1. Introduction
Pseudomonas aeruginosa is a gram-negative, rod-shaped, mobility bacterium that reproduces at temperatures between 37 and 42 °C. The cytochrome-oxydase enzyme is present and does not ferment lactose. It produces a blue-green-coloured pigment, pyocyanin, and a black and green pigment, pyoverdin. It is an opportunistic pathogen that can lead to blood-circulation infections, respiratory, urinary, bone and joint infections and skin and subcutaneous tissue infections [
1,
2,
3].
P. aeruginosa is a common pathogen in environmental areas, but rarely colonizes in healthy people [
4].
In the treatment of
P. aeruginosa, penicillins such as piperasiline and tikarsiline, cephalosporins such as seftaidim and sefepim, aztreonam in the monobactam group, and carbapenems, imipenem (often used in conjunction with silastatin), meropenem and doripeneem are used [
5]. Another group used in antipsedomonal therapy are fluoroquinolones, the most commonly used being ciprofloxacin and levofloxacine [
6]. Amikacin, gentamicin, and tobramicin are aminoglycosites with antipsedomonal effects [
7]. In the treatment of
P. aeruginosa, which is resistant to commonly used antipsedomonal drugs, new antibiotics are used, such as seftalosan-tasobactam, seftasidim-avibaktam, imipenem-silastatin-relebactam and sefiderokol [
8].
In
P. aeruginosa infections, the main mechanisms that play a role in developing resistance (chromosomal) resistance to antimicrobials are acquired, including the production of enzymes that break down antibiotics such as beta-lactamase, including hyperexpressed exhaust pump and reduced external membrane permeability. As several of these resistance mechanisms often occur simultaneously, treatment options in drug-resistant
P. aeruginosa isolates are very limited [
3]. The most common mechanisms inducing intrinsic resistance in
P. aeruginosa isolates are inducable AmpC (cephalosporinase) expression, exhaust pump hyperexpression, and low level external membrane permeability. The production of injectable beta-lactamase, along with cephalosporins, has resulted in reduced sensitivity to aminopenicillins and imipename [
9]. The most commonly observed mutation-induced beta-lactam resistance mechanism in
P. aeruginosa isolates is the overexpression of AmpC cephalosporinase [
10]. Another common resistance mechanism is the excess expression of the exhaust pump. The expression of the MexAB-OprM exhaust pump leads to resistance to all beta-lactams, fluoroquinolones, tetracyclines, macrolytes, chloramphenicol and novobiosis, with the exception of imipenem, including aztreonam. MexXY-OprM and MexCD-OperJ exhaust pumps also show substrat specificity to other beta-lactam antibiotics, with the exception of beta-lactam antibodies such as aztreonam, imipenem and saftazidim [
11,
12].
To prevent increased antibiotic resistance, studies have been conducted showing synergies between metformin, antihistamines, anti-inflammatory drugs or neuroleptics and antibiotics [
13,
14,
15]. Patients taking metformin due to diabetes have been shown to have fewer cases of infection [
16]. Studies on the combination of metformin and different antibiotics have shown synergies between metformine and antibiotics [
13,
17].
P. aeruginosa can lead to serious infections and develop resistance to antibiotics used to treat the infections it causes, using different mechanisms. In this study, P. aeruginosa isolates were investigated using a chequerboard test for synergy between metformin and ceftazidime, cefepime, imipenem, ciprofloxacin and levofloxasin.
2. Results
The results of the liquid microdilution test of P. aeruginosa isolates included in our study showed that 31 isolates (62%) were resistant to ceftazidime, 37 isolates (74%), were resistant to cefepime, 48 isolates (96%) were resistant to imipenem, 30 isolates (60%) were resistant to ciprofloxacin, and 23 isolates (46%) were resistant to levofloxacin. No antibacterial effect of metformin alone has been detected in any of the tested dosage ranges.
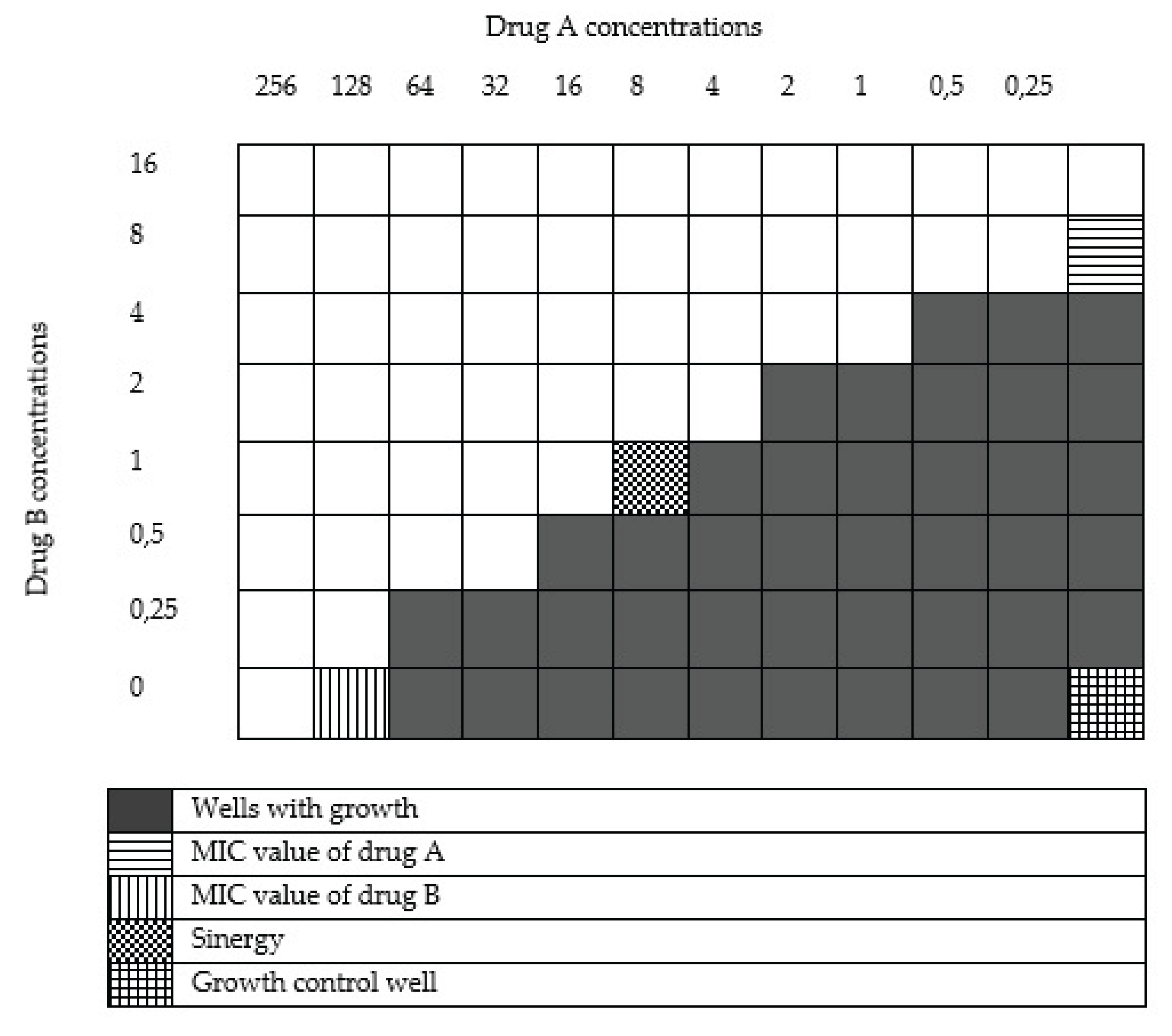
The results of the chequerboard synergy test between ceftazidime and metformin showed a synergistic effect in three isolates (6%), a partial synergetic effect in 11 isolates (22%) and an indifferential effect in 36 isolates (72%) (
Table 2) (
Supplementary Table S1). Prior to combination with metformin, 19 isolates (38%) were sensitive to high doses of ceftazidine, while 26 isolates (52%) were highly sensitive after combination. (
Table 3).
The results of the chequerboard synergy test between cefepime and metformin showed no synergistic effect on any isolate, partial synergic effect in 23 isolates (46%) and indifferential effect in 27 isolates (54%) (
Table 2) (
Supplementary Table S2). Prior to combination with metformin, 13 isolates (26%) were sensitive to high doses of cefepime; after combination, 23 isolates (46%) were susceptible to higher doses. (
Table 3).
The results of the chequerboard synergy test between imipenem and metformin showed a synergistic effect in two isolates (%4), a partial synergetic effect in five (%10) and an indifferential effect in 43 isolates (%86) (
Table 2)(
Supplementary Table S3). Before combination with metformin, two isolates (%4) were sensitive to imipening at high doses; after combination, three (%6) isolates were found to be susceptible to imiphening at higher doses. (
Table 3).
The results of the chequerboard synergy test between ciprofloxacin and metformin showed synergistic effect in three isolates (%6), partial synergies in three isolates (%6) and indifferential effect in 43 isolates (%84) and antagonism in one isolate (%2) (
Table 2)(
Supplementary Table S4). The only isolates with antagonist effect have been found to have a partial synergistic effect with cefepime and levofloxacin. Prior to combination with metformin, 20 isolates (40%) were sensitive to high doses of ciprofloxacin, while 23 isolates (46%) were highly sensitive after combination. (
Table 3).
Based on the results of the dama board synergy test between levofloxacin and metformin, a synergistic effect was observed in one isolate (%2), a partial synergetic effect in six isolates (%12) and an indifferential effect in 43 isolates (%86) (
Table 2) (
Supplementary Table S5). Prior to combination with metformin, 27 isolates (54%) were sensitive to high doses of levofloxacin, while after combination, 29 isolates (58%) were susceptible to higher doses. (
Table 3).
In our study, the value of MBC for antibiotics and metformin has also been investigated. The MBC value for metformin has not been studied since reproduction occurred in all stacks containing only metformine. After microdilution and chequerboard synergy testing for antibiotics and combinations of antibiotic and metformin, transition from non-microplate-reproducing pins to SBA, no reproduction was observed. Therefore, the MIC values determined by the microdilution method were also found to be equal to the MBC value.
3. Discussion
One of the most important health problems today is that microorganisms develop short-term resistance to the used antimicrobials [
20]. Microorganisms that are resistant to antimicrobials have a high mortality and morbidity rate because of the very severe infections they cause [
21]. The study investigated the presence of synergies between sephtazidim, sefepim, imipenem, ciprofloxacin and levofloxasin, commonly used in the treatment of
P. aeruginosa infections, and metformin, a pathogen that can develop short-term resistance to antimicrobials.
In our study, no antibacterial effect was shown on any isolates in the tested dose ranges for metformin (512-8 μg/ml). In a study by Masadeh and ark.[
11], the antibacterial effect of metformin was studied on strains of
P. aeruginosa ATCC BAA-2114 and
Staphylococcus aureus ATSC 33591 at concentrations of 500 μM or less, but no antibacteric effect was found on metformine alone. A study conducted by Zuo and ark.[
23] with
Streptococcus suis isolates found no inhibitory or bactericidal effect on metformin isolates.
In our study, the presence of synergies between metformin and septazidim, sefepim, imipenem, ciprofloxacin, and levofloxasin was studied on 50
P. aeruginosa isolates. A synergistic effect in three isolates for ceftazidime, a partial synergetic effect in 11 isolates; a synergic effect in zero isolate for cefepime, and a partial synergistic effect in 23 isolats; a synergic effect for imipenem in two isolates, a partial synergetic effect of five isolations; a synergic effect for ciprofloxacin in three isolates, a synergic effect on three izolates; and a synergic effect for levofloxase in one isolate and a partial synergic effect at six isolates were observed. There are studies in the literature that investigate the antibacterial synergic effects of metformin and various chemicals. The presence of synergies between metformin and levofloxacin, chloramphenicol, ampicillin, rifampicine and doxycycline was investigated by Masadeh and ark.[
11] on the multi-drug-resistant
P. aeruginosa ATCC BAA-2114 isolate and the methiciline-resistent
S. aureus ATCC 33591 isolate. The study showed a synergistic effect on
P. aeruginosa between metformin and all medicines. On
S. aureus, all drugs except rifampicin have been found to have a synergistic effect with metformin. However, although synergies were found in this study, the antibiotic's MIC values were not resistant to high doses, a subcategory of the resistant category. In a study conducted by Wu and ark. [
15] using metformin and silver ions (Ag+) on
Enterococcus faecalis isolates, colonies were found to be fewer than when used alone. He and ark. [
17] investigated synergies between metformin and Triton X-100 in the antibacterial effects of
E. faecalis isolates. When metformin and Triton X-100 were combined, they were found to be well below the MIC values detected when used alone. Another study by Liu and ark. [
16] investigated the synergic effects of metformin and tetracyclines on isolates of
Escherichia coli that are resistant to tetracicline. When used in combination with metformin and doxycycline, the MIC values of doxycyline were found to decrease.
Studies in the literature have shown that metformin increases the antibacterial effectiveness of various antibiotics. And in our study, there are isolates that have detected synergy and partial synergy when metformin is combined with antibiotics. In the light of this information, metformin still retains its potential antibacterial properties. However, it is believed that different chemical modifications may be required to reveal the potential antibacterial effect of metformin. In addition, unlike other studies in the literature in our study, when antibiotics are combined with metformin, the number of isolates passing through high-dose sensitive profiles from resistant profiles has been investigated. Seven for ceftazidime (14%), 10 for cefepime (20%), one for imipenem (2%), three for ciprofloxacin (6%), and two for levofloxasin (4%), while the isolate was resistant before combination with metformin; it passed a high-dose sensitive profile when combined with antibiotics.
In our study, metformin has not been investigated by what mechanism or mechanisms it increases the effect of antibiotics. In the literature, studies investigating the antibacterial effectiveness of metformin have shown synergistic effects with antibiotics through similar mechanisms. A study that investigated the synergistic effect of metformin and tetracyclines on
E. coli found that the dose of doxycycline in the bacterium increased after metformine was administered. The study showed that metformin interacts with the hydrophobic part of the phospholipid double layer of the bacterial cell membrane, increasing the permeability of the external membrane to antibiotics [
16]. A study that measured the effectiveness of metformin and Ag+ ions on
E. faecalis isolates also showed that metformine increases cell membrane permeability, leading to the accumulation of Ag+ ion in the cell [
15]. Although it is unclear exactly by what mechanism or mechanisms metformin increases the effectiveness of antibiotics, it is generally accepted that it increases intra-cellular antibacterial concentration by disrupting the external membrane of the bacterium [
11].
The most important mechanisms that contribute to antibiotic resistance on
P. aeruginosa isolates, primarily the beta-lactam group antibiotics and fluoroquinolones, are the inhibition of antibiotic penetration into the bacteria, causing changes in the structure of the outer membrane, and the exhaust pump that the bacterium has [
6,
9,
23]. In the light of this information, the synergy test with metformin and various antibiotics on the
P. aeruginosa isolates included in our study suggests that the isolates have different characteristic structures, such as synergistic, partial synergetic or indifferential effects. In isolates with a synergistic effect with metformin, the outer membrane of the bacterium was degraded on the metformine side, allowing the antibiotic to pass into the bacteria at sufficient concentrations; in isolates without a synergies effect, the antibiotic concentration in the bacteries did not increase even after the application of the external membrane.
It is also noted that the contribution of the exhaust pump to imipenem resistance at the head of the mechanisms that most commonly play a role in the resistance of beta-lactam antibiotics on
P. aeruginosa isolates is very limited [
10]. Our study also found that the antibiotic with the least synergy between metformin and beta-lactam antibiotics is imipenem (two synergies, five partial synergies). This suggests that the passage of imipenem bacteria through the outer membrane will not make a major contribution to immunodeficiency. This finding is also consistent with studies in the literature where the mechanism of action of metformin increases bacterial external membrane permeability.
In our study, the cytotoxic effect of metformin alone or in combination with antibiotics has not been investigated. In one study, the reliability of metformin was measured by treating it with mammary VERO cells. As a result of the study, even cells treated with high doses of metformin retained their vitality and were found to be quite safe for mammal cells [
11]. Another study investigated the cytotoxic effect of metformin on cells, but no significant cytotoxin effect was observed. Another study, in which the synergy between metformin and Triton X-100 was investigated, investigated the cytotoxicity of metformine, and found no significant cytotoxin in the control group [
17]. Another study using Ag+ and metformin on
E. faecalis isolates found no significant difference in cell proliferation from the control group to the metformine-treated group [
15]. Although the toxicity of metformin has not been investigated in our study, studies in the literature have not found a significant cytotoxic effect of metformine. These findings suggest that if metformin has a synergistic effect with antibiotics, it can be used safely.
In this study, various studies have investigated the synergic effect of metformin, which is described as a potential antibiotic, with various antibiotics. Electron microscopic examination or molecular tests to clarify the mechanism of action of metformin have not been carried out. The lack of research into the cytotoxic effect of metformin alone or in combination with the tested antibiotics is also a shortcoming of our study.
As a result, in our study, synergy and partial synergy between metformin and antibiotics have been observed on some isolates. It is believed that chemical modifications on metformin, even if not in its present form, could increase its antibacterial activity. Therefore, further research is needed to investigate the antibacterial effectiveness of metformin, with its present form and various chemical modifications to be produced synthetically. Furthermore, more extensive studies need to be undertaken to clarify the genotypic and phenotypic differences between the mechanism or mechanisms by which metphromine increases the effectiveness of antibiotics, the isolations that are effective, and the isolates that are not, the issues of whether it produces a cytotoxic effect when used in conjunction with antibiotics and the pharmacokinetic and pharmacodynamic effects that we will encounter if used in animals.