7b Data Using Interactomes
Figure 1 shows the circular network of human ORF7b-interacting proteins calculated by BioGRID. Since not all physical interactions flow into a real biological function, the concentric representation of the nodes shows different levels of reliability. Therefore, we used the densest layers as functional seeds. The nodes selected in this study have proven physical interaction through at least two different physical methods. The interaction should be non-redundant and high-throughput with optimal statistical significance between BioGRID levels 6 and 4. These options allowed us to select nodes with curated unique interactions.
In
Figure S1 (Supplements), we show an ARBOR representation of the network calculated by BioGRID with a minimum evidence value of 4, which illustrates the level/association relationships very well. An interactome shows the one-to-one mapping of all interactions, which turns the interactome into an information system [
18]. The goal is to decode the functional information of this biological map, the macroscopic properties of which are unpredictable and emergent properties of the system [
19,
20]. Its inherent complexity makes it difficult, if not impossible, to decode individual hidden molecular information. The datasets curated by BioGRID for each SARS-CoV-2 protein represent a suitable starting material. The list of 75 ORF7b interactors with significant levels ranging from 6 to 4 is available in
Table S1. Through the STRING platform [
21] we calculated the corresponding interactome (
Figure S2 in Supplements) with a score of 0.9 and with all 7 data source channels active, to gain as much information as possible. But the graph shows 54 proteins (72%) unconnected. So, we added 500 first order proteins to enrich the interactome and increase the functional relationships (
Figure S3 in Supplements). In this new graph, we also had to eliminate some parental proteins that were still disconnected, leaving 51 final parental proteins that were the basis of our enriched interactome. Network pruning helps eliminate artifacts due to noisy information [
22] while enrichment helps amplify those biological processes that are difficult to define because of their poor representation.
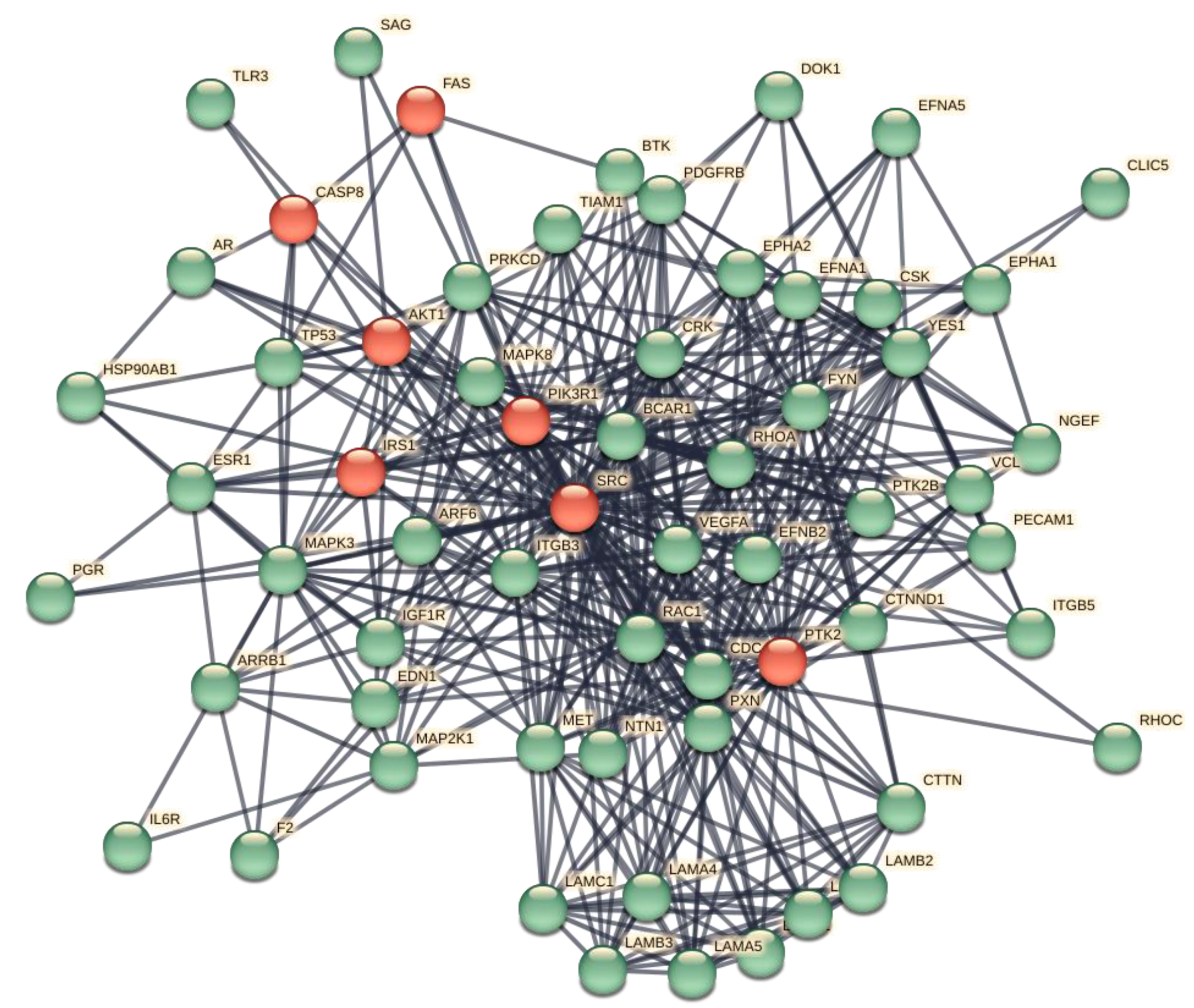
Figure 2 shows the interactome got after pruning and enrichment. The interactome now appears compact, with all nodes connected.
Typically, proteins that share similar functional information should appear as a compact set of nodes and edges (sub-graphs) performing one or more macroscopic functions. Subgraphs contain molecular partners that have relational links and perform similar functional activities. Analyzes of metabolic processes with Gene Ontology or KEGG allow us to evaluate the increase in functional annotations.
Many and rather compact peripheral modules with a large and very compact central module characterise this interactome. The peripheral modules suggest functional protein complexes. For example, the module at the top of the figure is very rich in ribosomal subunits and, very close to it, many proteins belonging to the translocon complex can be identified. While the complex on the right is rich in ATPase subunits characteristic of the proton-transporting vacuolar protein pump (V)-ATPase, required for acidification of secretory vesicles. These complexes represent the set of metabolic machinery necessary for normal cellular life. Surprisingly, the large central component shows nodes intra-connected, representing a significant fraction (37%) of the network's nodes. Components with these characteristics are called Giant Connected Components (GCC) [
23]. This type of component is often present in scale-free networks of which it is an important substructure. GCCs control the topological growth of the network, and so its evolution [
24]. Its capacity to aggregate new nodes and functions makes it a very compact system with a notable increase in the interaction turnover rate of new proteins [
24].
We can find a demonstration of this compactness in
Table S2 and Figure S4 (in Supplements). The figure shows the distribution graph of the mean shortest paths as a function of the degree of the single nodes. The 30 nodes with the highest ranks, i.e., with the greatest connectivity in the network, are those with the lowest average shortest path-length. These nodes are all concentrated in the GCC. Thus, this network has a "giant component", where almost every node is easily reachable from almost every other node in GCC, through a dense net of interactions. New nodes will massively join the GCC in a non-linear and unpredictable way to create biological functions, as GCC is a set of functionally very attractive metabolic nodes. This helps create the set of functions of this metabolic module [
24]. Typically, as the network grows, the giant component will continue to incorporate a significant fraction of incoming nodes. This means that we should find the main and crucial functional activities integrated into this subgraph.
3.3. Principal Characteristics of the Interactome
We transferred the interactome to Cytoscape [
25] and analyzed it with the help of CentiScaPe (v2.2), Analyze Network [
26], and STRING-app [
27,
28], which generated a Table of Nodes containing various columns with the quantitative values of many topological and functional parameters. This allowed the evaluation of characteristic topological and functional features for each node of the interactome.
The value of parameters in
Table 1 tells us we are considering a network made up of many independent and compact peripheral modules, which exchange relationships with fewer connections between them, albeit essential. The large diameter, network heterogeneity, and low-density support this view [
29]. The diameter also suggests components quite distant from the central module. While the shortest average path length, which gives the distance between two connected nodes, is a metabolic advantage because small average lengths minimize transition rates between metabolic states in response to external stress. The clustering coefficient also supports this topology. It is a basic index for local density in a network and is a measure of the degree to which nodes in a graph group together. It takes values 0 ≤ C ≤ 1, thus a value of 0.549 shows a tendency to form clusters, where each node shows an average of 16.817 neighbors. This coefficient of aggregation, according to Barabasi [
30], decreases with the increase in nodes.
| Summary Statistics of network* |
Notes |
| Number of nodes |
551 |
|
| Number of edges |
4648 |
** |
| Avg. Number of neighbors |
16,871 |
Average connectivity of the nodes
|
| Network diameter |
9 |
|
| Characteristic path length |
3.666 |
|
| Clustering coefficient |
0.549 |
0 ≤ C ≤ 1 |
| Network density |
0.031 |
|
| Network heterogeneity |
1.057 |
Tendency to contain hub nodes
|
| Network centralization |
0.259 |
The extent to which certain nodes are far more central than others
|
| Connected Component |
1 |
*** |
The
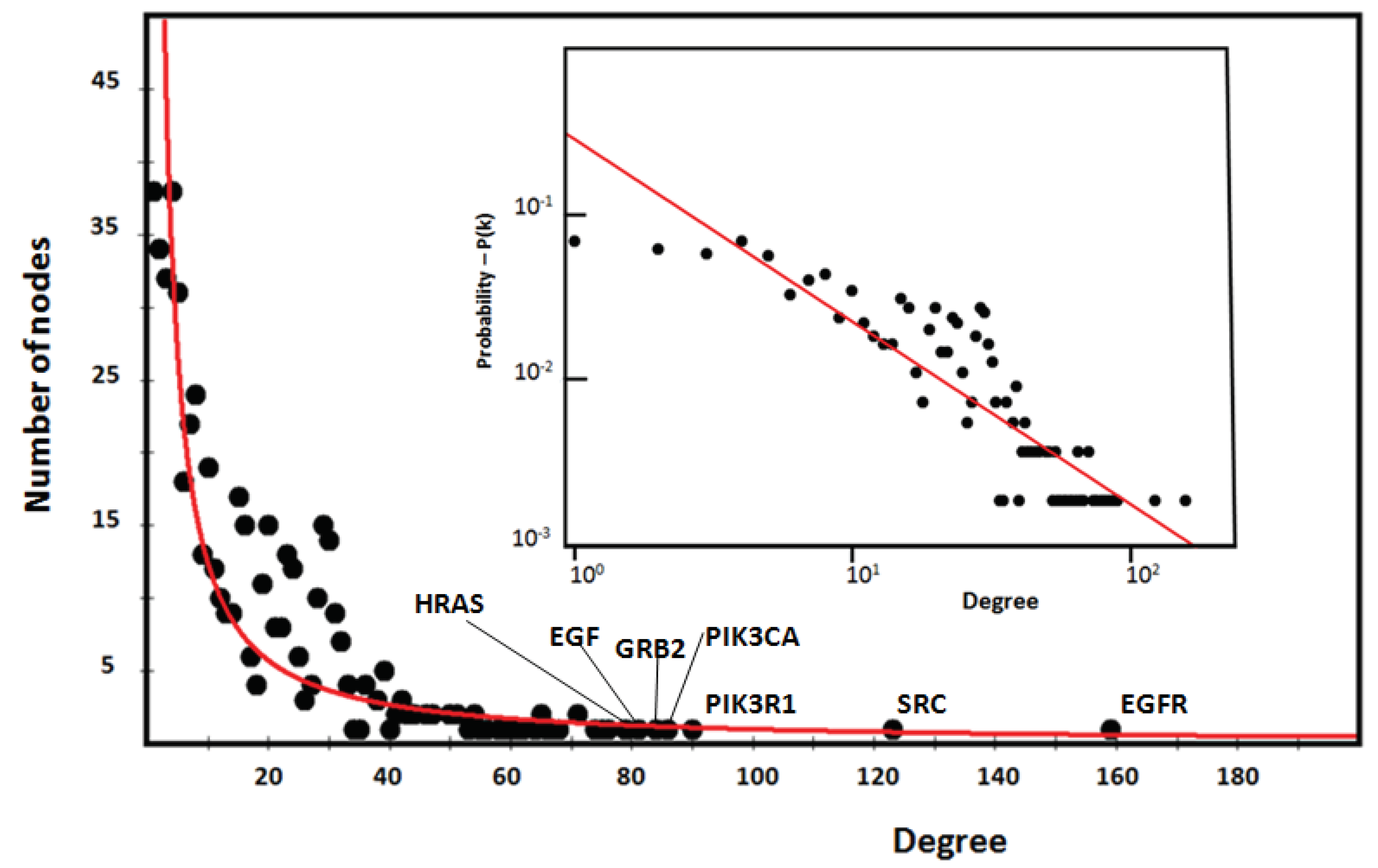
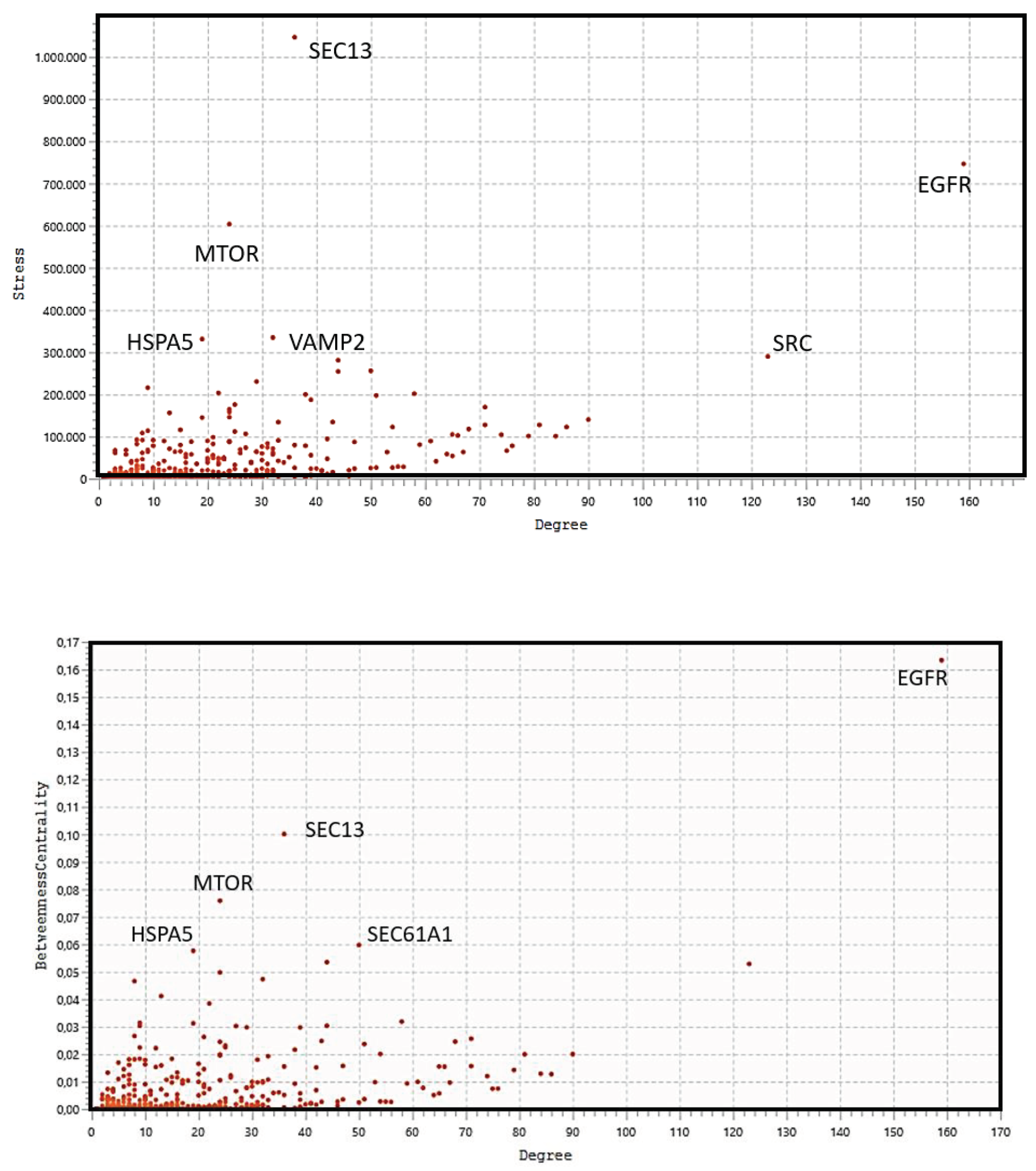
Figure 3 shows the characteristic power distribution of nodes of a scale-free network, where the vast majority of nodes have very few connections, and only few (HUBs) have a very large number of connections [
31]. This distribution is a defining characteristic of the biological network regardless of the experimental approach [
32] and is important in understanding the system's behavior. The power law exponent highlights a configuration for scale-free networks that minimize the number of nodes needed to control the entire system [
33,
34]. In the figure, we highlight the seven HUB nodes (EGFR, SRC, PIK3R1, PIK3CA, GRB2, and HRAS), which have superior ranks compared to all others, also remembering that the GCC includes the top 30 nodes with the highest ranks. Hub nodes model the architecture of metabolic modules and EGFR, which serves multiple critical functional roles in the cell, is the highest degree interactomic hub node also because of its exceptional capacity for PTMs (see
Figure S5).
We need alternative tests to prove the accuracy of our observations and hypotheses and to decode the information due to the actual functional activities in which ORF7b2 is involved. The following tables will show the most significant contents of some important functional categories. To evaluate the importance of each functional property, we will use the p-value as the evaluation criterion [
5] for the main significant processes. STRING calculated the tables with the methods and techniques of GO analysis.
3.4. Quantitative Evaluation of the Biological Functionalities in the Interactome
Table 2 shows the overall picture of the many functional activities performed by the entire network. Over 10,000 significant PubMed publications were used to provide coherent information on the 5,057 functional terms. STRING calculated the entire interactome using this knowledge base. This assures us that the functional relationships taken into consideration are very robust and that the pruning operation reflected real knowledge gaps in the considered node properties. The spectrum of biological activities induced by ORF7b2 appears remarkably broad in 15 categories and, therefore, both difficult to define and to study thoroughly. We have evaluated and selected the functional activities from time to time, as each of the 5,057 terms reported in
Table 2 has a statistical value (p-value) that is always less than 0.05, ensuring their significance. In this study, we will try to give a comprehensive view of the metabolic and molecular activity induced by ORF7b. Future studies will try to go into more detail.
Table 3 shows the most significant biological functions (GO-Biological Processes) among the 1,690 related to the human proteome following the action of ORF7b. The principal activities involve the control of intracellular transport, also by vesicles, and the control of their localization in the cell. The set of cellular processes includes the transportation, binding, and holding of a protein complex or organelle in a specific position. A transporter or group of transporters facilitates the directed movement of molecules or cellular complexes into or out of a cell, or between cells, to effect transmembrane, microtubule-based, or vesicle-mediated transport. A significant value ranging from a p-value of 1.0e-77 down to 0.05 marks all 1,690 activities. Enzymes and signaling pathway receptors also appear to be possible prime targets, also considering the large number of human proteins involved. In particular, the series of molecular signals started with an extracellular ligand binding to a receptor with tyrosine kinase activity on the surface of the target cell and ending with regulating a downstream cellular process. The statistical significance of these biological actions is very high, as is the number of proteins involved. However, the table shows a comprehensive picture of 1,650 functional activities that belong to both the virus and the cell in performing their respective strategies of attack or defense. A part of these activities also refers to the basal metabolic activities for the maintenance of normal vital functions (housekeeping functions). As we will see later, it will be possible to extract the specific activities of the virus.
The
Table 4 depicts the location in the cell where the most statistically significant functional activities (as presented in
Table 3) occur. Many cell membranes, cytoplasm, as well as protein complexes, are metabolically involved. Of particular interest is the significant activity performed by the SNARE complex, specifically involved in driving vesicles and endosomes towards the correct cellular target, also providing for the correct docking. SNARE proteins (
SNAp
REceptor, i.e., Soluble
N-ethylmaleimide-Sensitive Factor Attachment Proteins) are a family of cytosolic proteins involved in vesicular fusion with the target membrane during intracellular transport and exocytosis [
35]. SNAPs interact with proteins of the SNARE complex during the recycling of the fusion complex components [
36]. We know that interference with the function of SNAP proteins is associated with many pathological processes, such as colorectal cancer [
37], epilepsy [
38] or Huntington's disease [
39]. However, it is the post-translational process by which a PTM protein (a proteoform) trans-locates from the ER to its final destination, which drives function. This process also includes tethering and docking steps that prepare vesicles for fusion.
Table 5 (Reactome) shows the most statistically significant molecular mechanisms in which ORF7b might involve the human proteome. It contains biomolecules that perform precise metabolic and signaling activities and their relationships organized into biological pathways. Beyond the various interferences on important metabolic pathways, it is interesting to note the metabolic functions shown, such as, Nervous system development, Immune System, Infectious disease, Hemostasis, Innate Immune System, Platelet activation, Insulin receptor Signaling, Viral mRNA Translation and Cell-Cell communications. Although they are normal vital metabolic functions with high statistical significance, the parallelism with the known clinical effects of COVID-19 on the human organism [
40,
41] should not be overlooked, which is surprising.
The spectrum of possible viral interference also might involve intracellular transport mechanisms and cell-cell communications. Many of these "actions" have a deep impact on human biology and inter-organ signaling, according to recent research on the effects of COVID-19 on the human organism [
42,
43]. In particular, we relate the most significant one to signaling by receptor tyrosine kinases (RTKs), a family of proteins that act as cell surface receptors for various factors, such as cytokines and hormones. These receptors control many cellular processes but have also a crucial role in the development and progression of many types of cancer [
44,
45]. It is also interesting to highlight the high significance in this interactome of some activities, such as "Cell surface interactions on the vascular wall", "Platelet activation”, “Insulin receptor recycling", "Viral mRNA translation", "Cell-cell communication".
By using proteins directly involved with ORF7b, we extracted relevant activities in this interactome selectively from the human proteome. The symptoms in COVID patients, including thrombophilic alterations [
46], hyperglycemia [
47], and systemic spread of infected cells [
48], may not be independent, as their underlying mechanisms, as found in Reactome, all appear to have the involvement of ORF7b, which may be the underlying cause.
The number of human tissues and organs that are potential targets of ORF7b is also staggering.
Table 6 shows these tissues/organs, which are important constituent of human body through many cell types.
These tissues/organs share many of the previously described metabolic activities to varying degrees. Therefore, even if not all, they are potential targets of the virus where it finds the optimal metabolic conditions for its replication [
49]. The need to expand the list of terms in this table arises from the need to show the many target tissues of the virus with a significant potential. It is amazing how a tiny protein like ORF7b could induce so wide effect. This also means that the protein appears to be an authoritative candidate for altering the molecular mechanisms that keep cells in contact with each other [
50,
51,
52]. Dysregulating these mechanisms might free the cells to spread without a programmed death [
53,
54].
This TABLE shows a long list of the various organs in the abdominal cavity, which are potential targets of the action of this protein, and validates the clinical observations that covid is a systemic disease. The high statistical values suggest the enormous potential of the strategy implemented by SARS-CoV-2 in hitting the human body. Some objectives are of particular interest. Nervous system (central and peripheral), human reproductive system (male and female), placenta and fetus, blood and hematopoietic system should alert us to the consequences encountered in the long-covid. Long-covid is showing symptoms that suggest the involvement of these specific organs and tissues as well.
An important index is also the high total number of proteins involved in each of the multiple functional activities represented in the tables previously reported. Considering the finite number of proteins in the interactome and the large number of them involved in many and different metabolic activities, this suggests that there is a high probability that single proteins may be involved in numerous functions. But all this also suggests that, in the event of a viral infection, a single human protein can perform many functional activities, some for the benefit of the cell and others for the benefit of the virus. KEGG pathways can infer higher-level functions and metabolic utilities of the human system from genomic and proteomic data. It groups genes and/or proteins into "pathways" as lists of genes/proteins taking part in the same metabolic process. Thus, KEGG is very useful for computational analyzes, including metabolic modeling and simulation according to systems biology, and translational research in disease development. KEGG's results show a wide range of activity. The breadth and diversity of the responses (195 pathways) and their statistical significance would require more space to highlight many of them. However, we have included the most probable in
Table 7. These pathways reflect precise connections with the functions reported in the previous tables, identifying and endorsing their metabolic role. We can only identify the most significantly represented functions, but we cannot at this stage establish a direct correlation to viral activity.
So far, we have examined the spectrum of functional/molecular activities present in an infected cell and, in particular, those involved by ORF7b. Once we have defined the principal functions, we need to highlight which single proteins favor the virus by "playing a double game".
3.5. Exploring the Physical Basis of Cytoskeletal Alterations Caused by ORF7b
The propagation of a virus to uninfected cells makes up a crucial phase in its life cycle, achieved through the liberation of novel viral particles from the infected cell. The ability of ORF7b to induce changes in the cytoskeleton that could promote the spread of infected cells is not coincidental. As we have previously discussed, these changes seem to derive from dysregulations induced at the cytoskeleton level. These results, however, suggest different biological events from those already known, not only a spread of viral particles after cell rupture but also a spread of entire infected cells to distant tissues, exactly like tumor metastases. Therefore, this aspect needs a greater attention. The key processes for modifications of the cell membrane, or that of cellular compartments, should pass through direct deformations caused by specific proteins that interact with the membrane [
165], or even through indirect deformation by the cytoskeletal structures [
166]. Therefore, the cytoskeleton is one of the key driving forces, with a close association to these events [
167].
Unfortunately, understanding the influence of these molecular processes on the physical structure of the membrane is still an unsolved challenge, despite a slight improvement in our understanding of the underlying physical basis. Until now, it has been difficult to quantify the forces present in living cells within these processes. However, we now have a first, albeit crude, quantitative understanding of force production and distribution at the molecular level using clathrin-mediated endocytosis as a model [
165,
166]. During endocytosis, the actin cytoskeleton generates forces that are transmitted to the plasma membrane through a multi-protein coat, leading to membrane deformation. Although the exact extent of these forces remains uncertain, we can highlight a phenomenon of accumulation and redistribution of force within the endocytic mechanism. This has led to the widespread belief that the EPNs and Hip1R proteins transmit the force generated by the assembly of the actin to the plasma membrane [
168,
169]. As both protein types also attach to clathrin and other coat proteins, it is plausible that the transmission of forces to the membrane might occur through multiple pathways [
170,
171].
However, we know which eukaryotic genes/proteins actively engage in these processes, serving as either components or regulators of the cytoskeleton, while an intricate interplay between lipids and proteins controls the membrane remodeling during intracellular trafficking [
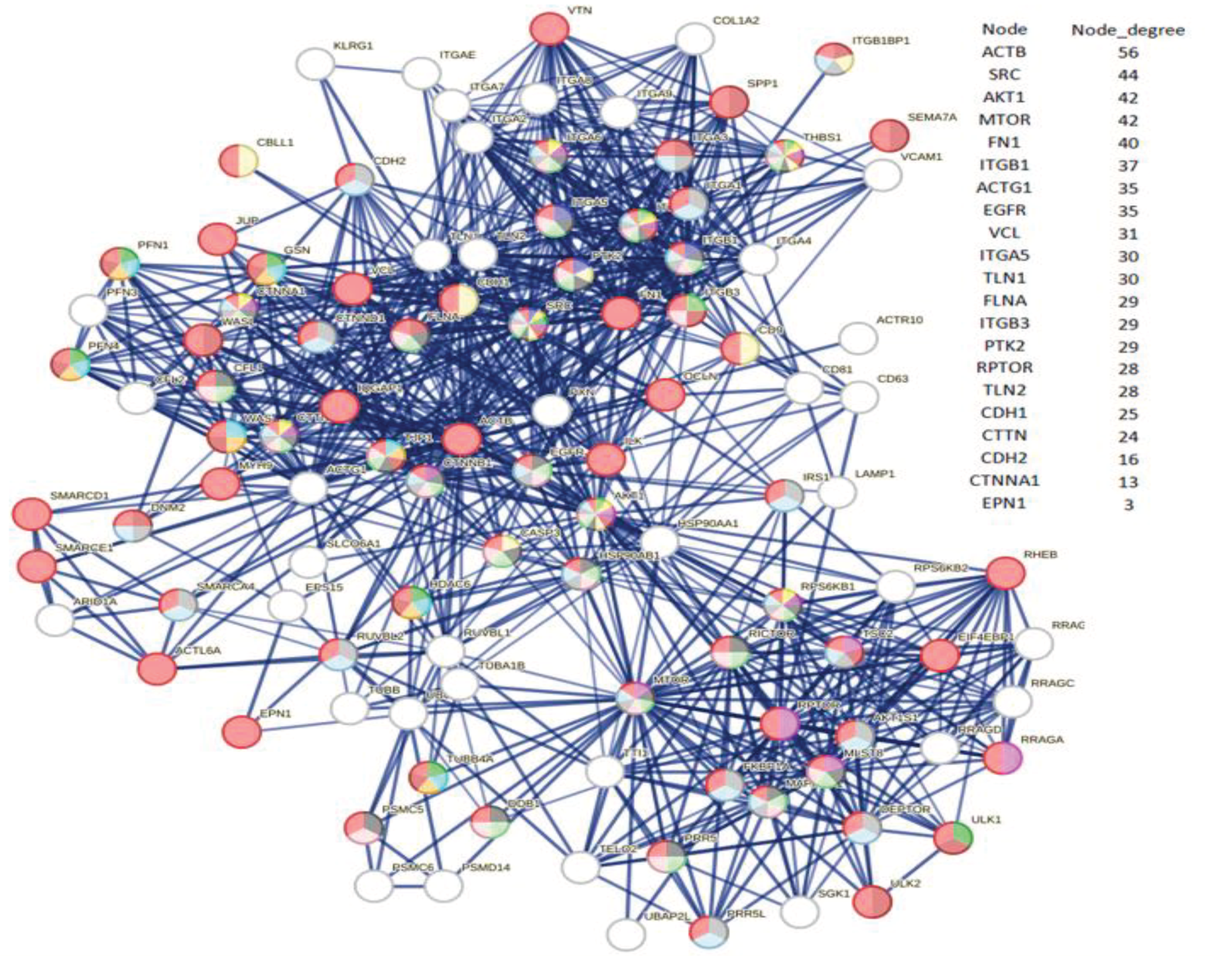
172]. Noteworthy examples include MTOR, CTNNA1 (alpha 1 catenin), CTTN (cortactin), ITGBs (integrins), CDH1, CDH2 (cadherins), ACTB (actin B), and EPNs (Epsin family). A check of the interactome in
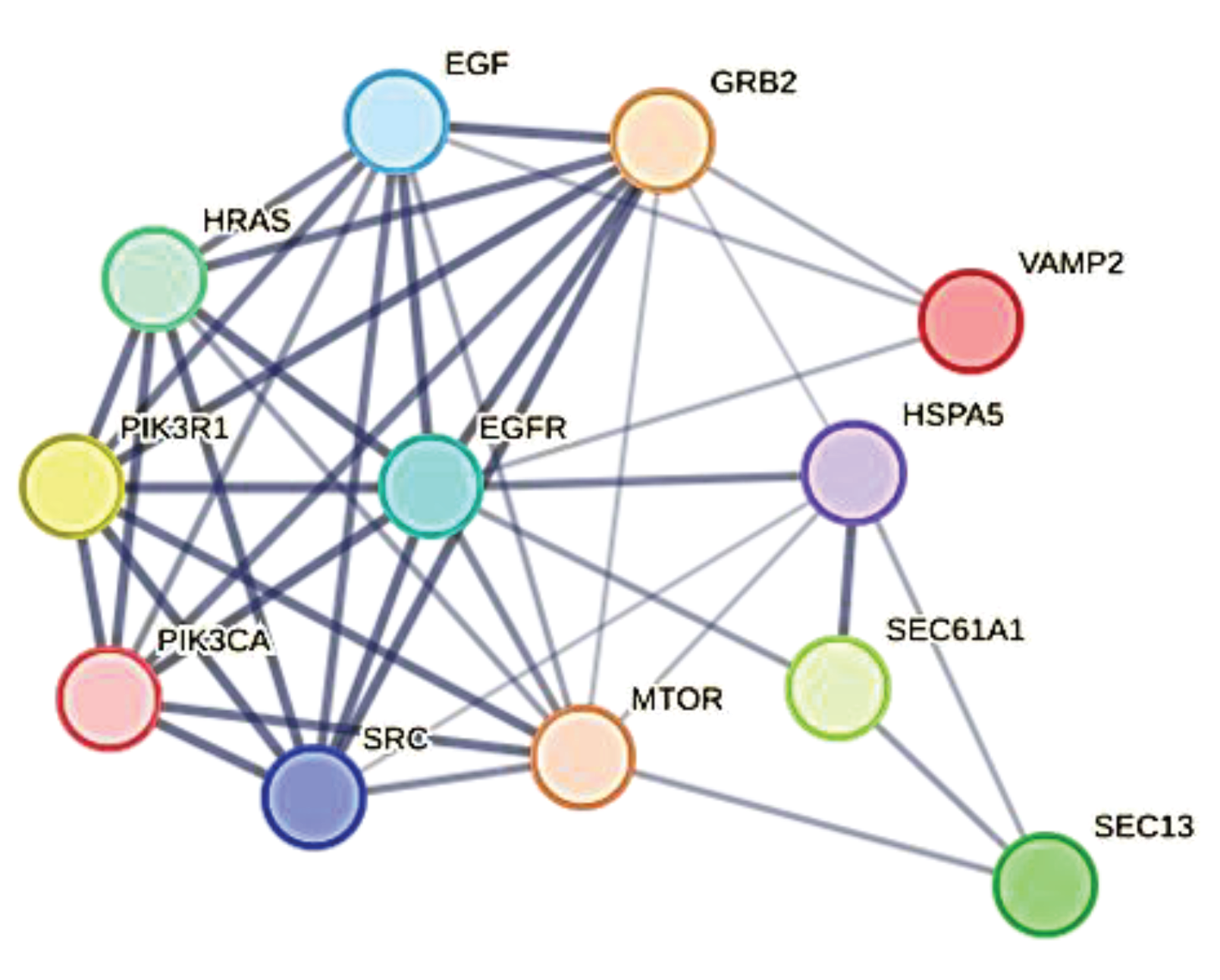
Figure 2 identifies all eight proteins and various members of their families (please also refer to the accompanying excel file for the comprehensive list and node degrees). This observation drew our attention to the intriguing possibility regarding the potential involvement of specific human proteins, in particular those associated with cytoskeletal modifications and negative regulation processes, in the mechanism of SARS-CoV-2 spread to non-infected cells and tissues. We used these proteins as seeds to tease out their functional relationships within the human proteome.
Figure 4 illustrates the specific and close relationships between them during their involvement in the processes that impact the organization of the cytoskeleton. Using a specific feature of STRING, the proteins involved in the same biological process were highlighted and colored (see Methods).
The network comprises all the human proteins involved in cytoskeleton dynamics. Since they are all reported in BioGRID as actively interacting, this suggests direct physical and/or functional associations. Among those of high rank, some, such as ACTB, are involved in a single dysregulated process (one color), others, such as MTOR, are involved in the management of multiple dysregulated processes (various colors). However, these interactions imply that SARS-CoV-2 exploits the host cell's proteins involved in processes regulated by CDH1, CDH2, EPN1, EPN2, CTNNA1, ITGB1, MTOR, ACTB, CTNNA1, and CTTN. This certainly affects cellular functions related to cell adhesion, signaling pathways, cytoskeletal organization, and programmed death through a Viral Hijacking of Cellular Machinery. But, these specific interactions also suggest potential roles for these cellular proteins in stages of the viral life cycle. In fact, their presence shows that these host proteins contribute to SARS-CoV-2 infection dynamics and pathogenesis, thus becoming appropriate therapeutic targets. However, further observations are important. Structural models of protein interfaces and the potential impact of post-translational modifications are crucial to understanding molecular mechanisms based on interactions because alteration of these characteristics might change protein-protein interactions and related biological functions. Many of the cytoskeletal proteins possess disordered structural domains and many phosphorylation sites. MTOR, serine/threonine protein kinase, in presence of RPTOR (Regulatory-associated protein of mTOR) and RICTOR (RICTR, Rapamycin-insensitive companion of mTOR) and through mTORC1 and 2 complexes, directly or indirectly controls the phosphorylation of at least 800 proteins and actin cytoskeleton is specifically MTOR sensitive [
173,
174,
175]. DEPTOR (DEP Domain Containing MTOR Interacting Protein) is a negative regulator of TOR signaling and of mTORC1 and 2 pathways, inhibiting activity of both complexes [
176,
177]. This leads to negative regulation of cell size, and negative regulation of protein kinase activity. MTOR, DEPTOR, RICTOR, and RPTOR are all part of the interactome and communicate extensively. Thus, the relationships between them validate the various dysregulations in
Figure 4 and
Table 8. A last consideration is that another viral protein also interacts with the cytoskeleton, it is the N protein, which plays various roles in the life cycle of the coronavirus [
178]. Here we want to underline that the N protein physically interacts with ACTB [
179], reconfiguring and manipulating the cytoskeleton as also happens for other viruses. This protein, as we will see in Table 10, also physically interacts with ORF7b. The N protein was mentioned because it is the SARS-CoV-2 protein that is involved in the formation of liquid droplets (see in "Discussion" for more details), a little discussed issue in the infection of this virus.
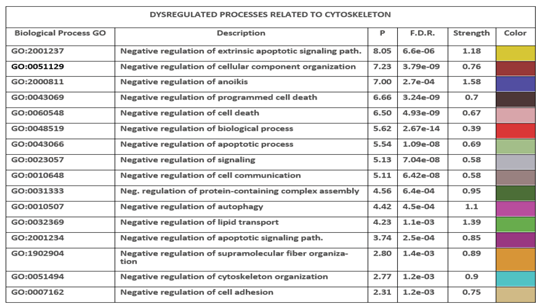
Table (right side) - the table shows the nodes with the highest degree. In the table there are also reported CDH2, CTNNA1, and EPN1 just to show all seeds. The number of colored segments of each protein node shows in which of the dysregulated processes shown in
Table 8 it is involved.
We can conclude that interaction of SARS-CoV-2 ORF7b protein with host cell proteins, especially those involved in cytoskeletal modifications, plays a role in the virus's ability to propagate infect cell to target distant tissues. Structural disarrangements or metabolic dysregulations induced at the cytoskeleton level impact the cell's ability to counteract viral infection, aiding in viral spread, or facilitating intracellular transport of viral components, so contributing to its long-distance diffusion.
3.6. Topological Analysis
When a virus infects a cell, viral proteins represent the attackers and seek vulnerabilities in the network. Vulnerabilities introduce uncertainties into the network as a loss of original metabolic performance, even by changing information flows. Examining the network topology allows us to study both vulnerability and functional uncertainty, and to seek any architectural or functional changes. Crossing pathways between metabolic pathways or between signaling pathways are among the most vulnerable topologies, while hub-and-spoke topologies have the least uncertainty of destabilization. Therefore, topological data analysis is a powerful biological network analytic method [
55]. To extract meaningful information from interactomic data, it is essential to understand the correlation between topological parameters and the mechanisms of biological functions [
56]. Centrality metrics measure the importance of nodes by trying to quantify the idea that some nodes are more "important" than others.
We can roughly divide topology scoring metrics into two groups, the local one to evaluate individual nodes and the global one to evaluate the network. Global metrics include Betweenness, Bottleneck, Eccentricity, Closeness, Radiality, Stress and more. It is a useful methodological approach to increase the efficiency in selecting, characterizing and classifying crucial proteins as both hub and/or bottleneck proteins. In particular, bottlenecks are key link proteins, almost always not HUBs, but hard-to-discover essential proteins which control and regulate metabolic cross-overs. In fact, in regulatory networks, being intermediate (i.e., "bottleneck") is an indicator of functional essentiality, which is often much more significant than degree (i.e., of being a hub) for understanding the direction of an information flow.
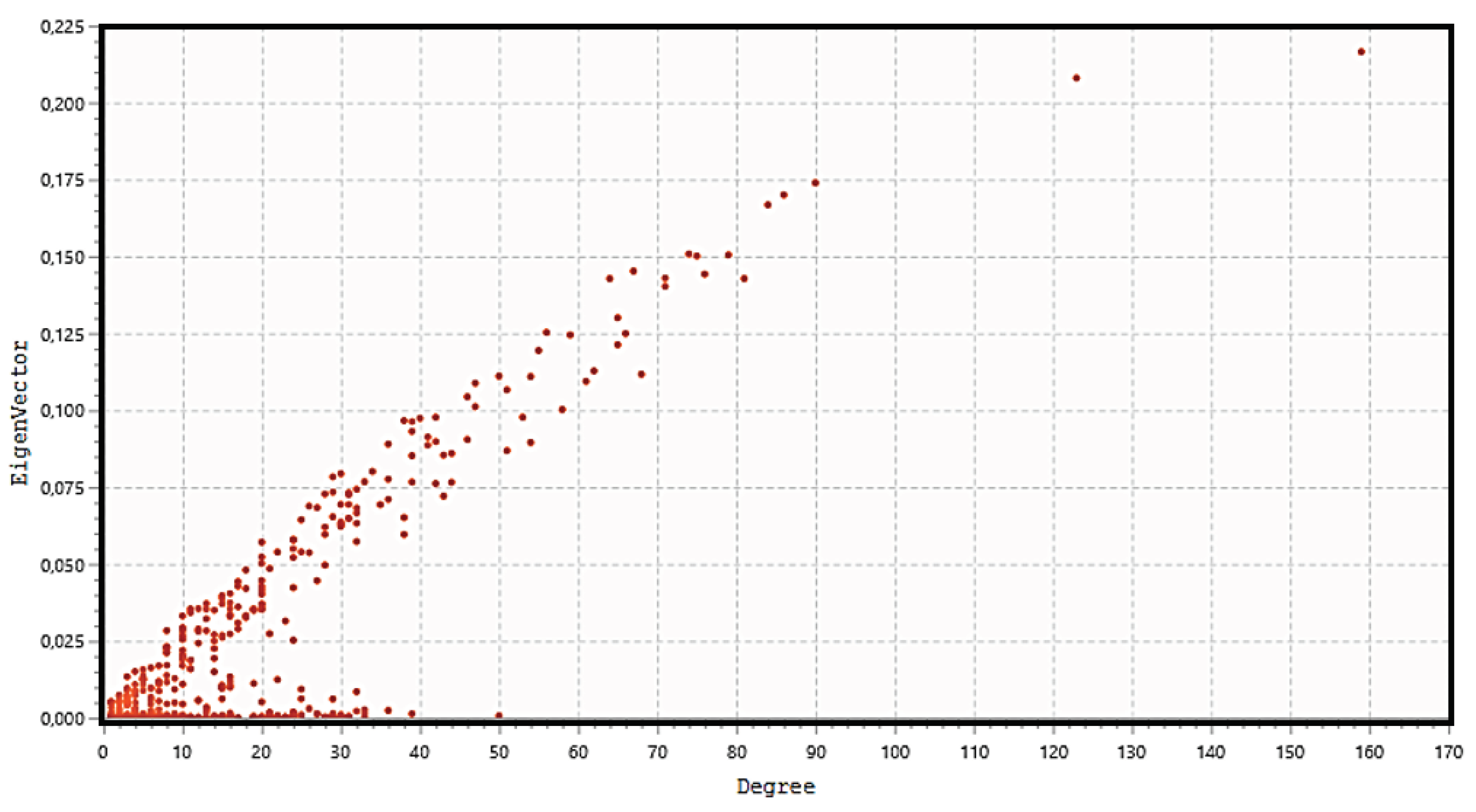
Eigenvector Centrality measures the transitive influence of nodes. Relationships originating from high-scoring nodes contribute more to a node's score than connections from low-scoring nodes. If a node has a high eigenvector score, it means that it is connected to multiple nodes that have high scores as well.
Figure 5 (top) shows the distribution analysis of the eigenvectors. The graph shows that the eight highest values have their degree value exactly matching that of the eight hub nodes previously selected, showing that all hub proteins also have the highest eigenvector scores. Stress is an index of node centrality. It represents the number of the shortest paths passing through a node. A high-stress node is a node traversed by a very large number of the shortest paths. In an interactomic network, it shows the relevance of a protein in keeping functionally communicating nodes together. We can consider such a protein as a "bottleneck" protein [
57,
58,
59]. The higher its value in the network, the more relevant the protein is in linking regulatory proteins of different pathways. However, because of the parametric significance of this index, it is sometimes possible that stress shows only a molecule involved in many cellular processes but not relevant for maintaining communication between other proteins [
60]. The
Figure 5 (middle) shows the stress distribution analysis where SEC13, EGFR, MTOR, HSPA5, VAMP2, and SRC are the major stress proteins. Betweenness [
56] is also an index of node centrality, similar to stress, but with more information. It is a measure to rank the relative importance of vertices or edges. It represents the total number of non-redundant shortest paths connecting a pair of nodes, a1 and a2, crossing the node a. The betweenness value of a node increases if it lies on a non-redundant shortest path between nodes a1 and a2. Therefore, a high Betweenness score characterizes a key node in maintaining connections and this type of nodes becomes the critical point that controls the communication between other distant nodes in the network. In biological terms, it characterizes the interactivity of a protein in an interactome, showing the protein's ability to link distant proteins. Thus, betweenness is a measure of how important the node is to the flow of information through a network. This feature of the node in a protein signaling network may also show the relevance of the protein to act as a bottleneck. It acts as a junction connecting metabolic pathways that can hold the communicating proteins of different pathways together. The higher the value, the greater is the relevance of the protein as a bottleneck molecule. The interdependence of a protein effectively shows the ability of this protein to link distant proteins. In reporting modules, intermediate relationships are crucial to maintain the functionality and consistency of the reporting mechanisms.
The analysis in
Figure 5 (bottom) confirms EGFR, SEC13, MTOR, HSPA5 as "bottleneck" proteins, also showing a new protein, SEC61A1. In the stress distribution, the SEC61A1 value was very close to that of VAMP2, while now is the VAMP2 value close to that of SEC61A1. Therefore, we can consider both proteins as bottlenecks.
Eigenvector, Stress, and Betweenness Centrality distributions were used in a multi-parametric approach to validate the 8 hub proteins and define the role of some proteins as bottlenecks. Among proteins selected as the most ranked bottlenecks (EGFR, HSPA5, MTOR, SEC13, SEC61A1, SRC, and VAMP2), EGFR and SRC show a dual role, both as a hub and as a bottleneck. Putting it all together, we have EGFR and SRC which are mixed (HUB/Bottleneck) proteins, HSPA5, MTOR, SEC13, SEC61A1, and VAMP2 which are pure bottleneck proteins, and PIK3R1, PIK3CA, GRB2, and HRAS which are pure hub proteins. These differences allow these proteins to be defined in three classes of molecular markers. In an eukaryotic protein interaction network, a node rarely represents the lone native protein because of alternative splicing [
61] and proteoforms [
62]. This may be a problem because in all databases (including STRING) it is customary to collapse all the dofferent functions of its isoforms and proteoforms onto the native protein, attributing to it a greater load of functions that it does not possess. In the interactome calculation, this anomaly produces biased nodes with higher and unreal connectivity.
Researchers have identified three different types of hubs in tissue-specific protein-protein interaction networks: few tissue-specific hubs, many tissue-preferred hubs that are formed by highly connected proteins, and housekeeping hubs that are involved in normal metabolic management [
63]. When we connect these features to their specific functional roles within different tissues, they exhibit distinct functional differences that are influenced by the structure/function relationships.
Disordered regions significantly enrich pure hub and hub/bottleneck proteins among the three previous classes, and as a result, these proteins harbor a significant number of predicted binding sites [
64]. They are also rich in splice variants, have longer peptide chains, and host a significant number of domains. This successful structural versatility drives their high propensity for interactions [
62]. Because they are involved in essential functions such as phosphorylation and mRNA slicing processes, they get tangled in multiple intracellular functional pathways. Pure bottleneck proteins are typically extracellular proteins that are connected to pathological conditions, such as cancer, and play a role in cell-to-cell signaling pathways. Defining the actual functional role of a node is challenging because of the convergence of multiple functions with varying spatio-temporal characteristics. Many researchers still use static and deterministic approaches to select their experimental design, which leads to these limitations.
The topological role of network hubs depends also on the exponent value of the power law [
65]. A value of <2 for the degree exponent b (see
Figure 3), however, very close to 2, suggests a hub-and-spoke architectural model. The hub-hub network of the entire interactome fits a hub-and-spoke model, as Perera [
66] and Barabasi [
67,
68] suggest. The largest hub (EGFR, 159 nodes) acts as a central coordinator and connects to a significant portion of nodes, which is shown in
Figure 3 and
Figure 5. These structures act as a backbone connecting different metabolic modules. In this topological context, we should also identify the top-hubs as significant centers of control over the entire network. This view is also in agreement with the topological parameters calculated by the Cytoscape Network Analyzer.
The
Figure 6 also shows the relationships and the particular topology involving both HUBs and bottlenecks nodes [
69].
Figure S6 (in the Supplements) shows how EGFR organizes in a topologically similar manner, even under normal conditions. Relationships between the HUB nodes are strong, while those with the bottleneck nodes are less intense, as the figure shows. All these significant nodes play a collective role in maintaining the stability of the hub-spoke system, albeit with varying functions and methods [
70]. Each of them controls many and different biological processes [
71]. The question remains: which node, regardless of its degree, is involved in the greatest number of functional processes? The question is not far-fetched. Because of the many metabolic crossroads, greater connectivity may not correspond to greater functional involvement [
72]. When designing a drug, it is important to have this information.
Table 9, while surprising for the very high number of functional involvements, shows how a HUB node is not always the main controller of the metabolic landscape. MTOR (degree = 24) and HSPA5 (degree = 19), although with lower connectivity, are involved in a very significant number of processes. The distribution of nodes and biological functions on the hub-and-spoke system, coupled with the ORF7b-induced interactome's complexity, handles this outcome. How functionally significant are the processes they regulate, would be the next inquiry. The answer would require a large analysis not covered by this study. Certainly, these same nodes, depending on their level of genomic expression, can both up-regulate and down-regulate a biological process [
73,
74,
75]. Down-regulated processes, or "negative biological processes" according to GO, are important to highlight because of their higher probability of resulting from viral strategy [
76]. Here, as we will see below, statistical significance is no longer the only parameter to follow.
3.7. The Functional Effects Depend Not Only on ORF7b but Also on the Integrated Action of Several Viral Proteins
The virus shows extraordinary strategic potential. Our previous results indirectly showed the specific impact of its proteins on crucial metabolic processes. About 200 symptoms of patients [
77] generated various hypotheses based on clinical impression found to be associated with long-covid. All this shows how broad and diversified the systemic action of the virus is. Thus, part of the broad spectrum of metabolic activities found in this interactome might be associated with the multitude of clinically observed symptoms. However, we should not think that the ORF7b protein alone is capable of so much. The proteome yields biological functions via target proteins, which result from specific one-to-one interactions between viral and human proteins. Other viral proteins could target human proteins present in metabolic modules where ORF7b also operates. The ORF7b circular interactome (
Figure 1) displays other viral proteins, ORF3a, and M, which may show their ability to target human proteins in the same metabolic modules as ORF7b. As of July 2023, we have organized a database called SHPID, which contains BioGRID interactions. In this database, we have collected 33,823 interactions between SARS-CoV-2 and human proteins. We analyzed the hub proteins highlighted in
Figure 3. The proteins EGFR, SRC, and PIK3R1 are the major HUB nodes of the ORF7b interactome with 159, 123 and 90 links, respectively. Although these proteins are involved in the ORF7b interactome,
Table 10 reveals that they also interact with other viral proteins.
The
Table 10 depicts how these high-degree human proteins are a common target for many viral proteins. Our analysis of the interactions between the thirty-one viral proteins and the human proteome, as reported by BioGRID, yielded this result. Even though viral proteins have co-evolved with their human host or other species, they seldom possess structurally detailed molecular interfaces for accurate and stable interaction. Only a few viral proteins exhibit strong interactions, akin to those observed in complexes. Most of the interactions have weak bonds, also because of the anisotropy of the contact areas [
79]. Viral proteins attempt to establish competition with normal binding proteins by mimicking interaction interfaces to the greatest extent possible, binding to target proteins with interaction constant values that typify weak processes. The interfaces mimicked by viral proteins compete through multiple and transient cellular interactions. They interact with hubs and bottlenecks in the human PPI network to control vital proteins in complexes and pathways. Proteins can overcome a structural difficulty by introducing an intrinsically disordered region (IDR) in the sequence, which can enhance the mimicry of contact surfaces. IDPs have IDR stretches that may be part of low affinity inter-molecular interactions [
80]. With the emergence of IDPs in eukaryotic proteomes [
81], the disorder becomes a crucial information for PPI evaluation.
Many of the interacting viral proteins in the
Table 10 show IDR (data not shown), thus, the probability of multi-targeting is high and this could explain the phenomenon (see Methods for details). After all, even the three human proteins analyzed have inherently disordered and highly mobile segments (data not shown). They are lipid-anchored proteins with the central body in the cytoplasm or outside the cell. Two long disordered and mobile tails are present in EGFR, which is found on several internal membranes (endosomes, ER, Golgi, nucleus) and on the surface. SRC also has long disordered and mobile tails and some mobile central segments and has multiple localizations, both on the surface and on intracellular organelles (endosomes, mitochondria, etc.). Finally, PIK3R1 too shows a long-disordered C term with many mobile intermediate segments and is on the cell surface. To this we should add that the disordered/mobile parts often show PTM sites. The presence of PTM sites expands the number of proteoforms for any single protein, increasing the probability of interacting with new molecular partners, establishing new functions.
A particular observation is that our database shows that ORF7b itself interacts with the viral N protein (see
Table 10). Among the various functional peculiarities of this protein, we find it is involved in the formation of liquid droplets [
178]. The liquid-liquid phase separation is considered the key mechanism for organizing macromolecules, such as proteins and nucleic acids, into membrane-free organelles [
184], and N protein can self-bind into spherical aggregates which can freely diffuse in the condensed phase with liquid-like behavior [
185,
186].
Although we had also examined other relevant human HUB nodes of the ORF7b interactome, such as PIK3CA, EGF, and HRAS, we did not find other direct targeting of viral proteins. Therefore, these seem nodes extracted specifically from the ORF7b functional enrichment and functionally connected with the other HUBs of this network. Thus, their presence in this interactome seems due to a specific functional requirement of ORF7b. After all, the human metabolic system responds intricately to the ORF7b protein, consistent with the multiple metabolic responses of multicellular eukaryotic systems. In particular cases, viral action may require the synergistic action of different viral proteins. Thus, to achieve its biological effect, the virus can also use complex and sequential interaction modes on a single protein. This analysis is in excellent agreement with the previous classification of hub and bottleneck proteins. Unfortunately, we currently do not know where, how, and when these interactions occur. Hence, our vision of a dynamic phenomenon is only static and somewhat unclear, which may also be spatio-temporally inappropriate or distorted in our reconstruction of it [
82]. Anyway, SARS-CoV-2 employs a known strategy of targeting the same human protein with multiple viral proteins [
83].
3.8. The Peculiar Case of GRB2, a Protein in the Service of ORF7b
GRB2 (Growth Factor Receptor Bound Protein 2 – UniProt: P62993) is a protein that according to BioGRID binds ORF7b, although with the low level 1. While, our observation within the BioGRID dataset reveals that this protein exclusively interacts with ORF7b. We excluded it from the seed proteins owing to its low significance, but found it recluted in the interactome. The enrichment suggests that this ORF7b interactor is essential for virus infection. It assumes the role of HUB with 84 connections and controls 233 biological processes (see
Table 9). GRB2 is an important protein that provides a critical link between the phosphorylated cell surface growth factor receptors (EGFR) and the PI3K-Akt signaling pathway. Both KEGG and Reactome Pathways reported its significant involvement in several signaling mechanisms (hsa04151, PI3K-Akt signaling pathway; HSA-1963640, GRB2 events in ERBB2 signaling; HSA-179812, GRB2 events in EGFR signaling; HSA-354194, GRB2:SOS linkage to MAPK signaling). Later on, we come to know that it often involves in various dysregulation processes that assist viral activity.
Table 9's proteins and GRB2's case show the sophisticated and diverse molecular strategy of SARS-CoV-2. The hubs listed in this table are proteins obtained through functional enrichment, but are not direct molecular interactors of ORF7b.
3.9. The Role of ORF7b
The diverse and sometimes contrasting metabolic properties of some of the interactome nodes are surprising. Among the 1,691 Biological Processes (GO) induced by ORF7b, there are 117 peculiar metabolic activities mentioned as negative activities (approximately 7%). Most of the HUB and bottlenecks proteins are also involved. According to AmiGO-2, the official web-based set of tools for searching and browsing the Gene Ontology database, negative activity means "any process that stops, prevents or reduces the frequency, rate or extent of metabolic functions". To identify which terms are most significant for these purposes, p-values alone cannot guide us. STRING measures the size of the enrichment effect using also the "Strength" score. The sole use of the p-value can produce an overrepresentation of the GO term, while the value of P (see methods) is useful for amplifying those underrepresented biological processes preferentially connected with a specific context [
85] through their expression. A limitation of this approach is that, in a complex interactome, many proteins are not specific to a single metabolic pathway, but are sometimes even part of multiple pathways. Here, the massive study of some of these pathways favors the assignment of the protein to the more studied GO pathways. In fact, the databases favor assigning the protein to the more studied GO pathways and obscure the emerging relationships towards different biological pathways that are not studied or poorly represented [
86]. Therefore, the analysis should select only the most reliable terms.
In addition, Hong et al. [
86] demonstrated that functionally linked gene pairs, even in different functional pathway types, as defined in KEGG pathways, show positively correlated expression levels. Therefore, these two genes (or their proteins), even in a functional pathway altered by a disease, are similarly up-regulated or down-regulated. This is because of their reciprocal and close functional relationships [
87]. So, when a disease affects a metabolic pathway, all the genes in the pathway will regulate their expression positively. Therefore, an over-representation of a GO process suggests an over-expression of the genes and their decoded products that make up the metabolic pathway, since they have close functional relationships with each other in regulating the expression [
86,
87].
We selected 17 terms with the highest possible strength value, paired with a very significant p value and listed according the value of P (see Methods).
Table 11 reports these terms according to the previously expressed rule. In the table, among the proteins involved in these negative functional activities, we can note (in bold) many of the proteins previously highlighted as HUB nodes, or as "bottlenecks" or involved in other important signaling pathways. Although all Biological Processes show positive values of enrichment (high strength), very many have minimal or negligible enrichments. It is necessary to exceed the value of 0.5 to have an enrichment of 3 times. We found that 32.28% of the processes have enrichments lower than 3 times and only 14.7% have enrichments greater than 10 times. The remaining 53.02% has intermediate enrichment values, between 3 and 10. This means that the most enriched fractions are very few and we can think the average enrichment of most biological processes as suitable for the normal metabolic function to be performed. The 17 selected terms therefore make up a very limited set, less than 1%, but the only one that can boast a statistically significant and even conspicuous enrichment. However, the negative term means over-enrichment and, therefore, suggests a gene over-expression. Some sets of proteins, enriching themselves, change their functional state, inducing changes in the pathways they control. Since the meaning of the negative term is loss of control, down-regulation, this means that by weakening their functions, they favor the activation, or deactivation, of the functional pathways they control. This is not new. We find a dysfunctional expression of genes with overexpression and deleterious functions during disease or even aging, in particular, of genes involved in pathways related to stress responses, antioxidant defenses, and DNA repair [
88,
89,
90].
Our examination of
Table 10 enables us to confidently affirm that many pathways show statistically significant dysregulation, and we may have successfully identified pivotal genes associated with these pathways. At present, accurately describing what occurs is challenging because of the lack of data to pinpoint causes, determine the opportune moment for the process, and establish the sequence of events, all because of the absence of space-time information. The strategy of ORF7b, in collaboration with other viral proteins, aims to create a viral microenvironment that helps infected cells minimize cell matrix rigidity and adhesion, increase intracellular oxidative stress, generate pro-survival signals, to trigger the epithelial-mesenchymal transition process, to inhibit intracellular transport and ER activity, starting widespread cellular metabolic deregulation. We should emphasize that the process of metastasis, characterized by the epithelial-mesenchymal transition (EMT) and its inverse, the mesenchymal-epithelial transition (MET), plays a crucial role in the metastatic spread of carcinomas [
91]. Likewise, these events appear to be among the primary targets in preventing programmed cell death mechanisms of infected cells, allowing survival after separation and systemic spread.
In particular, we can see the dysregulation of all protein tyrosine kinase receptor activities. This reduces the processes of internalization of external signals and the activities of receptors activated by growth factors. Integrin-mediated alterations of the intercellular matrix and loss of control over cell-extracellular matrix adhesion processes are also favored by integrated dysregulation of oxidative stress, unfolded protein response of the ER and lysosomal action [
92,
93]. The intention behind all these activities is to dysregulate programmed death processes such as apoptosis and anoikis, promoting the spread of infected cells in the body.
The systemic spread of infected cells explains well why the tissues and organs showed as infectible in
Table 6 are so numerous and all significant. In the presence of infected cellular material widespread in the body, the virus has also the potential ability to cause inflammatory processes in the brain, so it is important to pay particular attention to the dysregulation of blood-brain barrier permeability. Through altering endocytosis, endosomal trafficking, lysosomal degradation, blocking anabolic processes and lipid transport, this creates mitochondrial dysfunction, resulting in a heavy dependence on glucose for energy production. Numerous miRNAs work within the cell and could interfere with these procedures. However, distinguishing them individually through this type of analysis is not yet possible.
In a nutshell, this tiny protein is involved in controlling the intercellular communication of the virus. By suppressing intracellular signaling, it created a metabolic microenvironment that caused generalized metabolic dysregulation and blocked intracellular transport of cargos. Prevention of local programmed death mechanisms leads to viral shedding. Various viruses show comparable infection strategies [
95], such as extending particular stages of the cell cycle, managing programmed cell death, and using the nuclear membrane to transmit viral genetic material to and from the nucleus. These findings help to understand how SARS-CoV-2 can spread via cell-to-cell transmission [
95], where ACE2 is not required. Our assessment shows that viral mutations shared by different variants are unsuitable for evaluating disease mechanisms. This is due to the high metabolic interference capacity of the remaining information package of the virus. Attention to mutations in the Spike protein has distracted from the evaluation of the molecular mechanisms underlying the metabolic dysregulations induced by the virus.
3.10. Cluster Analysis
Cluster analysis allows us to extract protein interaction sub-networks that interact with each other in functional complexes and pathways to produce reliable hypotheses that can explain the various dysregulations of human metabolism induced by ORF7b. This also increases the likelihood of identifying candidate genes/proteins that can help us understand the rationale for viral action and the metabolic pathways involved.
Cluster analysis is a data analysis that explores the groups present within a dataset, known as clusters. We used Cluster K-means analysis, which does not need to group data points into predefined groups and is an unsupervised learning [
96] method. In unsupervised learning, insights come from data without predefined labels or classes. K-means is also an iterative partition algorithm and is a good clustering algorithm that ensures high similarity within cluster and low similarity between clusters. The clusters representing our entire population of interacting molecules in the ORF7b interactome derive from a base of significant experimental data and rigorous procedures for implementing the network. This should produce high-quality clusters, which means non-redundant and low-noise results, as they can reduce the quality and interpretability of the clusters. The value to be attributed to K is one of the major drawbacks of this algorithm. In our analysis, K is equals 10 (
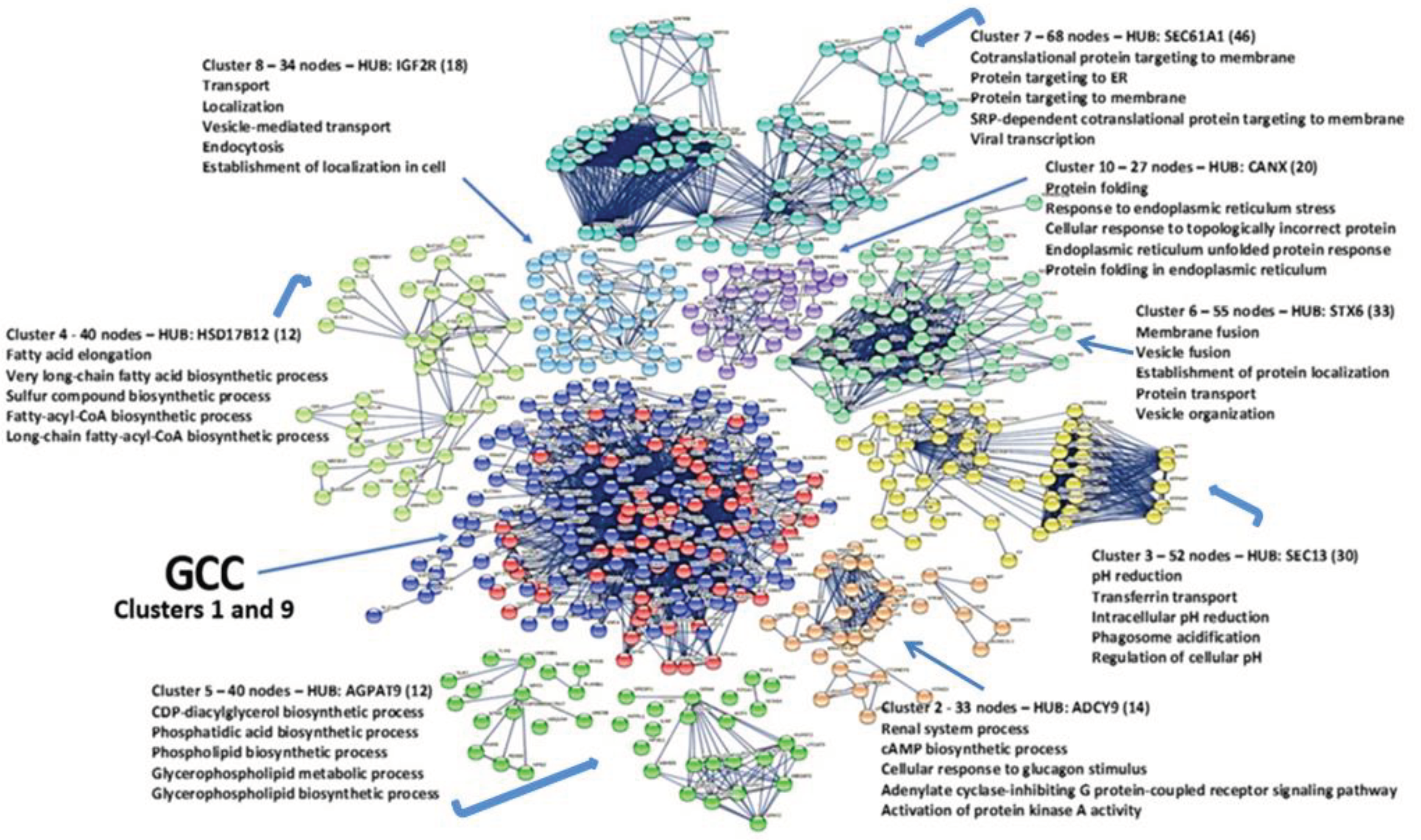
Figure 7).
This result, got after many attempts with lower K values, has to be considered as the best compromise. We used this K-value because it gave us the most compact clusters and statistically significant p-values (all p-values are always <1.0e-16). The ten metabolic modules are all functionally consistent, and in the
Figures S7 and S8, we also show the links existing between the clusters. The many metabolic relationships existing between the clusters, as shown in the figure, mostly represent the normal metabolic machineries necessary for cellular life. Only the GCC shows an overlay of two modules, but, as we shall see, they resolve into two independent sub-graphs. The greatest interest is precisely in these two sub-graphs because they contain most of the HUBs and bottlenecks nodes previously found and control crucial metabolic pathways. While the other sub-graphs seem to regulate typical metabolic activities, understanding the specific functions of these central modules and where their constituent proteins operate within the cell is essential. This is a core-periphery organization. Core-periphery is a characteristic we can find at group-level relationships in biological networks, but not only [
97]. The situation involves meso-scale dominance events [
98]. It describes a scenario where a group of core nodes captures an excessive number of contacts in the network. On the contrary, the nodes on the periphery possess fewer interconnections with one another, albeit they are connected to the core nodes. In networking, the mesoscale describes sub-cellular events on length scales ranging from that of a single cell, up to the size of molecular complexes, where groups of molecules self-organize relationally to form large, functional core structures [
99]. While individual nodes perform only local operations, their organization into clusters generates a richer and more diverse functional repertoire.
3.11. Analysis of GCC Core
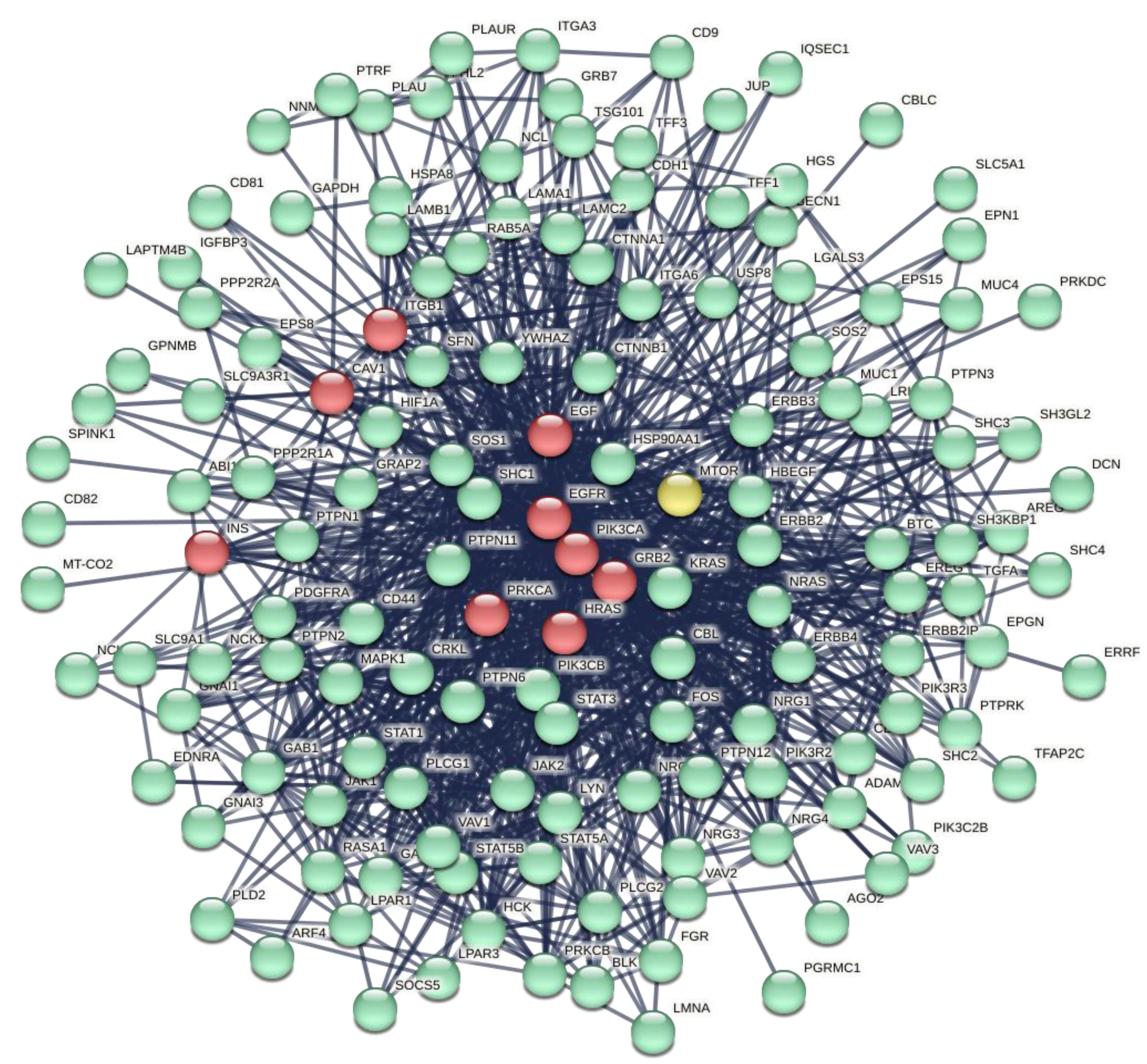
The cluster analysis extracted from the compact GCC area two clusters (1 and 9), both statistically significant and compact.
Figure 8 shows the cluster No.1. In the caption, there are the major topological parameters. This cluster is very compact. Its major role is to regulate the EGFR family signaling pathway (EGFR, ERBB, ERBB2) where the receptors' protein tyrosine kinases signaling show a p <6.85e-48. It is involved in regulation of the Jak-Stat pathway, ERBB and ERBB2 signaling (p <2.55e-40), and regulation of peptidyl-tyrosine (p <2.99e-27). We can find the key details in the following GO terms: GO:0007169, GO:0038127, and GO:1901184. But in the cluster No1 we find also ITGB1, CAV1, EGF, EGFR, PIK3CA, INS, GRB2, PRKCA, HRAS, MTOR, just to mention the major nodes. Thus, the role of this cluster is also to control cell migration, cell motility, immune response, phosphorylation, cell death, apoptotic cell process, cell adhesion, cell migration, stress, insulin path, phagocytosis, lymphocyte activation, blood coagulation, Cytokine-mediated signaling pathway with very high statistical significance, as it appears from the list calculated by STRING in the Biological Process (GO) category.
Proteins operate in their specific environments, therefore knowledge of where proteins are located is crucial to understanding the metabolic processes of which they are a part. We can perform this analysis with the help of Cytoscape. After transferring the cluster 1 to Cytoscape, we have with the help of STRING app and Nodes Table (Compartment analysis) selected the protein nodes with the highest statistical value (5.0) that operate in the various cellular compartments. Level 5 collects the most important proteins in defining the biological processes of which they are part.
In the
Table 11 we can see in which cellular compartment the cluster No 1 proteins operate, but we also see that there are various proteins already defined as dysregulated, so we can know where they operate. Nucleus and plasma membrane, as well as the cytoskeleton, are among the richest compartments of functional activities and proteins crucial for the progression of these activities. In the
Table 11 we find many of these proteins, for which symbolic notations have been used to distinguish them (see note to the Table). The table summarizes two important proteomic characteristics: a) there are numerous proteins that operate in a multipolar way, i.e., in several compartments (e.g., EGFR); b) there are many dysregulated proteins, in particular those involved in the fundamental processes of signaling and in favoring cell diffusion. Various proteins localize in multiple compartments, showing a shared protein pool even if apparently unrelated. However, each protein has its own level of expression and its own compartmental distribution.
Viewing all together, we could read this as indicative of functional progressions starting at the membrane and proceeding towards the nucleus. The limit is the absence of temporal information that statistically flattens the metabolic dynamics and makes it very difficult to have reliable sequential explanations. But this is not the only intricacy.
Figures S7 and S8 demonstrate how single nodes can take on multiple roles to engage in various functional processes. Even a single functional activity can have its nodes distributed in numerous modules. This is a straightforward demonstration of how difficult it is to describe the actual behavior of concurrent functional processes without a temporal chronology, but the entire network, i.e., the operational context, can help.
Not only the regulation of space and time but also the compartmentalization characterizes the cellular proteomes. The presence of similar proteins in different compartments suggests different local proteomes [
100], each performing its local metabolic activities, so it is difficult to identify any distortion. Nucleus and cytoplasm are among the most populated compartments. The proteomes of these compartments show a multipolar protein distribution, which makes them functionally very ductile. Therefore, attributing static and specific roles to the metabolism and to the proteins that operate within is a vision that does not correspond to reality. We cannot attribute a protein's metabolic function solely to its presence or absence. The function is also determined by the reactions happening at different omic levels and compartments [
101]. Reactions that are always the result of protein-protein interactions. Thus, interactomic level reflects what happens at the genomic or transcriptomic level, generating a network that differs from the underlying ones because displays a portion of the total functional mechanisms. The event in question has recently gained prominence [
102]. Some melanoma cells show a dependence on external sources of methionine for their growth. The authors describe the methylome, transcriptome and proteome of these cells. Only the multilevel contemporary study allowed the authors to understand the real metabolic behavior of methionine addiction because the study of the methylome alone led to trivial conclusions.
In short, we have the spatial distribution of proteins of the ORF7b interactome, but the temporal distribution is missing. Multi-localization of a protein increases the probability of interactions, generating possible new functional characteristics specific to the context. This expands the functional capabilities of the cell but makes any modeling that does not include all the parameters involved difficult.
Due to functionally important proteins, cluster No 9 has the potential to perform multiple functions (
Figure 9). This cluster controls the process that modulates the cell transport to, or maintained in, a specific location (GO:0032879 p = 2.30e-34); extent of addition of phosphate groups to a molecule (GO:0042327, p = 1.89e-29); cell migration (GO:0030334, p = 3.11e-29); regulation of cell migration (GO:0030334, p = 3.11e-29); The transmembrane receptor protein tyrosine kinase signaling pathway (GO:0007169, p = 1.78e-28). It is also associated with the negative regulation of cell death (GO:0060548, Strength = 0.92, p = 1.75e-17) and programmed cell death (GO:0043069, Str.= 0.90, p = 3.96e-16). 0.98, p = 2.26e-16) or in the Negative regulation of production of miRNAs involved in gene silencing (GO:1903799, Str. 1.78, p = 4.6e-4). Similar considerations also apply to cluster No 9.
Dysregulated proteins, such as CTNNB1, SRC, PTK2, ITCB3, or PRKCD, found in Cluster No 9 (see
Table 12), are present in many cellular compartments, including those that are distant from each other or different from a chemical-physical point of view, such as cytosol and plasma membranes. This means that they regulate temporally their expression and that they require post-translational modifications that depend on the context. Because most analysis platforms collapse this information into the native protein, nodes end up having more functional links than the context. This induces errors on the degree value and on the related topological evaluations, which can lead to alterations in the network.
An instance of this is the activation of the Human SRC (P12931, Proto-oncogene tyrosine-protein kinase Src), a Non-receptor protein tyrosine kinase that is triggered upon binding to various cellular receptors, including integrins and other adhesion receptors, regulating a wide range of biological processes. It belongs to the Src kinase family and is functionally redundant, making it challenging to identify its specific role in each compartment and determine which member is involved without the knowledge of its spatio-temporal characteristics in that specific context.
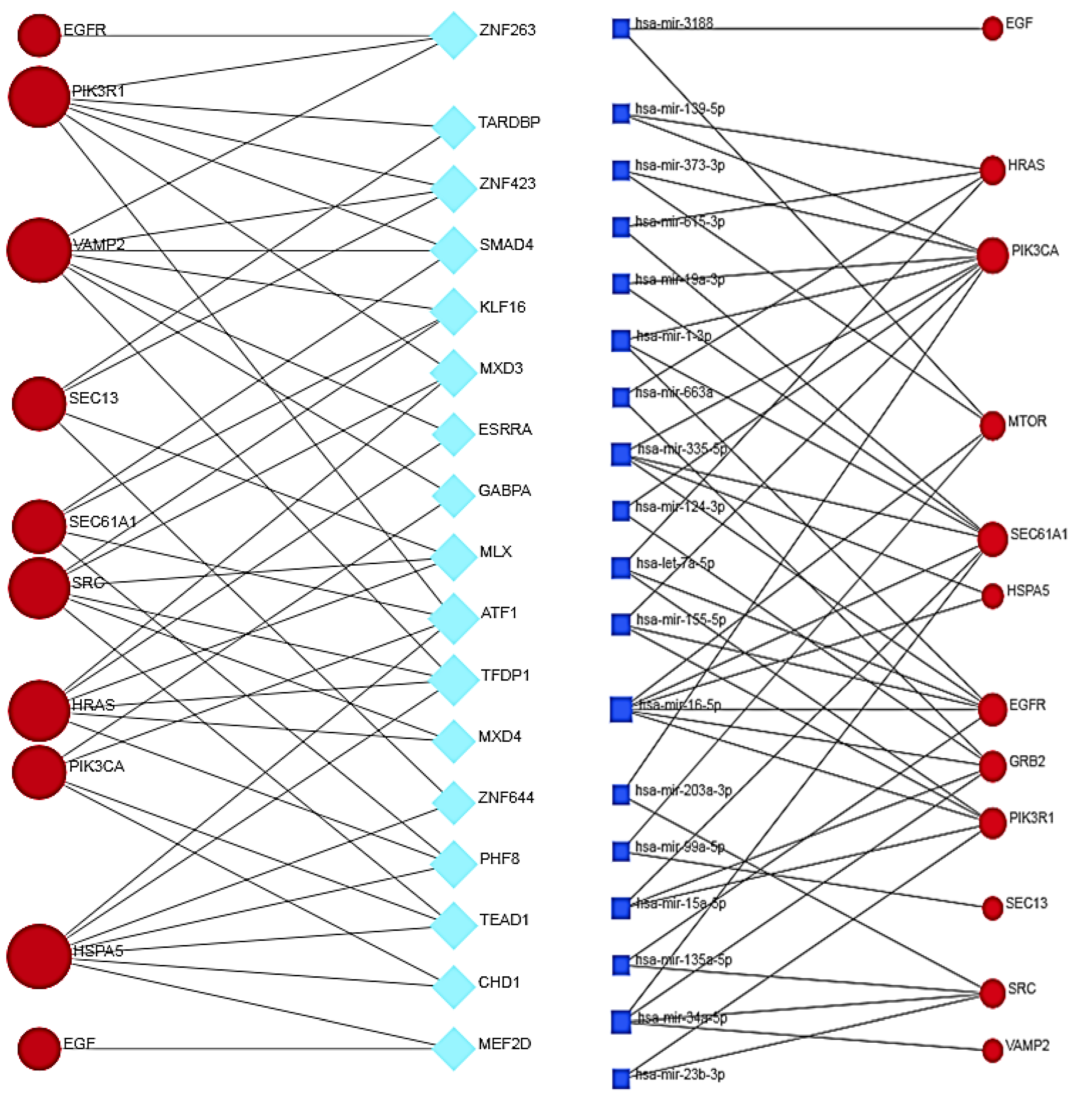
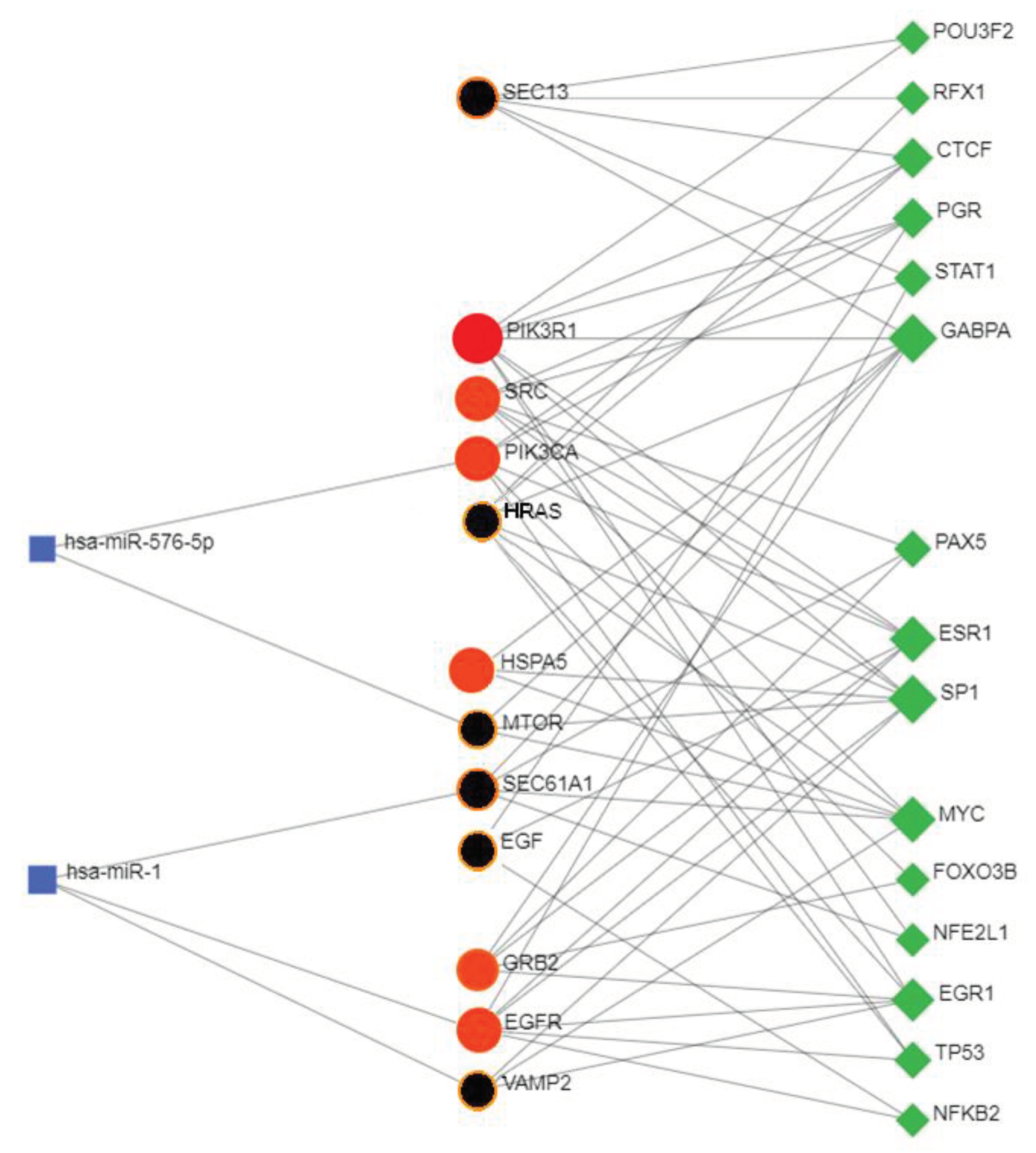
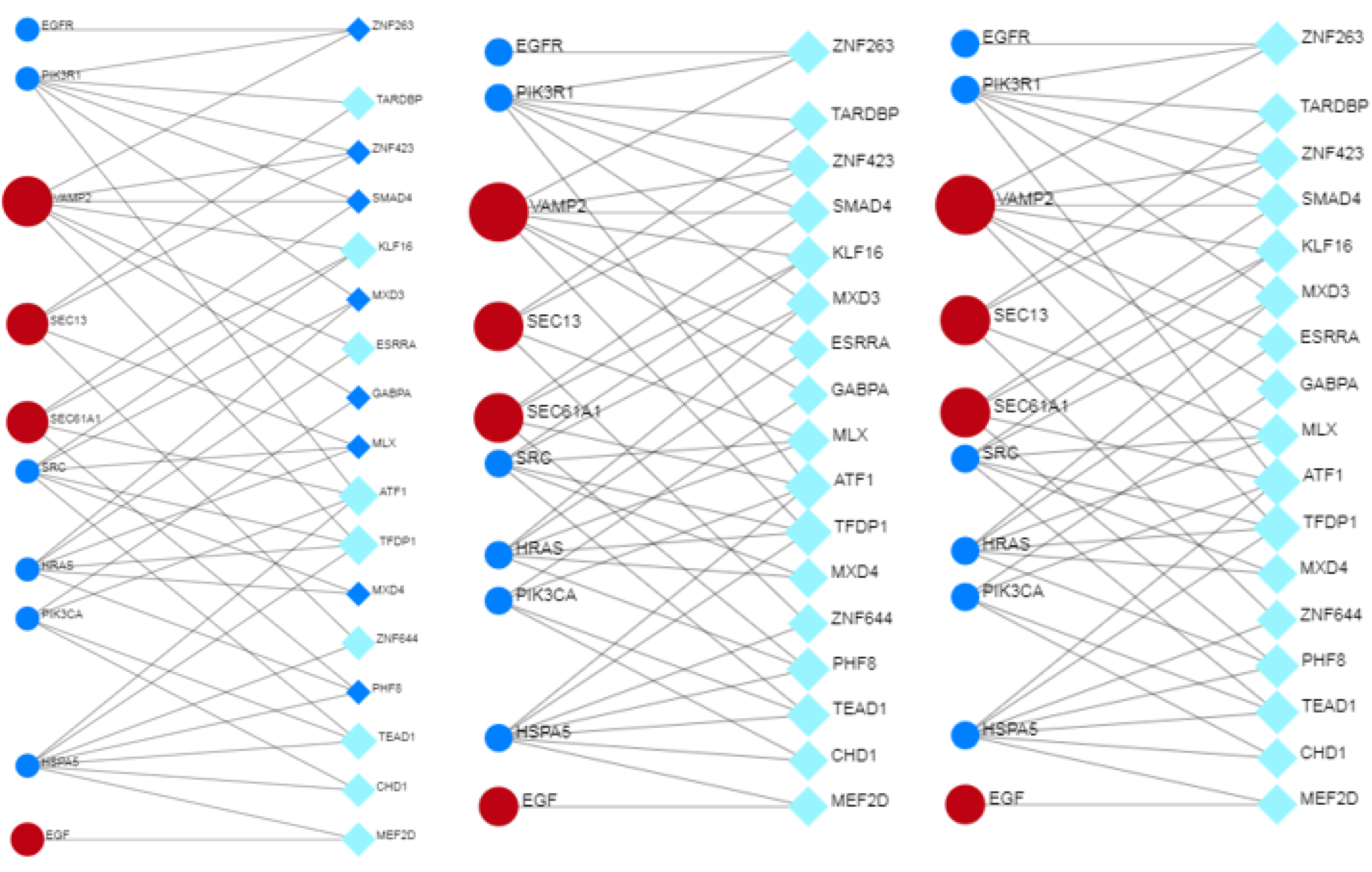
3.12. Co-Regulation between Hub and Bottleneck Proteins, Transcription Factors and miRNAs
Our findings thus far, have revealed a metabolic depiction that outlines the involvement of a specific group of significant high-ranking proteins in a series of dysregulated metabolic processes aimed at promoting the dissemination and spread of virus-infected cells throughout the body, because of the influence of the accessory viral protein, ORF7b. However, we still have limited vision because we can only glimpse the purposes, know some of the involved actors, but we still cannot understand who planned and performed the entire process.
Understanding the intracellular mechanism of complex biological processes driven by ORF7b also depends on deciphering its complicated co-regulatory network. The identification of Hub and Bottleneck proteins in protein groups dysregulated by viral infection prompts investigation to understand their co-regulation. Within the co-regulatory network, there are both post-transcriptional and transcriptional regulators that can regulate themselves and each other.
A limitation that should give pause for thought is the evidence that hub and bottleneck proteins control and regulate an enormous number of functional processes. Finding them involved in a particular process, even experimentally, does not mean that process is actual and existing in fact. Precise rules govern the occurrence of a functional process, primarily depending on the context of the events and the chemical-physical characteristics of the compartmentalized microenvironment where the event should occur. To ensure a functional event, the cell must program when, where, and how it should occur. The metabolic network is not solely dependent on proteins. To synchronize basic functional activities according to the circadian cycle or unexpected events, we need several other actors to accelerate or slow down an intricate and dynamic system. The comprehension of co-regulatory mechanisms that are fundamental to cellular identity and function requires the involvement of transcription factors (TFs) and microRNAs (miRNAs). TFs and miRNAs work together to regulate transcription and post-transcriptional processes, respectively [
102,
103].
Combining computational and experimental interaction data in network models can highlight functional mechanisms in TF- and miRNA-mediated gene regulation. These models can provide insight into the mechanisms that control gene expression at the system level, rather than at the individual gene level. Typically, TFs act as activators or repressors, increasing or decreasing transcription, while miRNAs are mostly repressors. We can visualize the distinct activities by using two separate networks: transcriptional networks and post-transcriptional networks. It is noteworthy that both networks are bipartite and direct. In each network, there are two distinct types of nodes interconnected by unidirectional edges. One network contains interactions between genes and transcription factors, which is known as a transcriptional regulatory network. The other network contains interactions between genes and miRNAs, which is known as a post-transcriptional regulatory network. We assume that, in post-transcriptional regulations, the regulatory actions of miRNAs towards targets are negative. However, it is possible to get integrated gene regulatory networks that include genes, TFs, and miRNAs, provided that the components are statistically more significant. The databases on TFs and miRNAs are quite recent and the data collected are both experimental and predictive because this area of research is still very young. Selective filtering is required to get statistically significant nodes. As explained in the Methods section, the reference databases of transcriptional and post-transcriptional networks comprise experimental data, whereas the integrated co-regulatory database comprises mixed data. This means that the comparison of the integrated co-regulatory network with the transcriptional networks may yield diverse interactions, which depend on the respective node rank in the two distinct systems.
3.13. Transcriptional and Post-Transcriptional Regulatory Networks
As a result, in transcriptional regulatory networks, FTs possess two types of action since it is the TF that binds to its target gene, rather than the reverse. The information comprises an in-degree, which signifies the number of transcription factors binding a gene, and an out-degree, which signifies the number of genes bound by a transcription factor. All this reflects the functional and biological aspects underlying these interactions. High-grade TFs (i.e., hub TFs interacting on many genes) have a high key character of biological functionality, while target genes bound by many TFs do not have a tendency to be functionally essential. Therefore, analyzing this type of network provides insights into biological systems that are not obtainable through single gene studies.
Both the networks containing TFs and miRNAs are represented in
Figure 10, illustrating the transcriptional and post-transcriptional networks of gene interactions, which include hubs and bottlenecks. The transcriptional network reveals that EGFR, the top-ranking hub node within the PPI network, possesses an in-degree value of 1 in relation to its interaction with ZNF263, whereas ZNF263 exhibits an out-degree value of 3. Therefore, within this network, ZNF263 holds greater biological significance in relation to EGFR. Its role in this transcriptional network involves functioning as a DNA-binding transcriptional repressor that specifically targets RNA polymerase II, resulting in the repression of EGFR, PIK3R1, and VAMP2. The TFs and miRNAs represented in the two networks are those of higher rank with a higher probability of interaction.
3.14. Co-Regulatory Network
Getting a co-regulated network requires integration of HUBs and bottlenecks with FTs and miRNAs. To determine the transcriptional regulatory relationships that these nodes may hold, we employed hub and bottleneck as enrichment seeds. This co-regulated network allowed us to pinpoint the 14 most reliable TFs and 2 miRNAs that were significantly associated with the expression of HUB and bottleneck genes.
The network (
Figure 11) shows that among bottlenecks SEC13 is one of the most regulated genes. The protein encoded by this gene belongs to the SEC13 family of WD-repeat proteins and is a component of several important complexes. It is a component of the nuclear pore complex (NPC), which regulates transport between the nucleus and cytoplasm and has a direct role in regulating gene expression [
104]. It is also a component of the COPII Coat Complex, where it plays a role in the formation of coated vesicles [
105]. Four of the transcription factors that regulate SEC13 also regulate PIK3R1, the gene responsible for encoding Human_P85A, a protein that modulates glucose uptake in insulin-sensitive tissues by binding to activated Tyr kinases on the cellular membrane. Due to its inhibitory action, it appears to be a significant factor contributing to the hyperglycemia observed in covid patients. EGFR is also controlled by several TFs. The governance of each of these genes is multifaceted and bolstered by two miRNAs, specifically hsa-miR-576-5p and hsa-miR-1. The role of miRNA expression levels in disease processes and physiological development is significant, as changes in microRNA copy number or expression are closely associated with the onset of various human diseases [
106]. miRNAs are present in a substantial number in humans [
107].
The correlation between miRNAs and human genes during SARS-CoV-2 infection is still an expanding research field with initial studies. Some preliminary evidence shows potential associations between miRNAs and genes that participate in the reaction to infection. It is essential to highlight that the analysis of this subject is still in progress. Despite this, miRNAs may be related to genes during SARS-CoV-2 infection to control inflammation. miRNA-155 [
108] links the regulation of genes involved in inflammation, such as tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6). According to previous research on COVID-19, miRNA-146a may be involved in regulating the innate immune response [
109], and its upregulation may contribute to the dysregulation of inflammatory pathways. miRNAs might exert direct control over the replication of SARS2, as well as its capacity to infect host cells [
110]. This could involve both the regulation of viral proteins and genes/proteins involved in human metabolism. Observations in cell lines and cancer patients led researchers to predict that miR-576-5p could down-regulate both PIK3CA and its mRNA [
111]. Meanwhile, their target mRNAs were up-regulated. Hsa-miR-1 is believed to be linked to regulating human genes, especially in cancer patients [
112]. Additionally, it has been noted that this specific miRNA also plays a role in the disturbance of glycemia for individuals with type 2 diabetes [
113].
The co-regulatory network provides a better picture of metabolic events that the simple identification of a gene or protein in a metabolic pathway cannot give, even more so when we study the molecular mechanisms involved in a pathology. Merely asserting the involvement of a protein or gene in a pathological state without comprehending the coordinated activity of genes, miRNAs, TFs, mRNAs, and proteins may not always culminate in accurate inferences. Co-regulatory networks offer more decisive direction by elucidating the general coordination of the aforementioned actors, besides the appraisal of the pathological consequences of ORF7b.
3.15. Comparative Analysis of Negative Regulations According to the GO
Figure 12 shows the set of negative regulations vital for cellular diffusion represented by three transcriptional networks, which, upon comparison, exhibit remarkable similarities. In all three networks, EGFR, HRAS, HSPA5, PIK3CA, PIK3R1, and SRC are the genes involved in the negative control of programmed death. Their transcription at the individual gene level is negatively controlled through DNA-dependent transcription.
Below is a brief illustration of the most intriguing transcription factors found in the networks. ZNF423 and ZNF263 (Zinc Finger Protein 423 and 263) can act as both transcriptional repressors and activator by binding to DNA, where ZNF423 plays a central role. MXD4 (Max-interacting transcriptional repressor MAD4) is a protein that in humans is encoded by the MXD4 gene. PHF8 (Histone lysine demethylase PHF8) is a transcription activator which acts on the epigenetically methylated Histone 3 but is a repressor for the methylated histone 4. Acts as a coactivator of rDNA transcription, by activating polymerase I (pol I) mediated transcription of rRNA genes and playing a role in the cell cycle. However, its role remains still unsure in vivo. GABPA (GA Binding Protein Transcription Factor Subunit Alpha) is a transcription factor interacting with purine rich repeats (GA repeats), so positively regulating the transcription of transcriptional repressor RHIT and of ZNF family such as ZNF205. MLX (MAX Dimerization Protein MLX), its decoded product (Max-like protein X) forms many sequence-specific DNA-binding protein complexes with various proteins. These complexes act as transcriptional repressors. Plays a peculiar role as a transcriptional activator of glycolytic target genes, thus it is involved in glucose-responsive gene regulation. Here, we have another pro-glycemic effect, common to covid patients.
While the
Figure 13 shows the relationships between genes and miRNAs involved in blocking programmed death at the post-transcriptional level the
Figure 14 shows its co-regulated network where we find the occurrence of Myc and TP53, two well-known transcription factors. MYC, (MYC Proto-Oncogene or BHLH Transcription Factor, which codes for P01106 · MYC_HUMAN) is involved in many diseases (114). The Gene Ontology (GO) annotations that concern MYC comprise DNA-binding transcription factor activity, and the ability to function with TAF6L to activate target gene expression through RNA polymerase II cis-regulatory region sequence-specific DNA binding.
TP53, also known as Tumor Protein 53 and encoding for P04637, Cellular tumor antigen p53, acts as a tumor suppressor, in response to cellular stresses to regulate the expression of target genes [
115]. However, in specific metabolic contexts, it can induce cell cycle arrest, apoptosis, and changes in metabolism [
116]. As a matter of fact, it has been discovered that SARS-CoV-2 infection leads to the stabilization of TP53 on chromatin [
117], contributing to a robust host cytopathic effect. Modification of chromatin accessibility, cellular senescence, and inflammatory cytokine release through TP53 is brought about by the involvement of this protein in various SARS-CoV-2 spike variant-induced syncytia formations. The protein appears to have a role in inflammation associated with cellular senescence [
117]. In addition, TP53 was discovered to be implicated in IFN-γ-mediated signaling, apoptosis, and proteasomal degradation of CD4 T cells [
118]. However, uncertainties regarding the functionality of miRNAs persist because of technical difficulties and the considerable number of miRNAs that are still subject to systematic profiling [
107]. Because of their low intrinsic stability and RNAses [
119], they are susceptible to degradation, and laboratory manipulations can have questionable effects on their measurements [
120,
121].
TFs are well-established proteins with reliable experimentally derived results, although miRNAs remain somewhat enigmatic. TFs are proteins that control the rate of transcription of genetic information from DNA to mRNA binding to DNA. Thus, their function is to regulate, switching on and off, genes. This functional activity is to address the gene expression to the exact target cells at the right time and in the right amount. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA polymerase to specific genes.
We have examined the various correlations between miRNAs, TFs and the components of the compact hub-and-spoke architectural system of the PPI network, getting information on the fundamental co-regulations operated by some TFs and miRNAs. These findings suggest that crosstalk motifs, comprising the direct and non-shared relationships between regulators and their target genes, can have downstream effects on diverse biological processes, in line with the features already highlighted in the interactome's analysis network. This analysis amplifies and substantiates our findings and deductions from the interactomic analysis. Our result, however, has limitations. The human genome contains thousands of coding and non-coding RNA genes. These genes express differentially, in diverse locations, at distinct times during normal homeostasis, or in response to environmental cues. The differential expression extends to TFs and miRNAs, too. The regulation of genes is specific to certain conditions and changes over time, meaning our findings only provide a static view of the molecular mechanisms affected by ORF7b. While our conclusions are valid, we can only show the presumed targets, not how they dynamically work.
4. Discussion
The guiding principle that underpins this research is that SARS-CoV-2 infection leads to changes in the deep metabolic activities of infected cells to favor the acquisition and maintenance of viral strategies, compared to normal cells. The virus causes a reprogramming of cellular metabolism by its proteins. The expression "metabolic reprogramming" refers to the recognition of normal metabolic pathways which are modified by viral proteins, when compared to those in normal tissue. This point is significant because the analysis of "metabolic normality" is often overlooked. Our training in cancer has taught us to search for mutations that can modify signaling processes. In viral infections, mutations are absent, as viruses achieve the same aim by up- or down-regulating normal signaling pathways or other metabolic processes.
Our results reveal the functional impact of the accessory protein ORF7b in SARS-CoV-2 infection and identify molecules that control metabolic processes dysregulated by this viral protein. Among the many functional activities highlighted, we focused on those that promote the spread of infected cells in the organism.
The release of virions into the extracellular space is a common event among numerous viruses, which has stimulated the study of virus egress/entry biology. Although some viruses spread through the shedding of infected cells [
180,
181], this is an understudied topic. Recently, several authors [
182,
183] have reported evidence by antibody experiments, that SARS-CoV-2 could spread through cell-to-cell transmission. However, no one has studied or hypothesized any related molecular mechanism. In this article, we confirm those authors' hypotheses and describe the deep molecular mechanism that underlies this feature of SARS-CoV-2. This discovery contributes to our understanding of the human immune response to the attack of this virus because cell-to-cell transmission is an effective means by which viruses evade host immunity.
It is still to be considered that our data on spread in some sense support the theory on the evolution of virulence which assumes that high growth rates of pathogens should both increase transmission between hosts and increase disease induced morbidity or mortality [
186,
187]. The spread of infected cells could fit into this logic, but the theory also suggests that through viral “tolerance”, virulence is mitigated without reducing viral load [
187,
188]. This also dictates that the host should allow the selection of the pathogen with a higher growth rate to gain a gain in transmission between hosts but without causing harm to the original host [
188,
189,
190]. Today's clinical data tells us that the virulence of Covid-19 is decreasing without any type of specific intervention. Our data does not explain the effects of diffusion on the virulence, but it paves the way for experimental designs with greater awareness of what actually happens.
We have analyzed by interactomics only functional and physical correlations between ORF7b and the entire human proteome determined by experiments. To obtain reliable interactomics results, we extracted from the set of interactors only those that were characterized by high significance. The investigation showed that the virus achieves its strategic goals by interacting with metabolic processes controlled by human proteins such as EGFR, SRC, HSPA5, MTOR, SEC13, SEC61A1, VAMP2, PIK3R1, PIK3CA, GRB2, and HRAS, which are important for human metabolism because they are high-ranking, HUB and bottleneck proteins. Through a series of analyses using transcriptional co-regulation networks, we have also validated our results by identifying regulatory actions conducted by transcription factors and miRNAs on genes that code for the previously identified key proteins.
Viruses do not perform metabolic processes but know how to interact with them to their own advantage. Although various attempts have been made to identify metabolic pathways and nodes under the control of the virus, to our knowledge, this is the first wide-ranging interactome map identified for a specific protein of SARS-CoV-2. We have identified some metabolic pathways under the control of ORF7b, but still, we have a limited knowledge of the comprehensive set of viral proteins involved, and by which specific mechanisms. Despite that, several authors have hypothesized some functional activities of ORF7b in the infected cell and its synergism with other viral proteins, but no one has attempted to study in depth the molecular and functional interactions within human metabolism implemented by ORF7b. In particular, they identified its involvement in SNARE-driven vesicular transport, exocytic processes, ERBB signaling, but without a functional characterization that identifies the actual role of ORF7b, possibly in synergy with other viral proteins [
122]. There have been other studies that have endeavored to juxtapose the mechanisms of diseases between SARS1 and SARS2 [
123,
124], but none of them have deciphered any common molecular mechanism. Both viruses lead to acute respiratory distress, but many phenomenological observations show differences. One study predicted that SARS-CoV-2 induces a systemic disease, which, unlike SARS1, damages various organs in the body, such as the heart, kidney, and brain [
125,
126]. These results suggest that the two viruses use different molecular mechanisms but also that we do not know which mechanisms they use. Out of curiosity, searching PubMed for "differences in molecular mechanisms of SARS-CoV-1 and SARS-CoV-2" or "molecular mechanisms of SARS-CoV-1 and SARS-C0V-2 (or similar terms), yielded no results.The continuous work conducted by the curators of BioGRID, in selecting and evaluating the statistical significance of each single experimentally characterized interaction between the viral proteins and the human proteome, has allowed us to design this study with the methods of Interactomics. Direct knowledge of the deep molecular mechanisms implemented by individual viral proteins is essential because only through this knowledge will we be able to design specific and effective antiviral drugs. A study at a deep molecular level, a research area still rather obscure in its modes of action in space and time, is an important approach if aimed at identifying those human proteins which, in viral infection, play crucial roles such as hub nodes or as a bottleneck. These proteins represent the crossroads of multiple biological activities and, therefore, are the best targets for disease control.
ORF7b is a tiny viral protein of 43 amino acids, a macro-polyanion with a net charge of –4 at neutral pH (four negative residues and no positive charge); the central part from 9 to 29 is helical, and the protein surface is negative [
6]. This protein does not appear to operate on its own (see
Table 9). What emerges from this study is the precise interference of ORF7b into various molecular mechanisms at the basis of our metabolism. ORF7b showed mostly diverse behaviors, in terms of localization, membrane recruitment, and metabolic dynamics. The results show its important role in conditioning cellular transport processes as well in some important signaling pathways (see
Table 3). The topological characteristics of this interactome reveal a group of proteins with structural and functional properties that are consistently implicated in multiple metabolic activities, some of which are dysregulated by ORF7b's action. These proteins are characterized by their high degree of functional relationships and by their high ability to regulate a multitude of significant metabolic and signaling pathways. The interactome shows certain metabolic modules that perform necessary functional activities for normal cellular metabolism. A large central core (GCC) comprising two closely connected clusters was identified through cluster analysis as the primary functional location of these proteins. The high number of tight connections favors a high metabolic rate, which accelerates any functional activity.
The activity of these proteins extends to very different places in the cell (see
Table 4) according to a hub-and-spoke topological model and potentially also to those tissues that have the molecular characteristics suitable for the entry of the virus (see
Table 6). All this suggests that ORF7b must have a remarkable ability to interact with different molecular partners, such as to allow it to operate practically everywhere, at the membrane level and in the cytoplasm. Indeed, the list of its main molecular interactors shows both membrane proteins and cytoplasmic proteins. Some authors have hypothesized a role for ORF7b as an intrinsic single-span membrane protein, in analogy with the 44 amino acid homolog ORF7b of SARS [
6]. This hypothesis is rather restrictive considering the wide spectrum of functions in which this protein is involved and the spatio-temporal characteristics that a biological object of this type must possess in order to be involved in the various intracellular transport processes (see GO:0006810, p-value 3.04e-67); or even having also to guide and regulate target localization (see GO:0008104 and GO:0045184, p-values 1.4e-58 and 2.85e-58, respectively). But, at the same time this protein must also have the characteristic to interact with different membrane systems (see GOCC:0016020, GOCC:0031090, GOCC:0031982 or GOCC:0098588, with p-values of 2.5e-92, 2.07e-77, 5.13e-62 and 3.17e- 58, respectively) and to interfere with metabolic signaling paths (see GO:0007169, GO:0007167; HAS-9006934, HAS-1227986, or HAS-6811558, with p-values of 1.23e-66, 7.95e-59, 4.44e-84, 2.43e-30, and 5.16e-24, respectively).
The regulated functional re-localization seems one of the most important characteristics of this protein [
127]. ORF7b shows coherent functional solutions with viable biochemical functional models. The closest class of proteins possessing these types of broad properties is called the “Peripheral Membrane Proteins”, a class of proteins that live at the membrane interface [
128,
129]. In 2002, Felix Goñi [
130] introduced the concept of "non-permanent membrane proteins", to encompass the wide variety of proteins that are not found in a stable membrane-bound form under physiological conditions, but interact with the membrane in certain phases of their specific course of action. Despite the fundamental biological meaning of these proteins, an experimental characterization of their structure has always been vague because attempts at structure prediction often fail. Therefore, this protein class has a poor representation of its 3D structures within the PDB because they are difficult to study [
131]. Wanting to represent them in a few words, they are soluble proteins that bind transiently to the surface of biological membranes or even to proteins on the outer side of the membrane, where they perform their functions. The reversible attachment of proteins to biological membranes shows how they can regulate cell signaling and many other important cellular events, through a variety of mechanisms [
132,
133]. Thus, the behavior of peripheral proteins, reversibly associated with the lipid bilayer [
132,
134], may also explain the behavior of ORF7b, coherently with its structural/functional properties. Therefore, this protein appears as a very reliable member of the class of "non-permanent membrane proteins" [
135].
Recent molecular dynamics simulations experiments provided molecular insights of the protein's dimerization [
191]. This study shows different dimerization models, parallel and antiparallel. Among the various structures modeled, the authors suggest the possibility that the parallel dimer may operate docked to the membrane, from 7 to 30, and floating in the cytoplasm from 31 to 43. According to authors, while simulations support the homodimerization of ORF7b, the analysis of genetic mutations of orf7b during the evolution of the pandemic suggests an unstable dimerization when associated with the regulation of IFN production, the apparent function attributed to this protein. They conclude that the lack of detailed structural information on lateral protein-protein associations hinders a thorough evaluation of packing, which means that there is not yet sufficient detail to define consistent structure-function relationships. This information adds to the previous considerations but every hypothesis made still remains valid.
Like other viruses, SARS-CoV-2 can cause reinfection/reactivation and persistent infection, as supported by several experimental studies [
136]. SARS-CoV-2 has the potential to activate or modulate oncogenic cancer-promoted pathways, leading to chronic low-grade inflammation and tissue damage, according to growing evidence [
137]. Several authors perceive oncogenesis as a potential long-term effect of SARS-CoV-2 infection, which could lead to the onset of cancer by inhibiting tumor suppressor genes [
138]. The utilization of similar tactics as EBV or HSV1 by SARS-CoV-2 to manipulate p53 is clear, as the virus takes over the protein using viral antigens, which lead to p53 deterioration [
139]. By deactivating both external and internal apoptotic pathways of host cells, SARS-CoV-2 may spread like cancer cells [
140,
141]. Our results suggest that the cancer-like effects of SARS-CoV-2 result from the virus capability to spread infected cells through the action of its proteins, mimicking cancer and its metastasis. The lack of adequate understanding of the mechanisms that govern the progression of the virus after the release of infected cells makes it impossible to make accurate predictions about the long-term implications of the long-covid.
However, we should make a last consideration given the recent advances in our understanding of the N protein of SARS-CoV-2. Phosphorylation of the central disordered region of the N protein forms dynamic, liquid-like condensates that control also the viral genome transcription [
142]. N protein contains three dynamic disordered regions that house putative transiently helical binding motifs and the protein undergoes liquid-liquid phase separation [
142,
143] thus phosphorylation regulates the accessibility and assembly of N protein to bio-condensate [
144]. Another critical function of N is to encapsulate the viral genome of ssRNA to evade immune detection and protect viral RNA from degradation by host factors [
144,
145,
146,
147].
Viral proteins form condensates for their molecular strategies, such as infection and signaling transduction [
148,
149]. Viruses regularly execute their molecular tactics in specific parts of cells. For instance, we should consider how phase separation in cell compartments affects important processes like viral transcription or viral spread [
148,
149]. The fact that ORF7b interacts with N (
Table 9) supports the involvement of ORF7b in viral diffusion phenomena, with mechanisms of alteration of the cytoskeleton, but, perhaps, also with more complex mechanisms involving liquid droplets. After all, phase separation is one of the basic molecular processes that govern multiple cellular activities, such as cancer progression, gene expression, and signaling transduction [
150].
In SARS-CoV-2, the properties of the liquid-like condensate that forms phase-separated compartments without a membrane and the transient nature of interactions within them, are determined by the interaction of N protein with viral RNA because of its intrinsic disorder properties [
151]. The threshold for phase separation decreases as the number of interacting sites of a molecule increases. This multivalency comes from structural domains, where each domain contributes to binding [
151]. Intrinsically disordered-regions (IDRs) often participate in phase separation, as they might provide a source of multivalency. In fact, the low affinity of their individual interactions can enable liquid-like properties [
151].
We are studying SARS-CoV-2-host interactions in a simplistic context because crucial information is missing from the databases. For example, there have been few studies evaluating virus-host molecular interactions considering the range of post-translational modifications. Without the enormous potential of the biological role of post-translational modifications, we run serious risks of having distorted information on the biology of this virus. Researchers have shown crucial roles of phosphorylation and ubiquitylation in other systems, but have not yet identified the corresponding proteoforms in SARS-CoV-2-host interactions.
However, many of the high-ranking proteins we have studied and selected show that they have all the characteristics necessary to act even through forms of bio-condensates. Therefore, we cannot exclude that, together with the co-regulation that we have highlighted, there may also be a further form of regulatory activity exerted by the liquid-like condensates. We cannot exclude it, considering their well-established presence in cells and the important roles they play.
Figure 1.
– Circular network of SARS-CoV-2-ORF7b and human host PPI (from BioGRID). Circles within circles representation show the layers closest to the center as more highly connected. BioGRID also suggests the likelihood of direct/indirect interaction of ORF7b (in dark red) with other viral proteins (ORF3a and M, in blue). The proteins used in the present analysis are among those of the most densely represented central area.
Figure 1.
– Circular network of SARS-CoV-2-ORF7b and human host PPI (from BioGRID). Circles within circles representation show the layers closest to the center as more highly connected. BioGRID also suggests the likelihood of direct/indirect interaction of ORF7b (in dark red) with other viral proteins (ORF3a and M, in blue). The proteins used in the present analysis are among those of the most densely represented central area.
Figure 2.
Interactome of 51 human proteins functionally involved with ORF7b2, enriched with 500 first order proteins. An overall look reveals the involvement of peripheral compact groups of nodes that can represent specific functional modules or even particular protein complexes. Network calculated by STRING and the score is 0.9. The number of edges we have is greater than the number of nodes in a similar random network we can calculate (PPI enrichment p-value < 1.06e-16). We show the topological parameters in
Table 1.
Figure 2.
Interactome of 51 human proteins functionally involved with ORF7b2, enriched with 500 first order proteins. An overall look reveals the involvement of peripheral compact groups of nodes that can represent specific functional modules or even particular protein complexes. Network calculated by STRING and the score is 0.9. The number of edges we have is greater than the number of nodes in a similar random network we can calculate (PPI enrichment p-value < 1.06e-16). We show the topological parameters in
Table 1.
Figure 3.
Node Distribution. The distribution follows a free scale distribution based on the power law. In the inset, we present the same nodes on a log-log scale, with the best fit of data shown in red. The function used for the fit is f(x) = a*xb, where the values of a, b, and R2 are 0.29, -1.89, and 0.62, respectively. A significant p-value of 1.0exp-16 of the interactome analysis and a good correlation index underscore a strong expectation of preferential relations or associations among nodes following their enrichment.
Figure 3.
Node Distribution. The distribution follows a free scale distribution based on the power law. In the inset, we present the same nodes on a log-log scale, with the best fit of data shown in red. The function used for the fit is f(x) = a*xb, where the values of a, b, and R2 are 0.29, -1.89, and 0.62, respectively. A significant p-value of 1.0exp-16 of the interactome analysis and a good correlation index underscore a strong expectation of preferential relations or associations among nodes following their enrichment.
Figure 4.
– Relationships among cytoskeleton related proteins. Network (top left side) - Score 0.7 (high confidence); all seven source channels are active; enrichment of the 8 basic proteins as functional seeds with 100 first-order proteins. Enrichment up to 100 proteins was necessary to achieve integration of all eight proteins into the network without expanding the number of functions too much. Topological data: number of nodes, 108; number of edges, 872; average node degree, 16.1; avg. local clustering coefficient, 0.697; enrichment p-value <1.0e-16.
Figure 4.
– Relationships among cytoskeleton related proteins. Network (top left side) - Score 0.7 (high confidence); all seven source channels are active; enrichment of the 8 basic proteins as functional seeds with 100 first-order proteins. Enrichment up to 100 proteins was necessary to achieve integration of all eight proteins into the network without expanding the number of functions too much. Topological data: number of nodes, 108; number of edges, 872; average node degree, 16.1; avg. local clustering coefficient, 0.697; enrichment p-value <1.0e-16.
Figure 5.
Eigenvector distribution (top); Stress distribution (middle); Betweenness Centrality distribution (bottom). We calculated distributions using Cytoscape with Analyzer and Centiscape. Cross-referencing parametric values have completed the selection of the best proteins in the Cytoscape Node Table for each protein.
Figure 5.
Eigenvector distribution (top); Stress distribution (middle); Betweenness Centrality distribution (bottom). We calculated distributions using Cytoscape with Analyzer and Centiscape. Cross-referencing parametric values have completed the selection of the best proteins in the Cytoscape Node Table for each protein.
Figure 6.
Hub-and-spoke organization of major HUBs in the ORF7b-induced human interactome. By removing all unnecessary nodes from the network in
Figure 2, we extracted this graph. Edge intensity is proportional to the interaction intensity between nodes (calculated by STRING).
Figure 6.
Hub-and-spoke organization of major HUBs in the ORF7b-induced human interactome. By removing all unnecessary nodes from the network in
Figure 2, we extracted this graph. Edge intensity is proportional to the interaction intensity between nodes (calculated by STRING).
Figure 7.
Clustering. The analysis shows ten clusters, all clearly identifiable except for the two central ones. All 10 clusters are statistically significant with p-values < 1.0e-16. In brackets, next to each key hub, there is its degree. We have not highlighted the links between the clusters to make them clearly visible. The Giant Connected Component (GCC) is made by two overlapping central clusters which add up to 206 total nodes, approximately 37% of the entire interactome. Except for clusters 1 and 9, which have distinctive features and require separate treatment, the most crucial parametric information is next to each cluster.
Figure 7.
Clustering. The analysis shows ten clusters, all clearly identifiable except for the two central ones. All 10 clusters are statistically significant with p-values < 1.0e-16. In brackets, next to each key hub, there is its degree. We have not highlighted the links between the clusters to make them clearly visible. The Giant Connected Component (GCC) is made by two overlapping central clusters which add up to 206 total nodes, approximately 37% of the entire interactome. Except for clusters 1 and 9, which have distinctive features and require separate treatment, the most crucial parametric information is next to each cluster.
Figure 8.
Cluster No.1 – 140 nodes, 1110 edges, p-value <1.0e-16. Average node degree 15.4, avg. Local clustering coefficient 0.622 (expected No of edges in a similar random network, 202), network diameter 3, network radius 2, Characteristic path length 1.91, network density 0.108. Main HUB node, EGFR (degree = 123). In red, HUBs previously found in the whole net; in yellow, a bottleneck node.
Figure 8.
Cluster No.1 – 140 nodes, 1110 edges, p-value <1.0e-16. Average node degree 15.4, avg. Local clustering coefficient 0.622 (expected No of edges in a similar random network, 202), network diameter 3, network radius 2, Characteristic path length 1.91, network density 0.108. Main HUB node, EGFR (degree = 123). In red, HUBs previously found in the whole net; in yellow, a bottleneck node.
Figure 9.
Cluster No.9 – 62 nodes, 437 edges, p-value <1.0e-16. Average node degree 14.097, avg. Local clustering coefficient 0.682 (expected No of edges in a similar random network, 83), network diameter 3, network radius 2, Characteristic path length 1.798, network density 0.231. Main HUB node, SRC (degree = 56). In red, some of the principal nodes of this cluster.
Figure 9.
Cluster No.9 – 62 nodes, 437 edges, p-value <1.0e-16. Average node degree 14.097, avg. Local clustering coefficient 0.682 (expected No of edges in a similar random network, 83), network diameter 3, network radius 2, Characteristic path length 1.798, network density 0.231. Main HUB node, SRC (degree = 56). In red, some of the principal nodes of this cluster.
Figure 10.
Transcriptional network (left) and post-transcriptional network (right) of interactions between genes (Hub and bottlenecks) and TFs and miRNAs, respectively. Red circles, genes; azure diamonds, TFs; blue rectangles, miRNAs. Rank of nodes in the networks is high as they undergo filtering based on degree and betweenness values. This is only a schematic view of the most significant molecules and their targets and where the size of the node is proportional to its rank.
Figure 10.
Transcriptional network (left) and post-transcriptional network (right) of interactions between genes (Hub and bottlenecks) and TFs and miRNAs, respectively. Red circles, genes; azure diamonds, TFs; blue rectangles, miRNAs. Rank of nodes in the networks is high as they undergo filtering based on degree and betweenness values. This is only a schematic view of the most significant molecules and their targets and where the size of the node is proportional to its rank.
Figure 11.
Integrated gene regulatory network associated with the dysregulated bottleneck and hub genes. Nodes: red orange circles, hubs; black circles, bottlenecks; green diamonds, TFs; blue rectangles, miRNAs. The figure shows the distribution of the potential gene–TF interactions (center and right side) and gene–miRNA interactions (center and left side). This is only a schematic view of the most significant molecules and their targets. We filtered the interacting network of miRNAs and TFs with betweenness centrality ≥ 100 and 45, respectively.
Figure S9 displays the log-log graph, which confirms a scale-free distribution and shows some topological parameters.
Figure 11.
Integrated gene regulatory network associated with the dysregulated bottleneck and hub genes. Nodes: red orange circles, hubs; black circles, bottlenecks; green diamonds, TFs; blue rectangles, miRNAs. The figure shows the distribution of the potential gene–TF interactions (center and right side) and gene–miRNA interactions (center and left side). This is only a schematic view of the most significant molecules and their targets. We filtered the interacting network of miRNAs and TFs with betweenness centrality ≥ 100 and 45, respectively.
Figure S9 displays the log-log graph, which confirms a scale-free distribution and shows some topological parameters.
Figure 12.
Comparison of three transcriptional networks related to negative metabolic controls because of ORF7b interference. GO analysis (genes in blue, bottlenecks in red). Left side - Negative regulation of transcription, DNA_dependent (p <1,56e-4) (EGFR, HRAS, HSPA5, PIK3CA, PIK3R1, SRC, and ZNF263, ZNF423, SMAD4, MXD3, GABPA, MLX, MXD4, PHF8) Middle - Negative regulation of apoptotic process (p <7.58e-4) (EGFR, HRAS, HSPA5, PIK3CA, PIK3R1, SRC) Right side - Negative regulation of programmed cell death (p <8.86e-4) (EGFR, HRAS, HSPA5, PIK3CA, PIK3R1, SRC).
Figure 12.
Comparison of three transcriptional networks related to negative metabolic controls because of ORF7b interference. GO analysis (genes in blue, bottlenecks in red). Left side - Negative regulation of transcription, DNA_dependent (p <1,56e-4) (EGFR, HRAS, HSPA5, PIK3CA, PIK3R1, SRC, and ZNF263, ZNF423, SMAD4, MXD3, GABPA, MLX, MXD4, PHF8) Middle - Negative regulation of apoptotic process (p <7.58e-4) (EGFR, HRAS, HSPA5, PIK3CA, PIK3R1, SRC) Right side - Negative regulation of programmed cell death (p <8.86e-4) (EGFR, HRAS, HSPA5, PIK3CA, PIK3R1, SRC).
Figure 13.
– Post-transcriptional networks related to negative metabolic controls because of ORF7b interference. GO analysis for Negative regulation of programmed cell death (p <8.28e-6) (EGFR, HRAS, PIK3R1, HSPA5, PIK3CA, SRC).
Figure 13.
– Post-transcriptional networks related to negative metabolic controls because of ORF7b interference. GO analysis for Negative regulation of programmed cell death (p <8.28e-6) (EGFR, HRAS, PIK3R1, HSPA5, PIK3CA, SRC).
Figure 14.
Co-regulated network related to negative metabolic controls because of ORF7b interference. GO analysis for Negative regulation of programmed cell death (p<1.82e-5). (TFs: TP53, MYC; Genes: SRC, EGFR, HRAS, PIK3R1, HSPA5, PIK3CA; miRNA: has-miR-1 and has-miR-576-5p).
Figure 14.
Co-regulated network related to negative metabolic controls because of ORF7b interference. GO analysis for Negative regulation of programmed cell death (p<1.82e-5). (TFs: TP53, MYC; Genes: SRC, EGFR, HRAS, PIK3R1, HSPA5, PIK3CA; miRNA: has-miR-1 and has-miR-576-5p).
| Action |
Enriched terms |
| Biological Process (Gene Ontology): |
1690 GO-terms |
| Molecular Function (Gene Ontology): |
166 GO-terms |
| Cellular Component (Gene Ontology): |
267 GO-terms |
| Reference publications (PubMed): |
>10,000 publications |
| Local network cluster (STRING): |
137 clusters |
| KEGG Pathways: |
195 pathways |
| Reactome Pathways: |
494 pathways |
| WikiPathways: |
259 pathways |
| Disease-gene associations (DISEASES): |
112 diseases |
| Tissue expression (TISSUES): |
186 tissues |
| Subcellular localization (compartments): |
249 compartments significantly |
| Human Phenotype (Monarch): |
1002 phenotypes |
| Annotated Keywords (UniProt): |
99 keywords |
| Protein Domains (Pfam): |
63 domains |
| Protein Domains and Features (InterPro): |
118 domains |
| Protein Domains (SMART): |
20 domains |
| All enriched terms (without PubMed): |
5,057 enriched terms in 15 categories |
Table 3.
Biological Functions.
Table 3.
Biological Functions.
| GO Term ID |
Term description |
Number of involved proteins |
p-value |
| GO:0051179 |
Localization |
378 |
2.01e-77 |
| GO:0006810 |
Transport |
320 |
3.04e-67 |
| GO:0007169 |
Transmembrane receptor protein tyrosine kinase signaling pathway |
124 |
1.23e-66 |
| GO:0051234 |
Establishment of localization |
322 |
7.72e-66 |
| GO:0015833 |
Peptide transport |
187 |
1.09e-62 |
| GO:0051649 |
Establishment of localization in cell |
230 |
3.37e-62 |
| GO:0051641 |
Cellular localization |
254 |
1.29e-60 |
| GO:0015031 |
Protein transport |
181 |
7.86e-60 |
| GO:0007167 |
Enzyme linked receptor protein signaling pathway |
131 |
7.95e-59 |
| GO:0008104 |
Protein localization |
213 |
1.46e-58 |
| GO:0045184 |
Establishment of protein localization |
183 |
2.85e-58 |
| GO:0016192 |
Vesicle-mediated transport |
189 |
1.18e-53 |
| GO:0032879 |
Regulation of localization |
229 |
2.95e-51 |
| GO:0009987 |
Cellular process |
546 |
4.49e-51 |
| GO:0046907 |
Intracellular transport |
168 |
1.19e-49 |
Table 4.
CELLULAR LOCALIZATION OF BIOLOGICAL FUNCTIONS.
Table 4.
CELLULAR LOCALIZATION OF BIOLOGICAL FUNCTIONS.
| GO Term ID |
COMPARTMENT |
Number of involved proteins |
p-value |
| GOCC:0016020 |
Membrane |
399 |
2.58e-92 |
| GOCC:0012505 |
Endomembrane system |
302 |
1.36e-91 |
| GOCC:0031090 |
Organelle membrane |
243 |
2.07e-77 |
| GOCC:0098796 |
Membrane protein complex |
189 |
7.35e-74 |
| GOCC:0005737 |
Cytoplasm |
437 |
1.41e-73 |
| GOCC:0031982 |
Vesicle |
213 |
5.13e-62 |
| GOCC:0098588 |
Bounding membrane of organelle |
174 |
3.17e-58 |
| GOCC:0005783 |
Endoplasmic reticulum |
133 |
6.29e-55 |
| GOCC:0098805 |
Whole membrane |
156 |
1.76e-53 |
| GOCC:0110165 |
Cellular anatomical entity |
531 |
2.59e-51 |
| GOCC:0005789 |
Endoplasmic reticulum membrane |
105 |
4.93e-51 |
| GOCC:0042175 |
Nuclear outer membrane-ER membrane network |
106 |
1.79e-50 |
| GOCC:0031410 |
Cytoplasmic vesicle |
177 |
1.29e-49 |
| GOCC:0032991 |
Protein-containing complex |
306 |
4.76e-44 |
| GOCC:0043226 |
Organelle |
437 |
1.20e-41 |
| GOCC:0043227 |
Membrane-bounded organelle |
406 |
5.80e-41 |
| GOCC:0005622 |
Intracellular |
462 |
8.33e-38 |
| GOCC:0043229 |
Intracellular organelle |
407 |
4.82e-34 |
| GOCC:0005829 |
Cytosol |
201 |
2.18e-32 |
| GOCC:0005886 |
Plasma membrane |
220 |
3.75e-30 |
| GOCC:0031201 |
SNARE complex |
34 |
3.79e-30 |
| GOCC:0043231 |
Intracellular membrane-bounded organelle |
349 |
6.22e-30 |
Table 5.
REACTOME.
| Term ID |
Molecular Mechanism |
Number of involved proteins |
p-value |
| HSA-9006934 |
Signaling by Receptor Tyrosine Kinases |
140 |
4.44e-84 |
| HSA-1643685 |
Disease |
189 |
2.66e-63 |
| HSA-422475 |
Axon guidance |
101 |
6.79e-45 |
| HSA-9675108 |
Nervous system development |
103 |
6.79e-45 |
| HSA-168256 |
Immune System |
176 |
6.06e-41 |
| HSA-5663205 |
Infectious disease |
115 |
6.30e-41 |
| HSA-162582 |
Signal Transduction |
204 |
2.84e-37 |
| HSA-5653656 |
Vesicle-mediated transport |
95 |
2.70e-34 |
| HSA-199991 |
Membrane Trafficking |
92 |
5.34e-34 |
| HSA-392499 |
Metabolism of proteins |
163 |
3.25e-33 |
| HSA-109582 |
Hemostasis |
89 |
1.02e-32 |
| HSA-1799339 |
SRP-dependent cotranslational protein targeting to membrane |
45 |
2.19e-31 |
| HSA-168249 |
Innate Immune System |
111 |
1.33e-30 |
| HSA-1227986 |
Signaling by ERBB2 |
35 |
2.43e-30 |
| HSA-74752 |
Signaling by Insulin receptor |
38 |
5.77e-29 |
| HSA-177929 |
Signaling by EGFR |
33 |
5.40e-28 |
| HSA-4420097 |
VEGFA-VEGFR2 Pathway |
39 |
5.35e-27 |
| HSA-202733 |
Cell surface interactions at the vascular wall |
42 |
3.19e-25 |
| HSA-76002 |
Platelet activation, signaling and aggregation |
52 |
4.89e-24 |
| HSA-6811558 |
PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling |
37 |
5.16e-24 |
| HSA-5683057 |
MAPK family signaling cascades |
52 |
1.19e-20 |
| HSA-5684996 |
MAPK1/MAPK3 signaling |
49 |
1.37e-20 |
| HSA-77387 |
Insulin receptor recycling |
21 |
1.07e-19 |
| HSA-192823 |
Viral mRNA Translation |
27 |
1.54e-16 |
| HSA-1500931 |
Cell-Cell communication |
30 |
1.91e-15 |
Table 6.
HUMAN TISSUES INVOLVED WITH ORF7b.
Table 6.
HUMAN TISSUES INVOLVED WITH ORF7b.
| TERM ID |
HUMAN TISSUES INVOLVED WITH ORF7b |
Number of involved proteins |
p-value |
| BTO:0000345 |
Digestive gland |
233 |
4.73e-56 |
| BTO:0001491 |
Viscus |
322 |
1.59e-54 |
| BTO:0001489 |
Whole body |
504 |
2.18e-45 |
| BTO:0000522 |
Gland |
356 |
2.76e-45 |
| BTO:0000759 |
Liver |
178 |
2.17e-44 |
| BTO:0001488 |
Endocrine gland |
323 |
1.15e-37 |
| BTO:0003091 |
Urogenital system |
341 |
3.03e-36 |
| BTO:0000227 |
Central nervous system |
303 |
1.41e-35 |
| BTO:0001484 |
Nervous system |
307 |
3.23e-35 |
| BTO:0000449 |
Fetus |
125 |
1.68e-32 |
| BTO:0001078 |
Placenta |
119 |
1.40e-30 |
| BTO:0000081 |
Reproductive system |
308 |
5.01e-30 |
| BTO:0003099 |
Internal female genital organ |
183 |
5.01e-30 |
| BTO:0000174 |
Embryonic structure |
159 |
7.75e-28 |
| BTO:0000203 |
Respiratory system |
127 |
9.81e-28 |
| BTO:0000083 |
Female reproductive system |
292 |
1.71e-27 |
| BTO:0000089 |
Blood |
136 |
1.39e-26 |
| BTO:0000570 |
Hematopoietic system |
172 |
6.39e-26 |
| BTO:0000763 |
Lung |
105 |
3.59e-23 |
| BTO:0000988 |
Pancreas |
72 |
6.06e-23 |
| BTO:0000431 |
Excretory gland |
106 |
8.44e-21 |
| BTO:0003092 |
Urinary system |
97 |
3.43e-19 |
| BTO:0001244 |
Urinary tract |
97 |
4.17e-19 |
| BTO:0000671 |
Kidney |
86 |
5.49e-19 |
| BTO:0001129 |
Prostate gland |
58 |
8.08e-19 |
| BTO:0000132 |
Blood platelet |
50 |
2.81e-18 |
| BTO:0000511 |
Gastrointestinal tract |
116 |
4.50e-17 |
| BTO:0000131 |
Blood plasma |
51 |
1.46e-16 |
| BTO:0000574 |
Hematopoietic cell |
77 |
1.21e-14 |
| BTO:0000082 |
Male reproductive system |
148 |
3.21e-14 |
| BTO:0000751 |
Leukocyte |
72 |
3.31e-14 |
| BTO:0000080 |
Male reproductive gland |
138 |
6.39e-13 |
| BTO:0000254 |
Female reproductive gland |
145 |
7.61e-12 |
| BTO:0005810 |
Immune system |
96 |
3.68e-11 |
| BTO:0003096 |
Internal male genital organ |
122 |
4.92e-11 |
| BTO:0000088 |
Cardiovascular system |
70 |
4.40e-10 |
| BTO:0000421 |
Connective tissue |
63 |
1.19e-09 |
| BTO:0000439 |
Eye |
59 |
1.33e-09 |
| BTO:0000706 |
Large intestine |
54 |
1.57e-09 |
| BTO:0000202 |
Sense organ |
69 |
1.98e-09 |
| BTO:0000855 |
Lymph |
25 |
4.56e-09 |
| BTO:0001085 |
Vascular system |
38 |
9.90e-09 |
| BTO:0001424 |
Uterus |
67 |
1.11e-08 |
| BTO:0000269 |
Colon |
46 |
3.05e-08 |
| BTO:0001363 |
Testis |
85 |
2.48e-05 |
| Pathway |
Description |
Number of involved proteins |
p-value |
| hsa04012 |
ErbB signaling pathway |
50 |
3.02e-41 |
| hsa04510 |
Focal adhesion |
64 |
2.27e-40 |
| hsa01521 |
EGFR tyrosine kinase inhibitor resistance |
46 |
4.29e-38 |
| hsa04151 |
PI3K-Akt signaling pathway |
74 |
1.16e-36 |
| hsa04141 |
Protein processing in ER |
55 |
1.44e-35 |
| hsa04015 |
Rap1 signaling pathway |
51 |
3.72e-28 |
| hsa04014 |
Ras signaling pathway |
52 |
3.76e-27 |
| hsa05206 |
MicroRNAs in cancer |
45 |
1.48e-26 |
| hsa04935 |
Growth hormone synthesis, secretion action |
40 |
3.80e-26 |
| hsa04130 |
SNARE interactions in vesicular transport |
27 |
1.38e-25 |
| hsa04062 |
Chemokine signaling pathway |
45 |
2.35e-24 |
| hsa04145 |
Phagosome |
40 |
9.33e-24 |
| hsa04360 |
Axon guidance |
43 |
2.15e-23 |
| hsa04072 |
Phospholipase D signaling pathway |
39 |
2.11e-22 |
| hsa04917 |
Prolactin signaling pathway |
30 |
2.84e-22 |
| hsa04150 |
mTOR signaling pathway |
39 |
4.38e-22 |
| hsa04810 |
Regulation of actin cytoskeleton |
42 |
3.07e-20 |
| hsa01522 |
Endocrine resistance |
31 |
4.10e-20 |
| hsa04915 |
Estrogen signaling pathway |
35 |
4.10e-20 |
| hsa04722 |
Neurotrophin signaling pathway |
33 |
4.29e-20 |
| hsa04919 |
Thyroid hormone signaling pathway |
32 |
9.72e-19 |
| hsa04664 |
Fc epsilon RI signaling pathway |
26 |
1.16e-18 |
| hsa04010 |
MAPK signaling pathway |
45 |
5.16e-18 |
| hsa04721 |
Synaptic vesicle cycle |
26 |
1.05e-17 |
| hsa04660 |
T cell receptor signaling pathway |
29 |
1.05e-17 |
| hsa04662 |
B cell receptor signaling pathway |
26 |
2.90e-17 |
| hsa04650 |
Natural killer cell mediated cytotoxicity |
30 |
7.46e-17 |
Table 8.
The table shows the dysregulated processes in which these proteins are involved. Strength is in logs and for P (see Methods). The colors are reflected in the metabolic characteristics expressed by the various nodes in
Figure 4.
Table 8.
The table shows the dysregulated processes in which these proteins are involved. Strength is in logs and for P (see Methods). The colors are reflected in the metabolic characteristics expressed by the various nodes in
Figure 4.
Table 9.
Involvement of HUBs and Bottlenecks in the control of Biological Processes (GO).
Table 9.
Involvement of HUBs and Bottlenecks in the control of Biological Processes (GO).
| HUB protein |
Number of GO Processes |
Bottleneck protein |
Number of GO Processes |
| EGFR |
408 |
EGFR |
408 |
| PIK3R1 |
328 |
HSPA5 |
234 |
| EGF |
646 |
MTOR |
413 |
| HRAS |
245 |
SEC13 |
83 |
| GRB2 |
233 |
SEC61A1 |
63 |
| SRC |
508 |
SRC |
508 |
| PIK3CA |
271 |
VAMP2 |
143 |
| Viral Protein |
Human target |
Viral protein features** |
| nsp4* |
EGFR |
Is involved in the assembly of virally induced cytoplasmic double- membrane vesicles necessary for viral replication. |
| M* |
EGFR |
Component of the viral envelope. |
| ORF3a* |
EGFR |
Homotetrameric potassium sensitive ion channels (viroporin) and may modulate virus release |
| ORF7b* |
EGFR |
This paper |
| S |
EGFR |
Spike or Surface glycoprotein |
| |
|
|
| nsp4* |
SRC |
See above |
| nsp5* |
SRC |
Is a cysteine protease, essential for the viral life cycle. |
| nsp6* |
SRC |
Plays a role in the initial induction of auto-phagosomes from host reticulum endoplasmic |
| nsp13* |
SRC |
Multi-functional helicase with a zinc-binding domain in N-terminus |
| nsp14* |
SRC |
3'-5' deoxyribonuclease |
| E* |
SRC |
Plays a central role in virus morphogenesis and assembly |
| M* |
SRC |
See above |
| ORF3a* |
SRC |
See above |
| ORF3b |
SRC |
Could be involved in immune evasion as interferon agonist (78) |
| ORF6* |
SRC |
Could be a determinant of virus virulence |
| ORF7a* |
SRC |
Non-structural protein, which is dispensable for virus replication in cell culture |
| ORF7b* |
SRC |
See above |
| ORF8 |
SRC |
Is a viral cytokine regulating immune responses |
| S |
SRC |
See above |
| |
|
|
| M* |
PIK3R1 |
See above |
| ORF7b* |
PIK3R1 |
See above |
| ORF3b |
PIK3R2 |
See above |
| M* |
PIK3R3 |
See above |
| S |
PIK3R3 |
See above |
| |
|
|
| N* |
ORF7b |
Responsible for wrapping viral RNA into a symmetric helical structure |
Table 11.
Common altered pathways.
Table 11.
Common altered pathways.
| Function |
Strength* |
p-value |
P |
Human Proteins involved in the process |
| Negative regulation of ERBB signaling pathway |
1.22 |
1.38e-18 |
22.13 |
HBEGF, EREG, PTPN12, TSG101, CBL, CBLB, EGF, ERBB2, CBLC, EGFR, TGFA, SOCS5, PTPN2, HGS, EPS15, ERRFI1, SNX5, SH3GL2, GRB2, BTC, AREG, SH3KBP1, CDC42, EPN1, EPGN |
| Negative regulation of EGFR signaling pathway |
1.23 |
3.24e-17 |
21.53 |
HBEGF, EREG, TSG101, CBL, CBLB, EGF, CBLC, EGFR, TGFA, SOCS5, PTPN2, HGS, EPS15, ERRFI1, SNX5, SH3GL2, GRB2, BTC, AREG, SH3KBP1, CDC42, EPN1, EPGN |
| Negative regulation of anoikis |
1.14 |
5.72e-05 |
6.56 |
PIK3CA, ITGA5, BCL2L1, CAV1, PTK2, SRC, ITGB1 |
| Negative regulation of extrinsic apoptotic signaling pathway |
0.76 |
9.26e-07 |
6.05 |
GCLC, LGALS3, BCL2L1, IGF1, CTNNA1, UNC5B, FYN, FAS, CASP8, LMNA, GCLM, SRC, AR, CTTN, NRG1, ITGA6, AKT1
|
| Negative regulation of protein tyrosine kinase activity |
0.99 |
9.13e-05 |
5.90 |
TSG101, CBL, CBLB, CBLC, SOCS5, PTPN2, CAV1, ERRFI1 |
| Negative regulation of epidermal growth factor-activated receptor activity |
1.18 |
1.7e-04 |
4.99 |
TSG101, CBL, CBLB, CBLC, SOCS5, ERRFI1 |
| Negative regulation of interleukin-6 production |
0.81 |
5.0e-05 |
4.61 |
CSK, SOCS5, GAS6, TLR9, VIMP, PTPN6, ARRB1, ENSP00000417517 |
| Negative regulation of peptidyl-tyrosine phosphorylation |
0.88 |
1.50e-05 |
4.55 |
TSG101, CBL, CBLB, CBLC, SPINK1, SOCS5, PTPN2, CAV1, ERRFI1, PRKCD, PTPN6 |
| Negative regulation of PERK-mediated unfolded protein response |
1.33 |
9.2e-03 |
4.12 |
NCK2, PTPN1, NCK1 |
| Negative regulation of endoplasmic reticulum unfolded protein response |
1.04 |
8.9e-03 |
4.11 |
NCK2, HSPA5, PTPN1, NCK1 |
| Negative regulation of blood-brain barrier permeability |
1.55 |
3.13e-02 |
3.88 |
SH3GL2, VEGFA |
| Negative regulation of response to oxidative stress |
0.81 |
4.1e-04 |
3.74 |
SLC7A11, MET, GGT7, CTNNB1, FYN, NFE2L2, INS, HIF1A, AKT1
|
| Negative regulation of protein tyrosine phosphatase activity |
1.39 |
4.67e-02 |
3.71 |
LGALS3, GNAI2 |
| Negative regulation of mesenchymal to epithelial transition |
1.38 |
4.77e-02 |
3.69 |
CTNNB1, STAT1 |
| Negative regulation of blood coagulation |
0.83 |
2.8e-04 |
3.69 |
PROC, PDGFRA, F2, PLAUR, PLAU, EDN1, CD9, PROS1, PRKCD |
| Negative regulation of primary miRNA processing |
1.38 |
4.67e-02 |
3.68 |
STAT3, IL6 |
| Negative regulation of lipid transport |
0.79 |
1.74e-02 |
1.77 |
EGF, PTPN11, SREBF2, AKT1, ITGB3 |
Table 12.
Operational cellular compartments of cluster No1 proteins.
Table 12.
Operational cellular compartments of cluster No1 proteins.
| COMPARTMENT |
PROTEINS* |
Protein number |
| EXTRACELLULAR |
AREG, BTC, CD81, CD9, EGF, EGFR, ERBB3, EREG, HBEGF, HSPA8, INS, LAMA1, LAMB1, MUC1, NRG1, NRG3, PLAU, SFN, TGFA, TSG101
|
20 |
| CYTOSKELETON |
CTNNA1, CTNNB1, GNAI1, GNAI3, LMNA, MAPK1, PPP2R1A, PTPN3
|
8 |
| PLASMA MEMBRANE |
ADAM17, ARF4, BTC, CAV1, CAV2, CD44, CD81, CD82, CDH1, CTNNA1, CTNNB1, EDNRA, EGFR, EPS15, ERBB2, ERBB2IP, ERBB3, ERBB4, EREG, GAB2, GNAI1, GNAI3, HBEGF, HCK, HRAS, ITGA3, ITGB1, ITGB4, JUP, KRAS, LAPTM4B, LPAR1, LPAR3, LYN, MUC1, NRG1, NRG3, PDGFRA, PIK3C2B, PLCG1, PLCG2, PPP2R1A, PRKCA, PRKCB, PTPN2, PTPN3, PTPRK, PTRF, SHC1, SLC9A1, SLC9A3R1, TGFA, TSG101, USP8
|
54 |
| CYTOSOL |
PIK3C2B, GRB7, ARF4, PPP2R1A, PLCG1, HCK, USP8, PRKCA, MAPK1, RAB5A, FOS, HSPA8, CTNNB1, HIF1A, GAPDH |
15 |
| MITOCHONDRION |
PPP2R1A, MAPK1, HSP90AA1,LGALS3, ERBB4, PTRF, MT-CO2 |
7 |
| GOLGI |
CAV2, CBL, CDH1, HRAS, LYN, MAPK1
|
5 |
| ER |
FOS, NCK1, PTPN2
|
3 |
| PEROXISOME |
No level 5 protein |
- |
| ENDOSOME |
CDH1, EGFR, CAV1, ERBB2, PTPN1, RAB5A, MAPK1, TSG101, GRB2, HGS, USP8, LPAR1, LAPTM4B, GRAP2
|
14 |
| LYSOSOME |
LAPTM4B, HSPA8, MTOR, HCK
|
4 |
| NUCLEUS |
CAV2,CTNNB1, EGFR, ERBB2, ERBB2IP, ERBB4, FOS, GRAP2, GRB2, HIF1A, HRAS, HSPA8, IGFBP3, JAK2, LGALS3, LMNA, LYN, MAPK1, MUC1, NCK1, NCL, NRG1, PLCG1, PPP2R1A, PRKCB, PRKDC, PTPN11, PTPN2, PTPN6, PTRF, STAT1, STAT3, STAT5B, TFAP2C |
34 |
Table 13.
Operational cellular compartments of cluster No9 proteins.
Table 13.
Operational cellular compartments of cluster No9 proteins.
| COMPARTMENT |
PROTEINS* |
Protein number |
| EXTRACELLULAR |
EDN1, F2, FAS, HSP90AB1, LAMA5, LAMC1, MET, NTN1, VEGFA
|
9 |
| CYTOSKELETON |
CDC42, CTNNB1, CTTN, LMNA, MAPK3, PTK2, PXN, YES1 |
8 |
| PLASMA MEMBRANE |
AKT1, ARF6, CASP8, CAV1, CDC42, CDH1, CDH2, CTNNB1, CTNND1, EFNA5, EFNB2, EPHA1, EPHA2, ESR1, FAS, HRAS, IGF1R, ITGB3, MET, NEDD4, PDGFRB, PECAM1, PRKCD, PTK2, PTK2B, PXN, RAC1, RHOA, SRC, TIAM1, TJP1, YES1 |
32 |
| CYTOSOL |
AKT1, ARF6, CASP8, CTNNB1, MAPK3, PRKCD, PTK2, RHOA, SRC, YES1, |
10 |
| MITOCHONDRION |
GJA1, HSP90AA1, MAP2K1, MAPK3, SRC
|
5 |
| GOLGI |
CBL, CDH1, ESR1, HRAS, MAP2K1, MAPK3, NEDD4, RAC1, YES1 |
9 |
| ER |
PRKCD, MAP2K1 |
2 |
| PEROXISOME |
No level 5 protein |
- - |
| ENDOSOME |
ARF6, CAV1, CDH1, MAP2K1, MAPK3, PRKCD, RAC1, SRC
|
8 |
| LYSOSOME |
PDGFRB, PRKCD, SRC
|
3 |
| NUCLEUS |
AKT1, AR, ARRB1, CTNNB1, ESR1, GJA1, HRAS, HSP90AB1, ITGB3, LMNA, MAP2K1, MAPK8, NEDD4, PGR, PRKCD, PTK2, PTK2B, RAC1, STAT3
|
19 |