Methods
In this simulation study, all dose escalation/de-escalation boundary tables were calculated using the
get.bounddary() function from R package
BOIN [
8].
tryCatch() function was used to handle
get.bounddary() errors such as “the probability deemed safe cannot be higher than or too close to the target!” (line 13-23 in the example script 1a). All
p.saf and
p.tox values that produce the same BOIN boundary table (
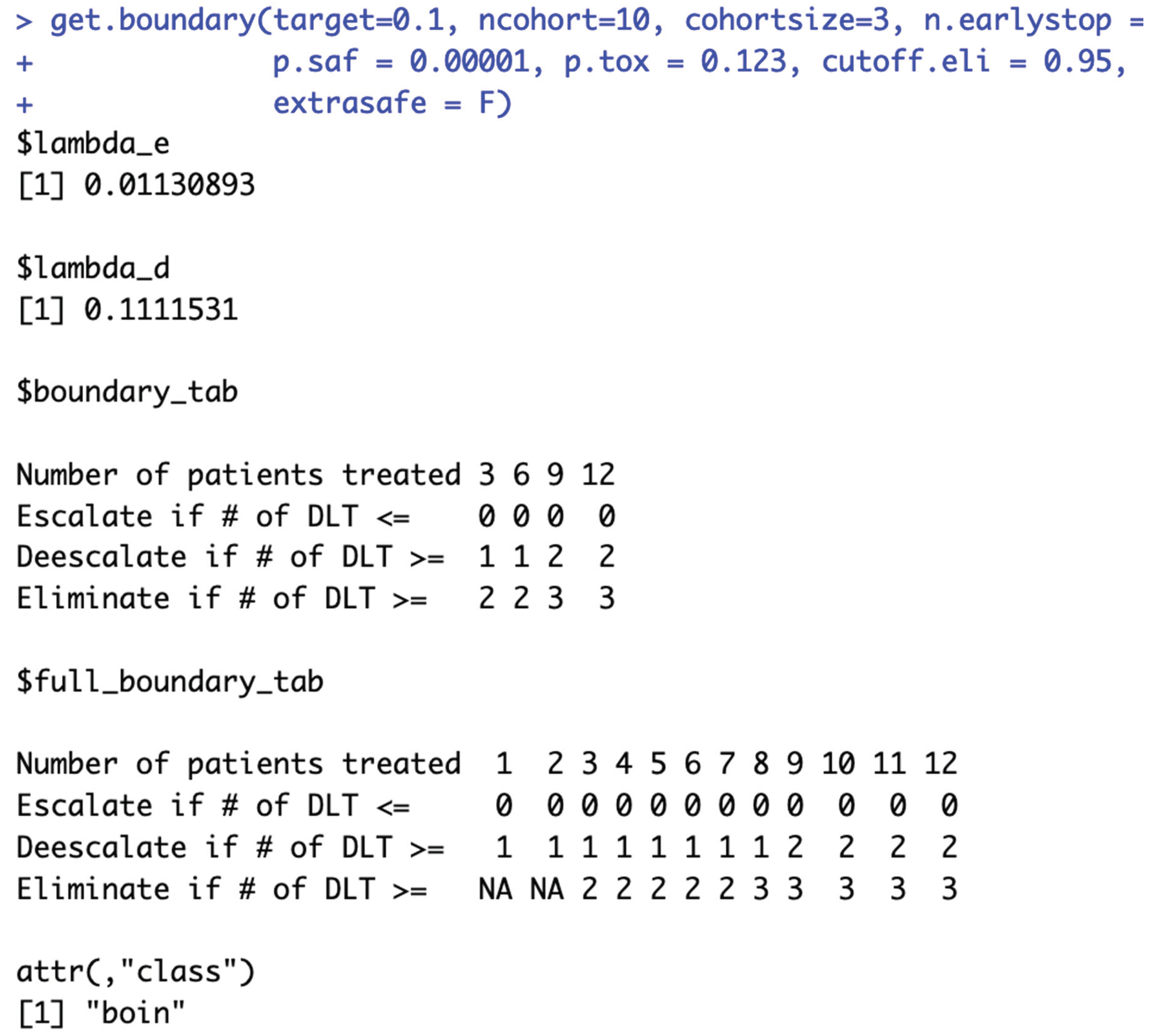
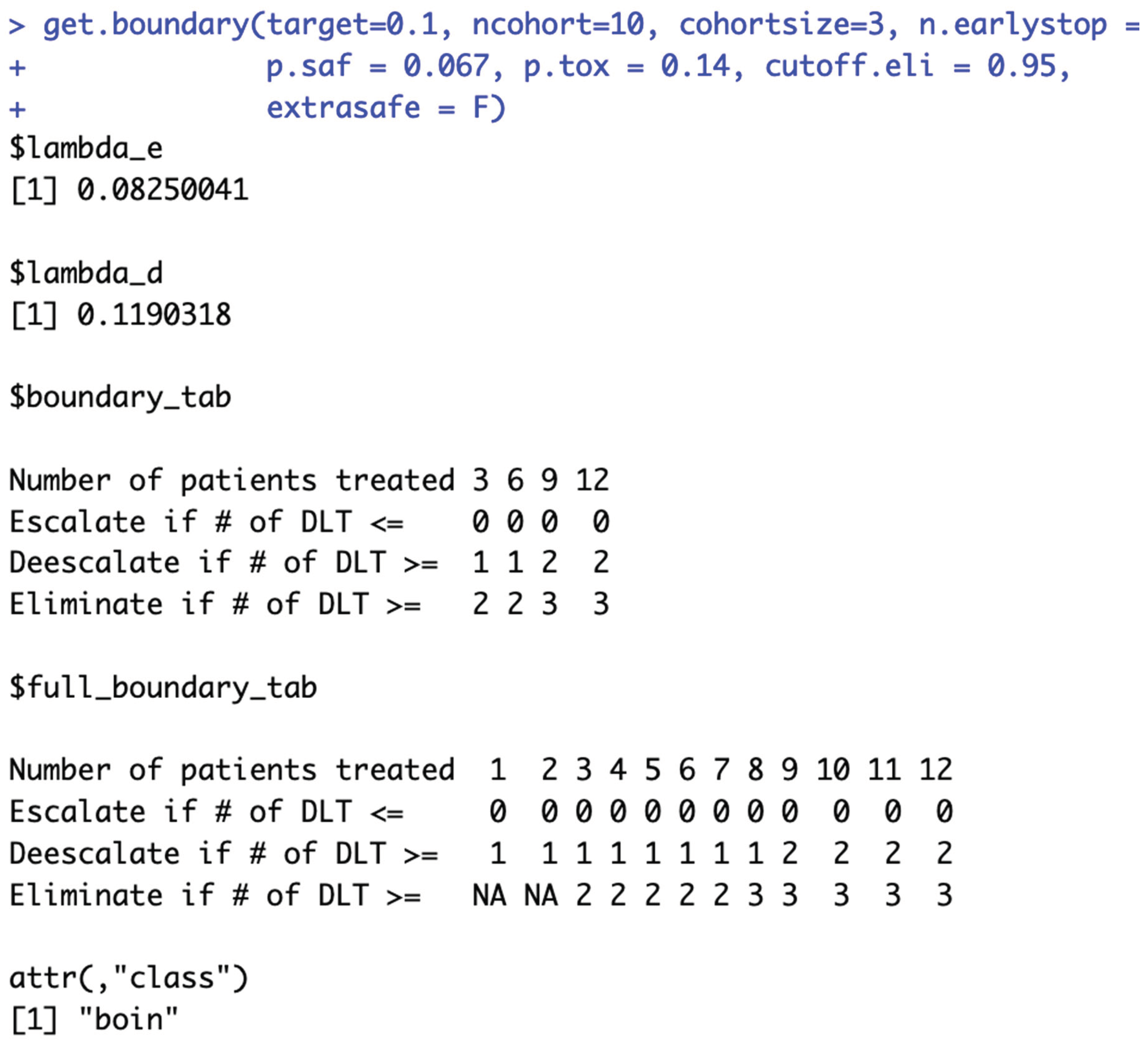
i.e., the same
$boundary_tab output) are considered equivalent (line 72-163 in example script 1b). It’s worth noting that while the
$boundary_tab output is the same, the
$full_boundary_tab outputs may be different (
Figure 2b).
Equivalent p.saf and p.tox under varying target DLT rates:
Equivalent values of p.saf and p.tox were explored via uniform search under varying target DLT rates: target = 10%, 15%, 20%, 25%, 30%, 35%, or 40% (line 26 in the example script 1a) and fixed values of following design parameters (line 15-17 in the example script 1a):
ncohort = 10
cohortsize = 3
n.earlystop = 12
cutoff.eli = 95%
extrasafe = FALSE
For each target DLT rate under evaluation, 100,000 pairs of p.saf and p.tox values were randomly drawn from the following uniform distributions (line 30-31, 55-56 in the example script 1a):
p.saf <- runif(1, min=0, max=target-0.0000001)
p.tox <- runif(1, min=target+0.0000001, max=1)
Equivalent p.saf and p.tox under varying n.earlystop:
Equivalent values of p.saf and p.tox were explored via uniform search under varying n.earlystop = 15, 18, 21, 24, 27, or 30 (line 26 in the example script 2a) and fixed values of following design parameters (line 15-17 in the example script 2a):
For each n.earlystop value, 100,000 pairs of p.saf and p.tox values were randomly drawn from the following uniform distributions (line 30-31, 55-56 in the example script 2a):
p.saf <- runif(1, min=0, max=target-0.0000001)
p.tox <- runif(1, min=target+0.0000001, max=1)
Equivalent p.saf and p.tox under varying cutoff.eli:
Equivalent values of p.saf and p.tox were explored via uniform search under varying cutoff.eli = 70%, 80%, 90%, 97%, or 99% (line 26 in the example script 3a) and fixed values of following design parameters (line 15-17 in the example script 3a):
For each cutoff.eli value, 100,000 pairs of p.saf and p.tox values were randomly drawn from the uniform distributions described in the previous sections (line 30-31, 55-56 in the example script 3a).
Equivalent p.saf and p.tox under varying cohortsize:
Equivalent values of p.saf and p.tox were explored via uniform search under varying cohortsize = 4, 5, 6, 7, or 8 (line 26 in the example script 4a) and fixed values of following design parameters (line 15-17 in the example script 4a):
For each cohortsize, 100,000 pairs of p.saf and p.tox values were randomly drawn from the uniform distributions described in the previous sections (line 30-31, 55-56 in the example script 4a).
Equivalent p.saf and p.tox under varying ncohort:
Equivalent values of p.saf and p.tox were explored via uniform search under varying ncohort = 5, 6, 7, 8, 9, 12, 20, 40, or 100 (line 23 in the example script 5a) and fixed values of following design parameters (line 12-14 in the example script 5a):
For each ncohort value, 100,000 pairs of p.saf and p.tox values were randomly drawn from the uniform distributions described in the previous sections (line 27-28, 52-53 in the example script 5a).
Equivalent BOIN parameter sets for generating the same 3+3 design table
Equivalent values of BOIN design parameters were explored via uniform search under varying offset = 0.01, 0.05, 0.1, 0.2, 0.3, 0.4, or 0.49 (line 27 in the example script 6a) and fixed values of following design parameters (line 16-18 in the example script 6a):
cohortsize = 3
ncohort = 10
n.earlystop = 6
extrasafe = TRUE
For each offset value, 100,000 sets of target (target DLT rate), p.saf, p.tox, and cutoff.eli values were randomly drawn from the following uniform distributions (line 31-34, 57-60 in the example script 6a):
target <- runif(1,min=0,max=0.5)
p.saf <- runif(1,min=0,max=target-0.0000001)
p.tox <- runif(1,min=target+0.0000001,max=1)
cutoff.eli <- runif(1,min=0,max=1)
All sets of
BOIN design parameters that can produce the 3+3 design table shown in
Table 1 are considered equivalent (line 8, 65-68 in example script 6a):
To help clarify all methodological details and ensure reproducibility of this work, all simulation scripts and the outputs of these scripts are made available in a publicly accessible code repository. These scripts can also be modified to facilitate simulation studies of other choices of BOIN design parameters. For example, simply replace the 3+3 design table in line 8 of example script 6a with any boundary table of interest, the updated example script 6a can be used to search all equivalent sets of BOIN design parameters that can produce this new boundary table of interest.
Results
Results of equivalent p.saf and p.tox under different target DLT rates
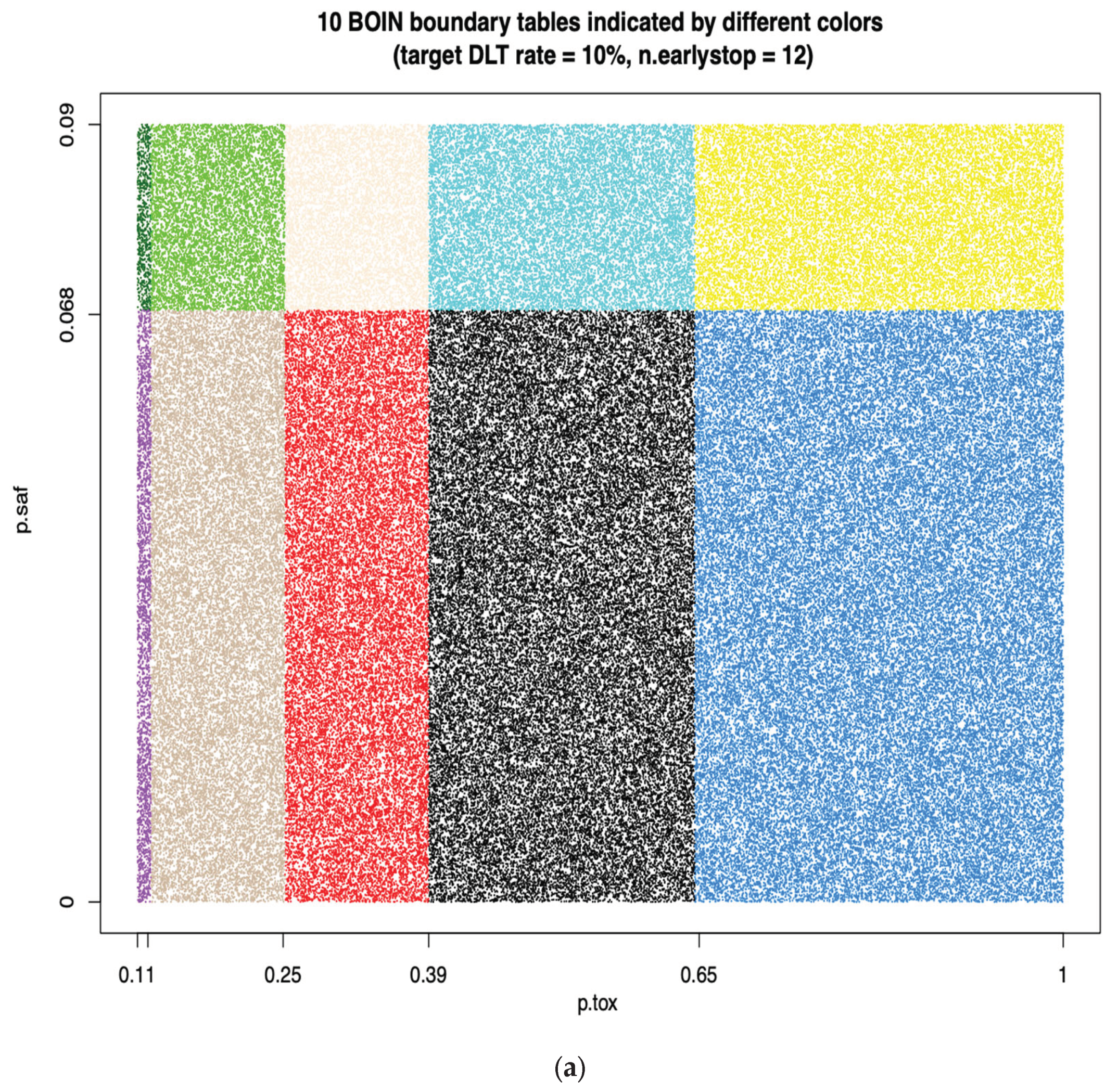
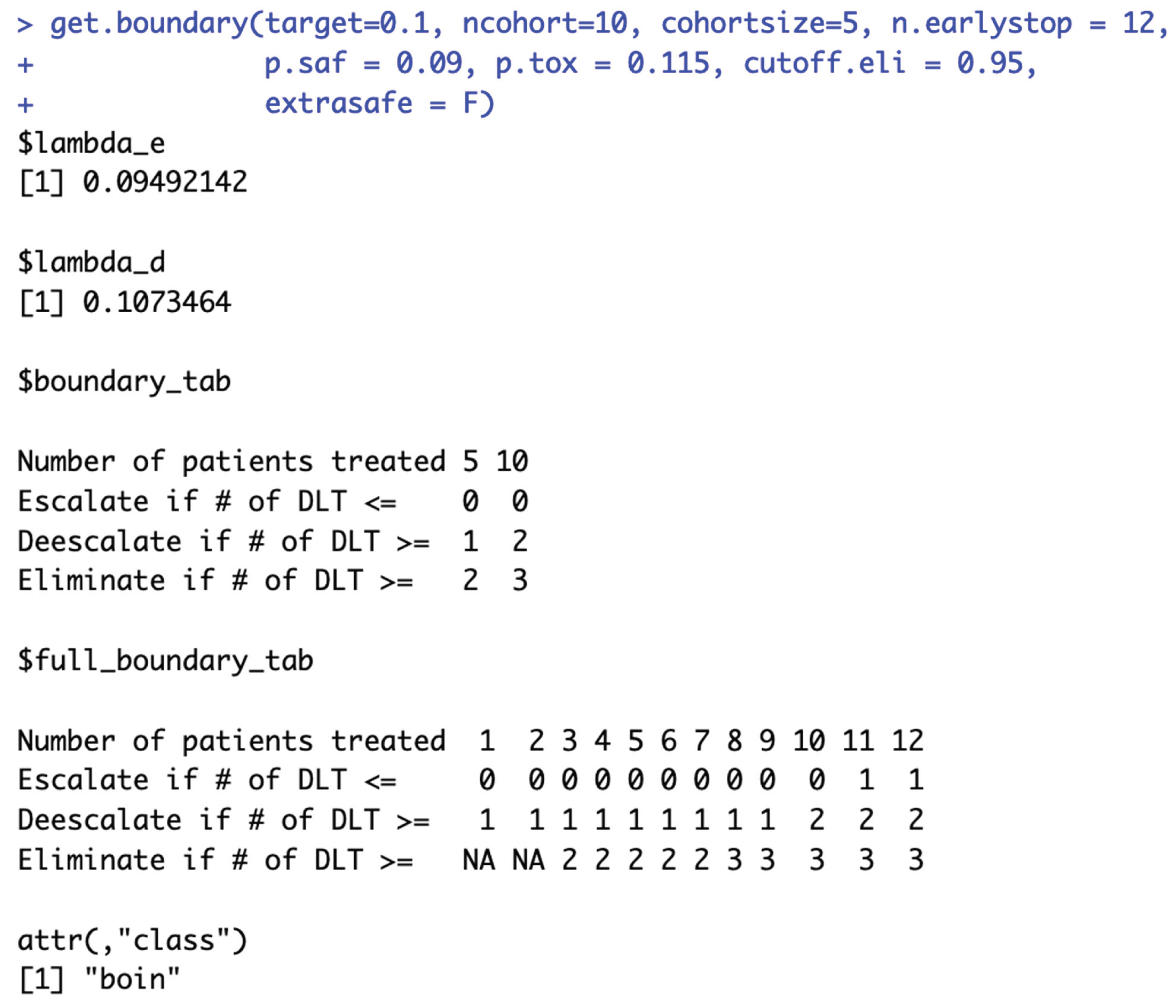
When target DLT rate = 10%,
cohortsize = 3,
ncohort = 10,
n.earlystop = 12,
cutoff.eli = 95%, and
extrasafe = FALSE, there are total 10 possible BOIN boundary tables, regardless of the choices of
p.saf and
p.tox. Equivalent intervals of
p.saf and
p.tox are visualized in
Figure 2a, using different colors to indicate different boundary tables. In total, there are 2 equivalent intervals of
p.saf in this setting: (0, 6.8%) and (6.8%, 9%), and 5 equivalent intervals of
p.tox: (11%, 12.3%), (12.3%, 25.2%), (25.2%, 39%), (39%, 64.6%), and (64.6%, 99.9%). Therefore, when target DLT rate = 10%,
cohortsize = 3,
ncohort = 10,
n.earlystop = 12,
cutoff.eli = 95%, and
extrasafe = FALSE, using
p.tox = 1.4 * target.DLT.rate to calculate BOIN boundary table is equivalent to using any p.tox
(12.3%, 25.2%), as long as
p.saf values fall into one equivalent interval of
p.saf.
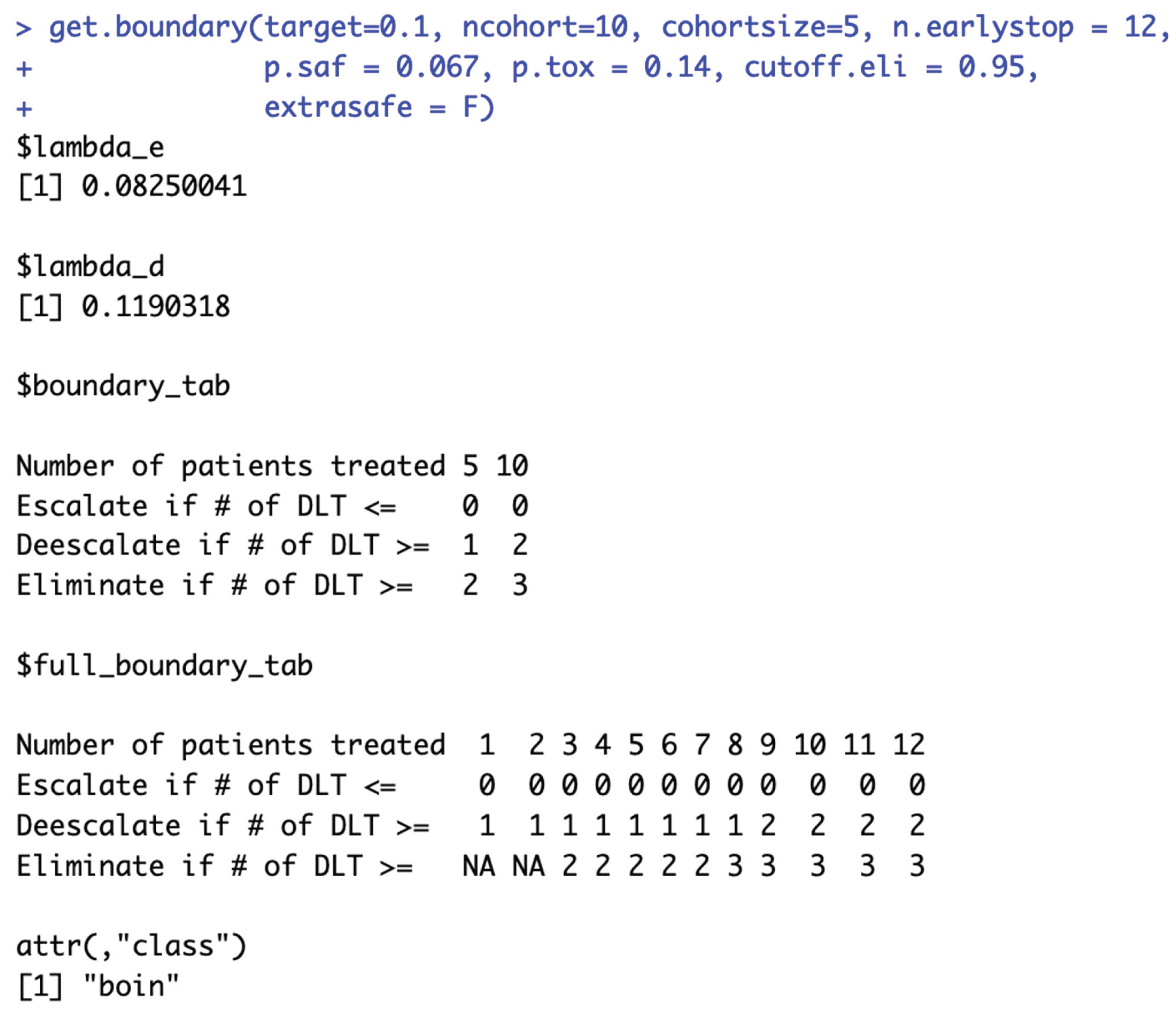
Figure 2b provides a few validation examples of the statement above. For detailed summary of the equivalent intervals reported above and their corresponding BOIN boundary tables, please see SupTable 2a.
When target DLT rate = 15%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 95%, and extrasafe = FALSE, there are total 15 possible BOIN boundary tables, regardless of the choices of p.saf and p.tox. Equivalent intervals of p.saf and p.tox are visualized in SupFig 2c, using different colors to indicate different boundary tables. In total, there are 3 equivalent intervals of p.saf in this setting: (0, 3.9%), (3.9%, 7.9%) and (7.9%, 13.5%), and 5 equivalent intervals of p.tox: (16.5%, 18.4%), (18.4%, 30.8%), (30.8%, 37.2%), (37.2%, 56%), and (56%, 99.9%). Therefore, when target DLT rate = 15%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 95%, and extrasafe = FALSE, using p.tox = 1.4 * target.DLT.rate to calculate BOIN boundary table is equivalent to using any p.tox (18.4%, 30.8%), as long as p.saf values fall into one equivalent interval of p.saf. And using p.tox = 33% to calculate BOIN boundary table is equivalent to using any p.tox (30.8%, 37.2%). For detailed summary of these equivalent intervals and their corresponding BOIN boundary tables, please see SupTable 2c.
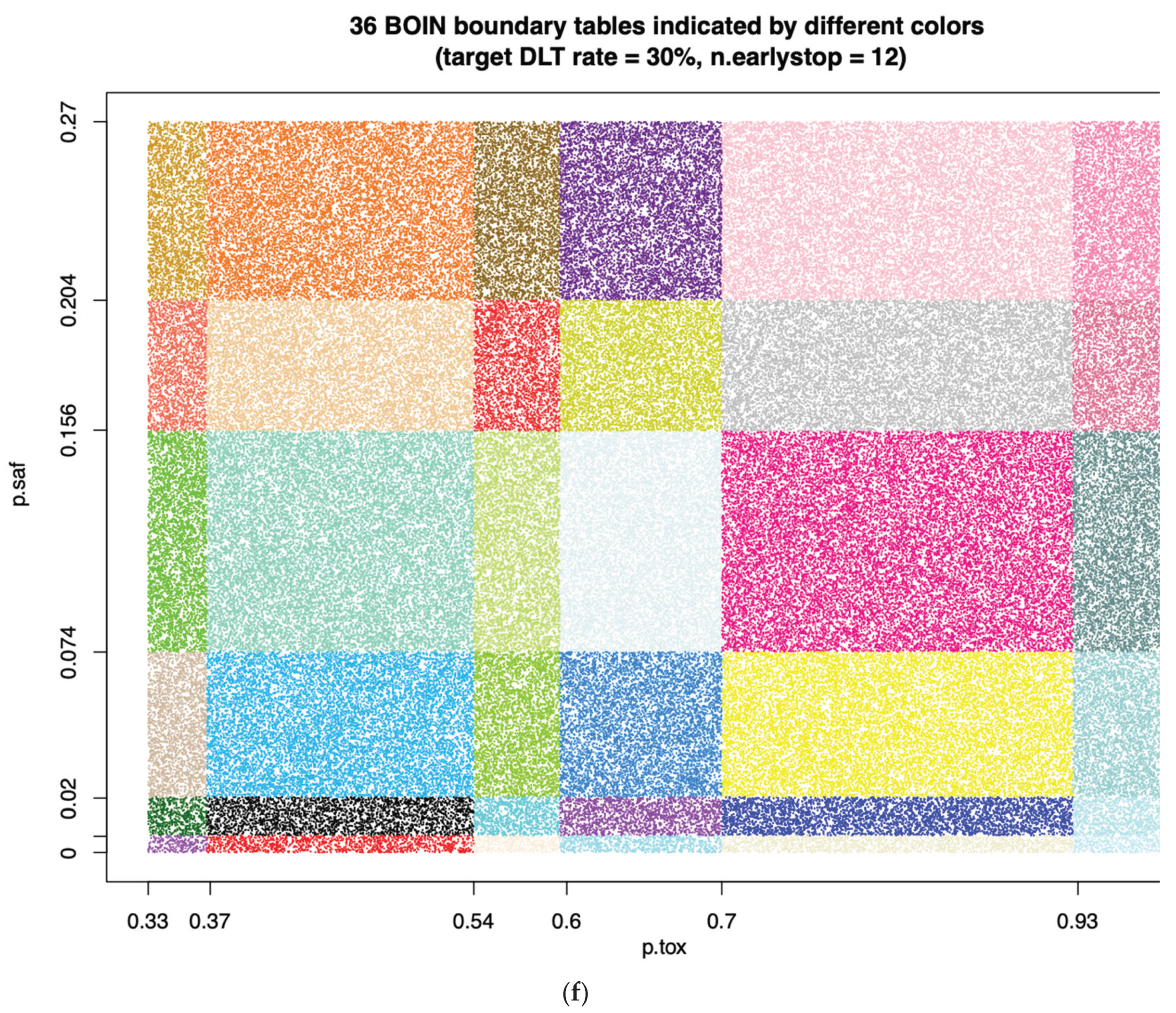
(12.3%, 25.2%), as long as p.saf values fall into one equivalent interval of p.saf: (0, 6.8%) or (6.8%, 9%). f: equivalent intervals of p.saf and p.tox under target DLT rate = 30%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 95%, and extrasafe = FALSE.
When target DLT rate = 20%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 95%, and extrasafe = FALSE, there are total 16 possible BOIN boundary tables, regardless of the choices of p.saf and p.tox. Equivalent intervals of p.saf and p.tox are visualized in SupFig 2d, using different colors to indicate different boundary tables. In total, there are 4 equivalent intervals of p.saf in this setting: (0, 2.2%), (2.2%, 5.1%), (5.1%, 13.7%) and (13.7%, 18%), and 4 equivalent intervals of p.tox: (22%, 24.6%), (24.6%, 30.5%), (30.5%, 48.8%), and (48.8%, 99.9%). Therefore, when target DLT rate = 20%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 95%, and extrasafe = FALSE, using p.tox = 33% to calculate BOIN boundary table is equivalent to using any p.tox (30.5%, 48.8%), as long as p.saf values fall into one equivalent interval of p.saf. For detailed summary of these equivalent intervals and their corresponding BOIN boundary tables, please see SupTable 2d.
When target DLT rate = 25%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 95%, and extrasafe = FALSE, there are total 30 possible BOIN boundary tables, regardless of the choices of p.saf and p.tox. Equivalent intervals of p.saf and p.tox are visualized in SupFig 2e, using different colors to indicate different boundary tables. In total, there are 5 equivalent intervals of p.saf in this setting: (0, 1.2%), (1.2%, 3.3%), (3.3%, 10.1%), (10.1%, 19.6%), and (19.6%, 22.5%), and 6 equivalent intervals of p.tox: (27.5%, 42.4%), (42.4%, 59.7%), (59.7%, 65.1%), (65.1%, 75%), (75%, 94.8%), and (94.8%, 99.9%). Therefore, when target DLT rate = 25%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 95%, and extrasafe = FALSE, using p.tox = 1.4 * target.DLT.rate to calculate BOIN boundary table is equivalent to using any p.tox (27.5%, 42.4%), as long as p.saf values fall into one equivalent interval of p.saf. For detailed summary of these equivalent intervals and their corresponding BOIN boundary tables, please see SupTable 2e.
When target DLT rate = 30%,
cohortsize = 3,
ncohort = 10,
n.earlystop = 12,
cutoff.eli = 95%, and
extrasafe = FALSE, there are total 36 possible BOIN boundary tables, regardless of the choices of
p.saf and
p.tox. Equivalent intervals of
p.saf and
p.tox are visualized in
Figure 2f, using different colors to indicate different boundary tables. In total, there are 6 equivalent intervals of
p.saf in this setting: (0, 0.6%), (0.6%, 2%), (2%, 7.4%), (7.4%, 15.6%), (15.6%, 20.4%), and (20.4%, 27%), and 6 equivalent intervals of
p.tox: (33%, 36.8%), (36.8%, 54%), (54%, 59.6%), (59.6%, 70%), (70%, 92.7%), and (92.7%, 99.9%). Therefore, in this setting, using
p.tox = 1.4 * target.DLT.rate to calculate BOIN boundary table is equivalent to using any p.tox
(36.8%, 54%), as long as
p.saf values fall into one equivalent interval of
p.saf. For detailed summary of these equivalent intervals and their corresponding BOIN boundary tables, please see SupTable 2f. For results of equivalent intervals of
p.saf and
p.tox under target DLT rate = 35% and 40% in similar settings, please see SupFig 2g, SupTable 2g, SupFig 2h, and SupTable 2h.
Results of equivalent p.saf and p.tox under different n.earlystop
When target DLT rate = 10%,
cohortsize = 3,
ncohort = 10,
n.earlystop = 15,
cutoff.eli = 95%, and
extrasafe = FALSE, there are total 21 possible BOIN boundary tables, regardless of the choices of
p.saf and
p.tox. Equivalent intervals of
p.saf and
p.tox are visualized in SupFig 3a, using different colors to indicate different boundary tables. In total, there are 3 equivalent intervals of
p.saf in this setting: (0, 4.1%), (4.1%, 6.8%) and (6.8%, 9%), and 7 equivalent intervals of
p.tox: (11%, 12.3%), (12.3%, 17.2%), (17.2%, 25.2%), (25.2%, 33.5%), (33.5%, 39%), (39%, 64.6%), and (64.6%, 99.9%). Therefore, in this setting, using
p.tox = 1.4 * target.DLT.rate to calculate BOIN boundary table is equivalent to using any p.tox
(12.3%, 17.2%), as long as
p.saf values fall into one equivalent interval of
p.saf. Compared to the results shown in
Figure 2a, 4.1% is the only new interval boundary point added for
p.saf; 17.2% and 33.5% are the two interval boundary points added for
p.tox. The remaining boundary points of the equivalent intervals calculated under
n.earlystop = 15 are the same as those calculated under
n.earlystop = 12, for both
p.saf and
p.tox. SupTable 3a contains detailed summary of these equivalent intervals and their corresponding BOIN boundary tables.
When target DLT rate = 10%, cohortsize = 3, ncohort = 10, n.earlystop = 18, cutoff.eli = 95%, and extrasafe = FALSE, there are total 28 possible BOIN boundary tables, regardless of the choices of p.saf and p.tox. Equivalent intervals of p.saf and p.tox are visualized in SupFig 3b, using different colors to indicate different boundary tables. In total, there are 4 equivalent intervals of p.saf in this setting: (0, 2.6%), (2.6%, 4.1%), (4.1%, 6.8%) and (6.8%, 9%), and 7 equivalent intervals of p.tox. Compared to the results under n.earlystop = 15 above, 2.6% is the only new interval boundary point added for p.saf. The 7 equivalent intervals of p.tox calculated under n.earlystop = 18 are the same as those calculated under n.earlystop = 15. SupTable 3b contains detailed summary of these equivalent intervals and their corresponding BOIN boundary tables.
When target DLT rate = 10%, cohortsize = 3, ncohort = 10, n.earlystop = 21, cutoff.eli = 95%, and extrasafe = FALSE, there are total 45 possible BOIN boundary tables, regardless of the choices of p.saf and p.tox. Equivalent intervals of p.saf and p.tox are visualized in SupFig 3c, using different colors to indicate different boundary tables. In total, there are 5 equivalent intervals of p.saf in this setting: (0, 1.7%), (1.7%, 2.6%), (2.6%, 4.1%), (4.1%, 6.8%) and (6.8%, 9%), and 9 equivalent intervals of p.tox: (11%, 12.3%), (12.3%, 17.2%), (17.2%, 19.4%), (19.4%, 25.2%), (25.2%, 31.1%), (31.1%, 33.5%), (33.5%, 39%), (39%, 64.6%), and (64.6%, 99.9%). Compared to the results under n.earlystop = 18 above, 1.7% is the only new interval boundary point added for p.saf; 19.4% and 31.1% are the two interval boundary points added for p.tox. The remaining boundary points of the equivalent intervals calculated under n.earlystop = 21 are the same as those calculated under n.earlystop = 18, for both p.saf and p.tox. SupTable 3c contains detailed summary of these equivalent intervals and their corresponding BOIN boundary tables. For results of equivalent intervals of p.saf and p.tox under n.earlystop = 24 in similar settings, please see SupFig 3d, and SupTable 3d.
Results of equivalent p.saf and p.tox under different cutoff.eli
When target DLT rate = 10%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 97%, and extrasafe = FALSE, there are total 12 possible BOIN boundary tables, regardless of the choices of p.saf and p.tox. Equivalent intervals of p.saf and p.tox are visualized in SupFig 4a, using different colors to indicate different boundary tables. In total, there are 2 equivalent intervals of p.saf in this setting: (0, 6.8%) and (6.8%, 9%), and 6 equivalent intervals of p.tox: (11%, 12.3%), (12.3%, 25.2%), (25.2%, 39%), (39%, 45.8%), (45.8%, 64.6%), and (64.6%, 99.9%). Therefore, in this setting, using p.tox = 1.4 * target.DLT.rate to calculate BOIN boundary table is equivalent to using any p.tox (12.3%, 25.2%), as long as p.saf values fall into one equivalent interval of p.saf. Compared to the results shown in Figure 2a, no new interval boundary point is added for p.saf; 45.8% is the only interval boundary point added for p.tox. The remaining boundary points of the equivalent intervals calculated under cutoff.eli = 97% are the same as those calculated under cutoff.eli = 95% for both p.saf and p.tox. SupTable 4a contains detailed summary of these equivalent intervals and their corresponding BOIN boundary tables. When target DLT rate = 10%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 99%, and extrasafe = FALSE, all equivalent interval results are the same as those calculated under cutoff.eli = 97% if other design parameters stay the same (SupFig 4a, SupTable 4a).
When target DLT rate = 10%,
cohortsize = 3,
ncohort = 10,
n.earlystop = 12,
cutoff.eli = 90%, and
extrasafe = FALSE, there are total 6 possible BOIN boundary tables, regardless of the choices of
p.saf and
p.tox. Equivalent intervals of
p.saf and
p.tox are visualized in SupFig 4b, using different colors to indicate different boundary tables. In total, there are 2 equivalent intervals of
p.saf in this setting: (0, 6.8%) and (6.8%, 9%), and 3 equivalent intervals of
p.tox: (11%, 12.3%), (12.3%, 25.2%), and (25.2, 99.9%). So, in this setting, using
p.tox = 1.4 * target.DLT.rate to calculate BOIN boundary table is equivalent to using any p.tox
(12.3%, 25.2%), as long as
p.saf values fall into one equivalent interval of
p.saf. Compared to the results shown in
Figure 2a, no new interval boundary point is added or reduced for
p.saf; 39% and 64.6% are the 2 interval boundary points removed for
p.tox. The remaining boundary points of the equivalent intervals calculated under
cutoff.eli = 90% are the same as those calculated under
cutoff.eli = 95% for both
p.saf and
p.tox. SupTable 4b contains detailed summary of these equivalent intervals and their corresponding BOIN boundary tables.
When target DLT rate = 10%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 80%, and extrasafe = FALSE, there are total 4 possible BOIN boundary tables, regardless of the choices of p.saf and p.tox. Equivalent intervals of p.saf and p.tox are visualized in SupFig 4c, using different colors to indicate different boundary tables. In total, there are 2 equivalent intervals of p.saf in this setting: (0, 6.8%) and (6.8%, 9%), and 2 equivalent intervals of p.tox: (11%, 12.3%) and (12.3%, 99.9%). So, in this setting, using p.tox = 1.4 * target.DLT.rate to calculate BOIN boundary table is equivalent to using any p.tox (12.3%, 99.9%), as long as p.saf values fall into one equivalent interval of p.saf. This is an example of why cutoff.eli < 90% should be used with caution. SupTable 4c contains detailed summary of these equivalent intervals and their corresponding BOIN boundary tables.
When target DLT rate = 10%, cohortsize = 3, ncohort = 10, n.earlystop = 12, cutoff.eli = 70%, and extrasafe = FALSE, there are only 2 possible BOIN boundary tables, regardless of the choices of p.saf and p.tox. Equivalent intervals of p.saf and p.tox are visualized in SupFig 4d, using different colors to indicate different boundary tables. The 2 equivalent intervals of p.saf in this setting are (0, 6.8%) and (6.8%, 9%), and the only equivalent interval of p.tox is (11%, 99.9%). So, in this setting, using p.tox = 1.4 * target.DLT.rate to calculate BOIN boundary table is equivalent to using any valid values of p.tox, as long as p.saf values fall into one equivalent interval of p.saf. This is again an example of why cutoff.eli < 90% should be used with caution. SupTable 4d contains detailed summary of these equivalent intervals and their corresponding BOIN boundary tables.
Results of equivalent p.saf and p.tox under varying cohortsize
When target DLT rate = 10%,
cohortsize = 4,
ncohort = 10,
n.earlystop = 12,
cutoff.eli = 95%, and
extrasafe = FALSE, there are total 8 possible BOIN boundary tables, regardless of the choices of
p.saf and
p.tox. Equivalent intervals of
p.saf and
p.tox are visualized in SupFig 5a, using different colors to indicate different boundary tables. In total, there are 2 equivalent intervals of
p.saf in this setting: (0, 6.8%) and (6.8%, 9%), and 4 equivalent intervals of
p.tox: (11%, 15.3%), (15.3%, 25.2%), (25.2%, 45.8%), and (45.8%, 99.9%). Therefore, in this setting, using
p.tox = 33% to calculate BOIN boundary table is equivalent to using any p.tox
(25.2%, 45.8%), as long as
p.saf values fall into one equivalent interval of
p.saf. Compared to the results shown in
Figure 2a, equivalent intervals of
p.saf are the same, but number of equivalent intervals of
p.tox reduced from 5 to 4. SupTable 5a contains detailed summary of these equivalent intervals and their corresponding BOIN boundary tables.
When target DLT rate = 10%,
cohortsize = 5,
ncohort = 10,
n.earlystop = 12,
cutoff.eli = 95%, and
extrasafe = FALSE, there are total 2 possible BOIN boundary tables, regardless of the choices of
p.saf and
p.tox. Equivalent intervals of
p.saf and
p.tox are visualized in SupFig 5b, using different colors to indicate different boundary tables. In total, there is only one equivalent interval of
p.saf (0, 9%), and 2 equivalent intervals of
p.tox: (11%, 33.5%) and (33.5%, 99.9%). Therefore, in this setting, using
p.tox = 1.4 * target.DLT.rate to calculate BOIN boundary table is equivalent to using
p.tox > 3 * target.DLT.rate in this setting.
Figure 5b provides 3 validation examples of the statement above. SupTable 5b contains detailed summary of these equivalent intervals and their corresponding BOIN boundary tables.
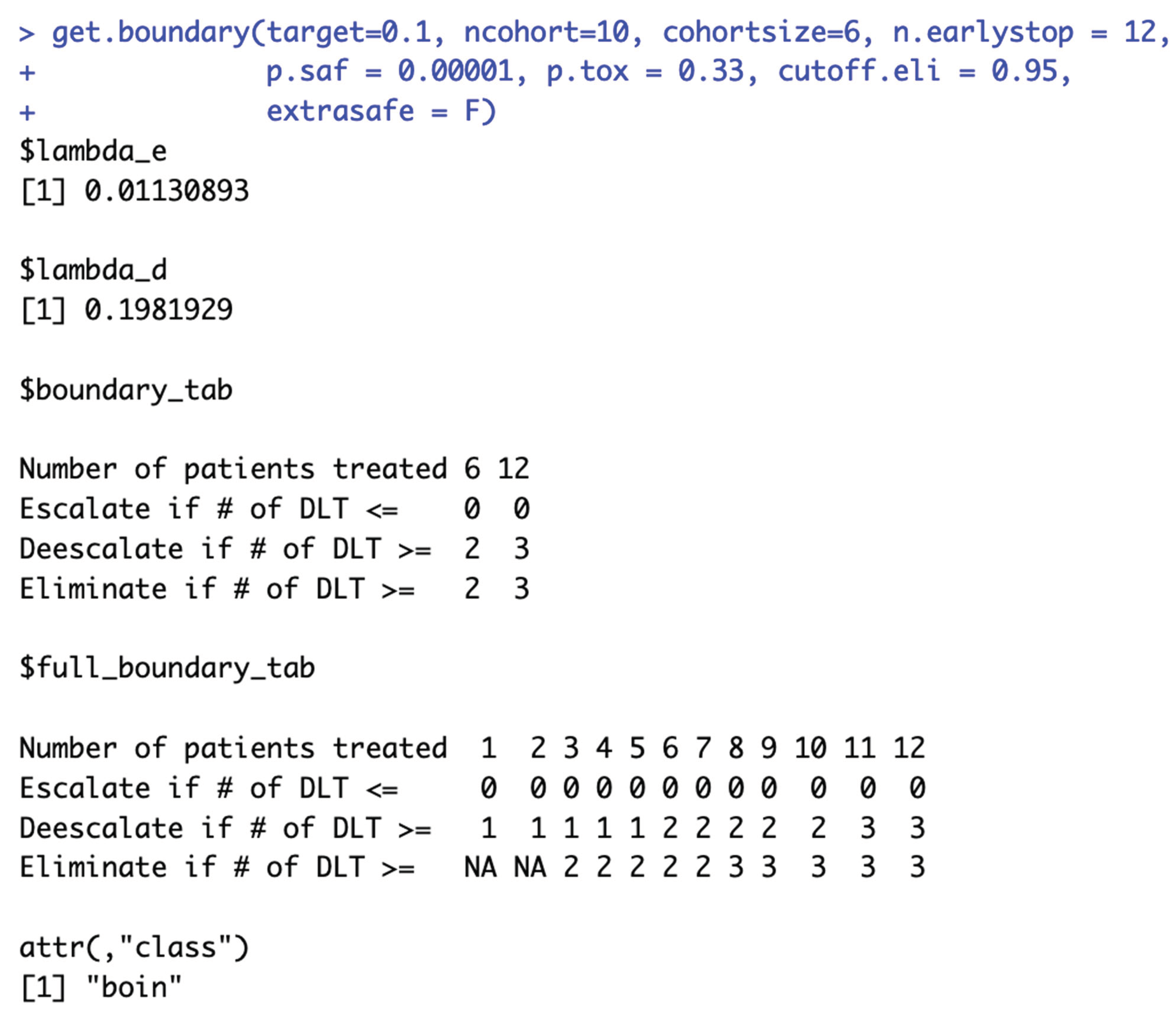
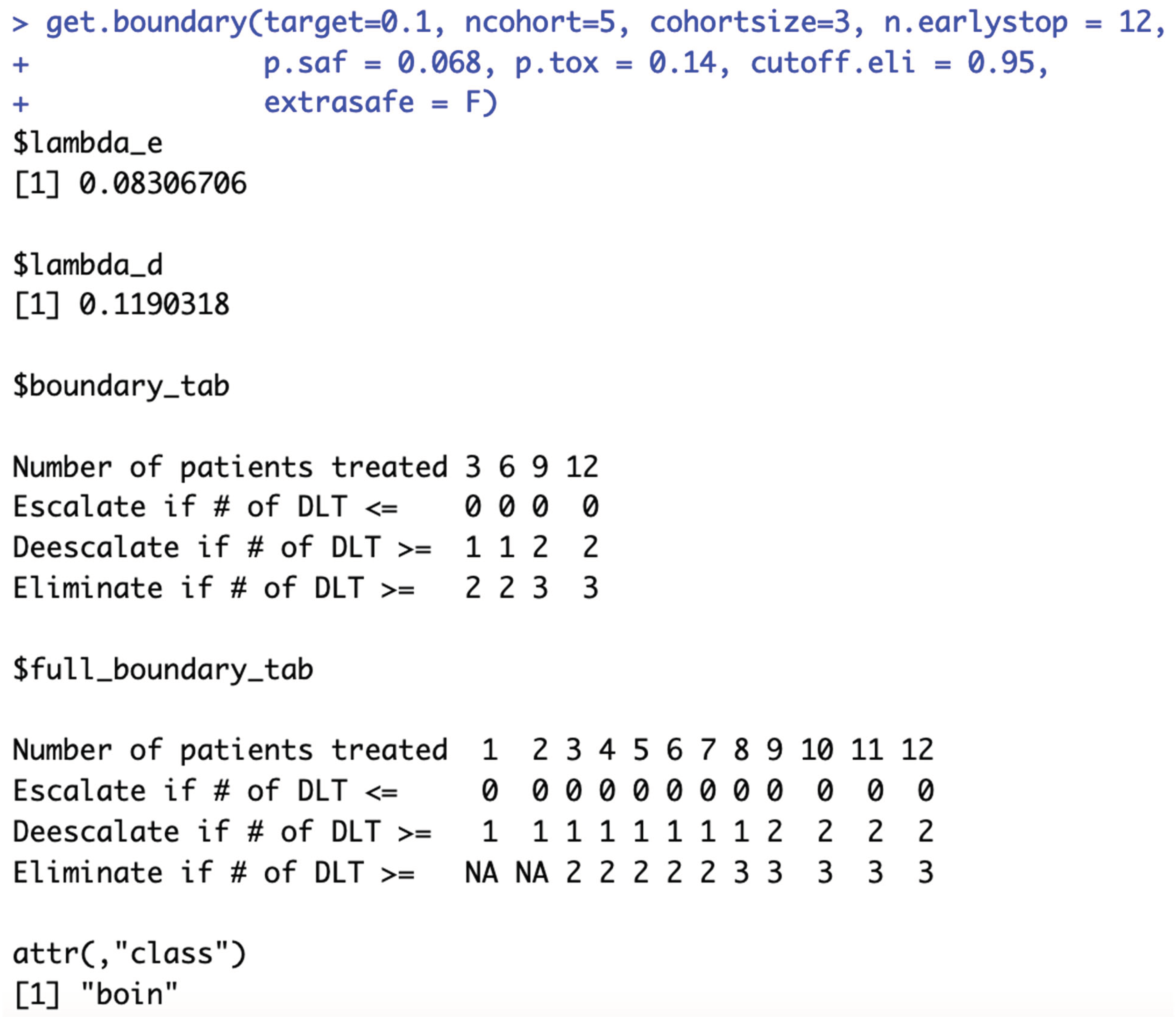
When target DLT rate = 10%, cohortsize = 6, ncohort = 10, n.earlystop = 12, cutoff.eli = 95%, and extrasafe = FALSE, there are total 4 possible BOIN boundary tables, regardless of the choices of p.saf and p.tox. Equivalent intervals of p.saf and p.tox are visualized in SupFig 5c, using different colors to indicate different boundary tables. In total, there are 2 equivalent intervals of p.saf in this setting: (0, 6.8%) and (6.8%, 9%), and 2 equivalent intervals of p.tox: (11%, 25.2%) and (25.2%, 99.9%). Therefore, in this setting, using p.tox = 33% to calculate BOIN boundary table is equivalent to using p.tox = 99%, as long as p.saf values fall into one equivalent interval of p.saf. Figure 5c provides 3 validation examples of the statement above. Compared to the results shown in Figure 2a, equivalent intervals of p.saf are the same, but number of equivalent intervals of p.tox reduced from 5 to 2. SupTable 5c contains detailed summary of these equivalent intervals and their corresponding BOIN boundary tables.
Results of equivalent p.saf and p.tox under different ncohort
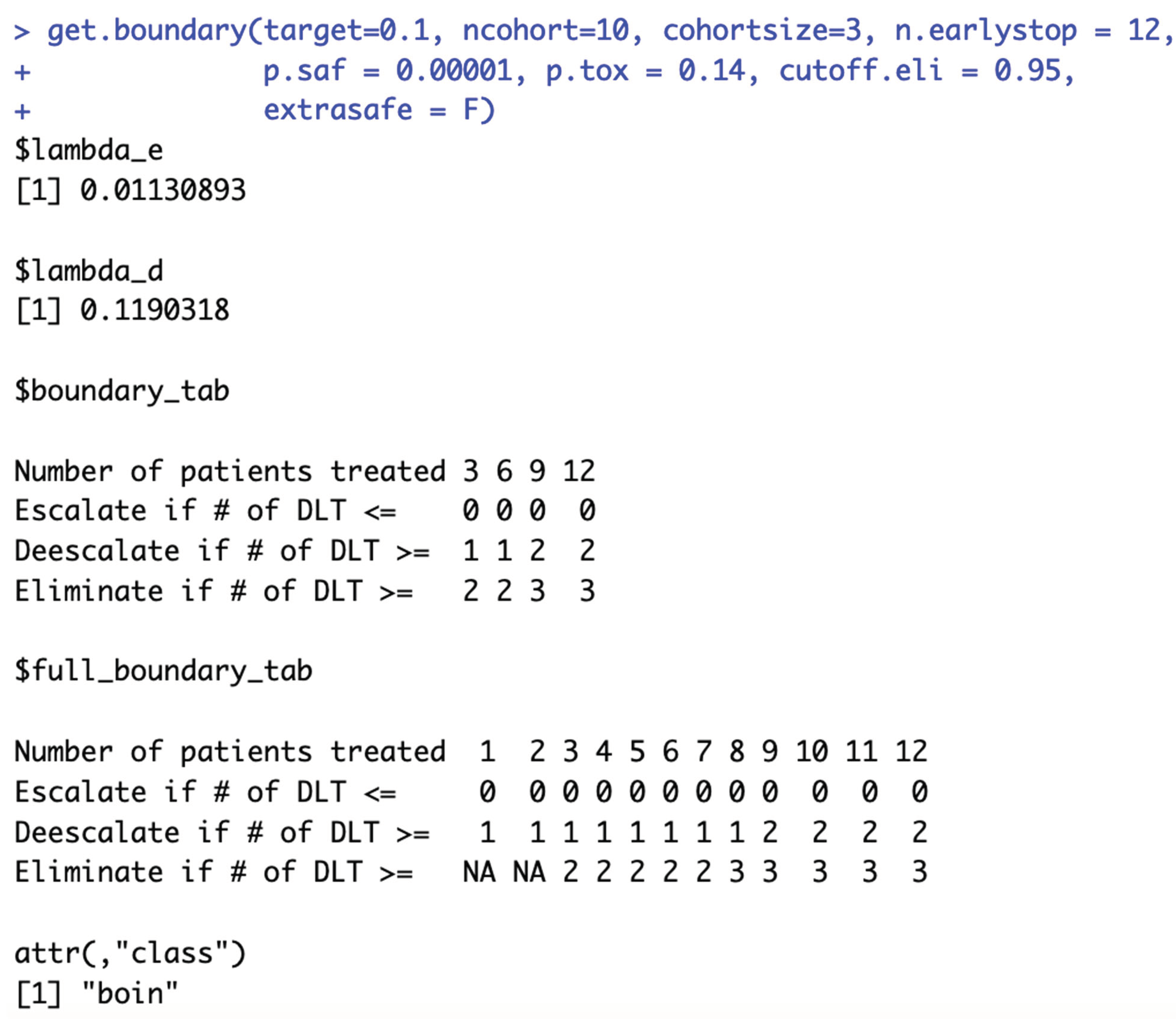
Although
ncohort is one of the required input parameter of
get.boundary(), the choice of
ncohort value has no effect on the calculation of BOIN boundary table. For example, when target DLT rate = 10%,
cohortsize = 3,
n.earlystop = 12,
cutoff.eli = 95%, and
extrasafe = FALSE, there are the same 10 possible BOIN boundary tables, regardless of the choices of
p.saf,
p.tox, and
ncohort {5, 6, 7, 8, 9, 10, 12, 20, 40, 100}.
Figure 6c also provides 3 validation examples. The main purpose of specifying
ncohort is to terminate dose finding process when the sample size budget is reached.
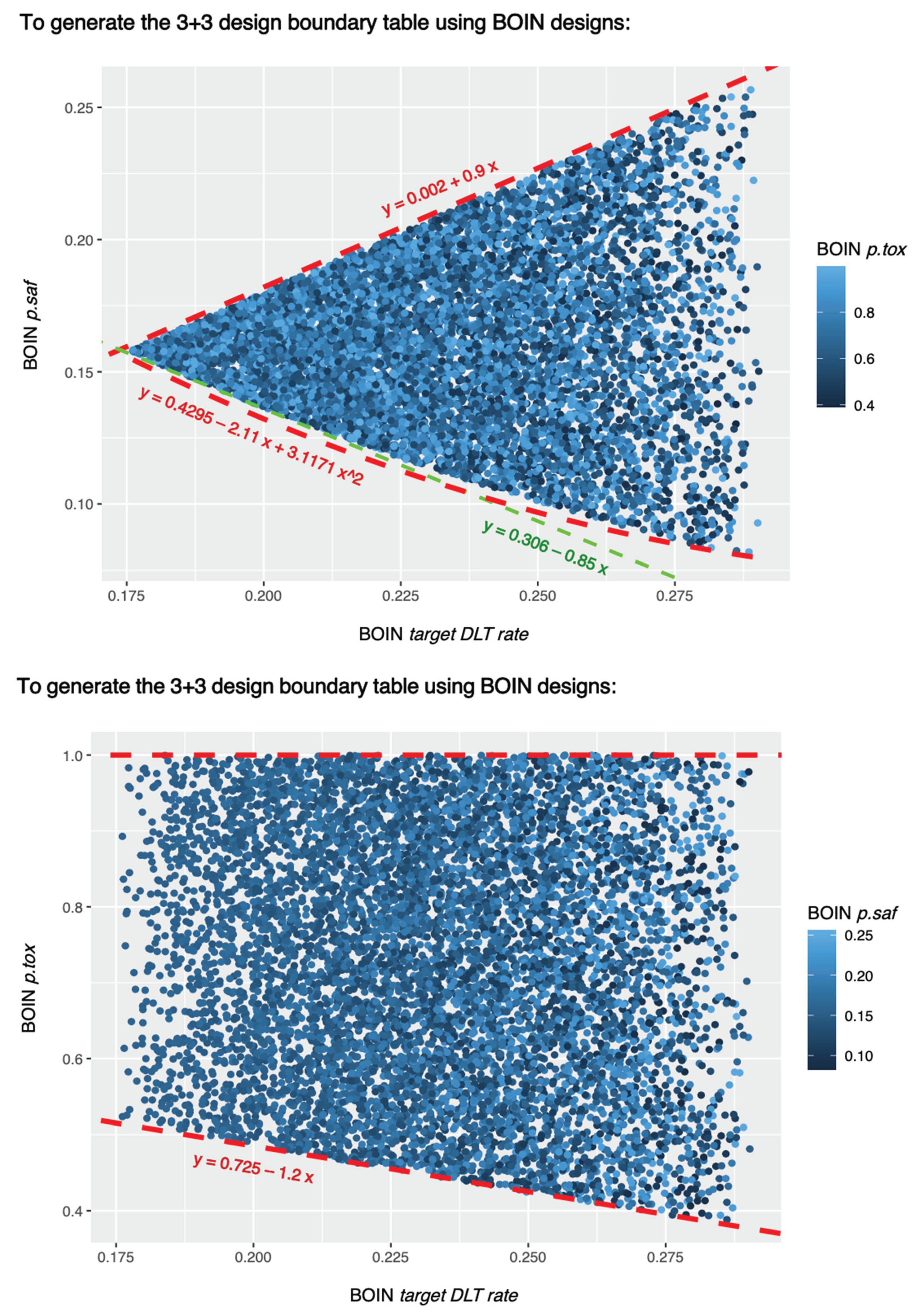
Results of equivalent BOIN parameter sets for generating the same 3+3 rules
The random search script described in the method section sampled 8,127 sets of BOIN parameters that can generate the 3+3 design table shown in
Table 1. These 8,127 sets of BOIN parameter values are all listed in SupTable 7. These 8,127 sets of BOIN parameter values satisfy following conditions:
.
These conditions are also visualized in
Figure 7.