1. Introduction
Gestational diabetes mellitus (GDM) is defined as any degree of glucose intolerance with onset or first recognition during pregnancy [
1]. It is considered as one of the most common complications of this period with different prevalence depending on the population and the diagnostic criteria. A recent meta-analysis provided a global standardized prevalence of 14.2% in women between 25-30 years using universal oral glucose tolerance test (OGTT) and the International Association of the Diabetes and Pregnancy Study Groups (IADPSG) diagnostic criteria [
2]. Unfortunately, figures continue to rise due to increased maternal obesity and advanced age at childbearing [
3].
GDM confers maternal, fetal and neonatal adverse outcomes [
4] and also increases the risk of type 2 diabetes (T2DM), obesity and cardiometabolic disease in the mother and the offspring later in life [
5,
6,
7]. In fact, women with GDM have nearly 10-fold higher risk of developing T2DM diabetes compared to women with a normoglycemic pregnancy [
7]. Therefore, pregnancy is a crucial period to decrease both perinatal and long-term medical complications.
Environmental factors interact with genetics in the development of GDM. Previous GDM, family history of diabetes, ethnicity, parity and higher maternal age and body mass index are the strongest clinical predictors for GDM [
8]. The role of genetics in GDM remains poorly defined [
3] and needs to be further explored. Genome-wide association studies (GWAS) have identified single-nucleotide polymorphisms (SNPs) associated with GDM [
9,
10] and some studies evaluated genetic data through genetic risk score (GRS) [
11]. However, their predictive value is sometimes limited and other indicators might be combined to enhance results.
Therefore, the aim of the present study is to build a prediction model of GDM based on a combination of genetic, clinical and demographic information. We hypothesized that genetics in combination with well-known environmental risk factors might improve prediction of GDM in pregnant women and thus, short- and long-term medical outcomes. Identifying the woman at risk as early as possible is the first step in designing preventive strategies
2. Results
2.1. Phenotype data
A total of 1588 pregnant women, 1069 (67.3%) Caucasian (CAU) and 519 (32.7%) Latin American (LAT) were studied. Phenotype characteristics of pregnant women at entry (< 12 gestational weeks (GW)) are represented in
Table 1.
GDM was diagnosed in 172 (16.1%) CAU and 83 (16.0%) LAT pregnant women. CAU and LAT pregnant women showed higher pre-pregnancy body mass index (BMI) and fasting plasma glucose levels (FPG) at 12 GW and more prevalent history of GDM than their counterparts without GDM. LAT women with GDM exhibited an older age at pregnancy, showing also a similar trend in the CAU group (
Table 1).
Based on the common differences between women with and without GDM observed in both races, age, pre-pregnancy BMI and FBG at 12 GW were studied for the association with GDM risk. Furthermore, these three variables were considered categorized regarding their risk in: age ≤ 35 years and age > 35 years; pre-pregnancy BMI < 25 kg/m2, pre-pregnancy BMI ≥ 25 kg/m2; FPG at 12 GW ≤ 83.5 mg/dL and > 83.5 mg/dL for CAU and FBG at 12 GW ≤ 82.5 mg/dL and > 82.5 mg/dL for LAT.
2.2. Genotype data
For each ethnicity, univariate logistic regression was performed for the 110 SNPs and 1588 samples. The genetic variants showing significant differences between women with and without GDM and/or previously identified in our cohort [
12] are showed in
Table S1 for CAU and
Table S2 for LAT.
To determinate if clinical variables provided better adjustment as continuous variables or categorical ones (with the calculated cut-offs) we used stepwise selection strategy on different models. This strategy consisted of eliminating those variables from the model that are found to be non-significant. The first model with the relevant SNP and quantitative age and pre-pregnancy BMI; the second model with the relevant SNP and quantitative age, pre-pregnancy BMI and FBG at 12 GW; the third model with the relevant SNP and categorized age and pre-pregnancy BMI; the fourth model with the relevant SNP and categorized age, pre-pregnancy BMI and FBG at 12 GW. In receiver operating curve (ROC) analysis, the area under curve (AUC) was higher when we considered the categorized independent variable.
2.2.1. Caucasian ethnicity findings
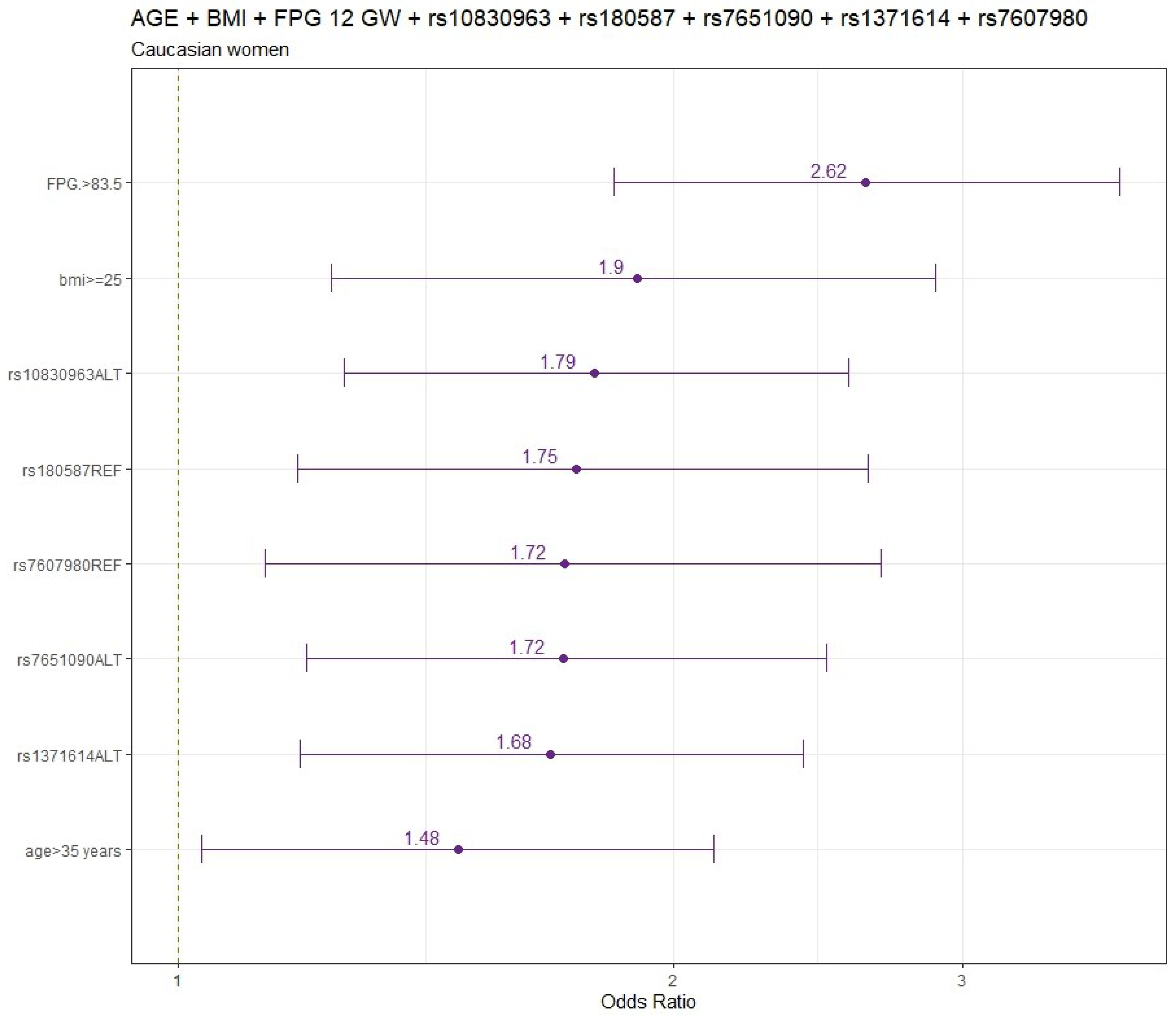
The final models for CAU pregnant women are summarized in
Figure 1.
Logistic regression model including age and pre-pregnancy BMI (
Table S3) and also considering FPG 12 GW for CAU women (
Table S4) are displayed in both tables. Adjusted by age and pre-pregnancy BMI, the genetic variants significantly associated with increased GDM risk were rs10830963 (OR:95%CI) (1.81:1.27-2.57), rs7651090 (1.73:1.21-2.49) and rs1371614 (1.66:1.18-2.35); whereas rs7607980 (0.58:0.37-0.88), rs180587 (0.59:0.40-0.87) and rs3783347 (0.67:0.45-0.98) variants were significantly associated with decreased GDM risk (Tables S3).
When FPG was included in the adjusted model (age, pre-pregnancy BMI and FPG at 12 GW), the genetic variants significantly associated with increased GDM risk were rs10830963 (1.79:1.26-2.56), rs7651090 (1.72:1.20-2.50) and rs1371614 (1.68:1.19-2.40) and variants significantly associated with decreased GDM risk were rs180587 (0.57:0.38-0.85) and rs7607980 (0.58:0.37-0.89) (
Table S4).
Receiver Operating Curve (ROC) analysis for the logistic regression for Caucasian women is summarized in
Table 2
In the models, the power of GDM prediction including only the age was (AUC;95%CI) (0.535:0.494-0.576) (
Figure S1 panel a), the age and the pre-pregnancy BMI (0.573:0.527-0.619) (
Figure S1 panel b) and the age, the pre-pregnancy BMI and the FPG at 12 GW was 0.644: 0.597-0.691) (
Figure S1 panel c). When the selected SNPs were considered the power of prediction increased to 0.714:0.672-0.756 (
Figure S1 panel d) including the age, pre-pregnancy BMI and FBG at 12 GW.
2.2.2. Latin American ethnicity findings
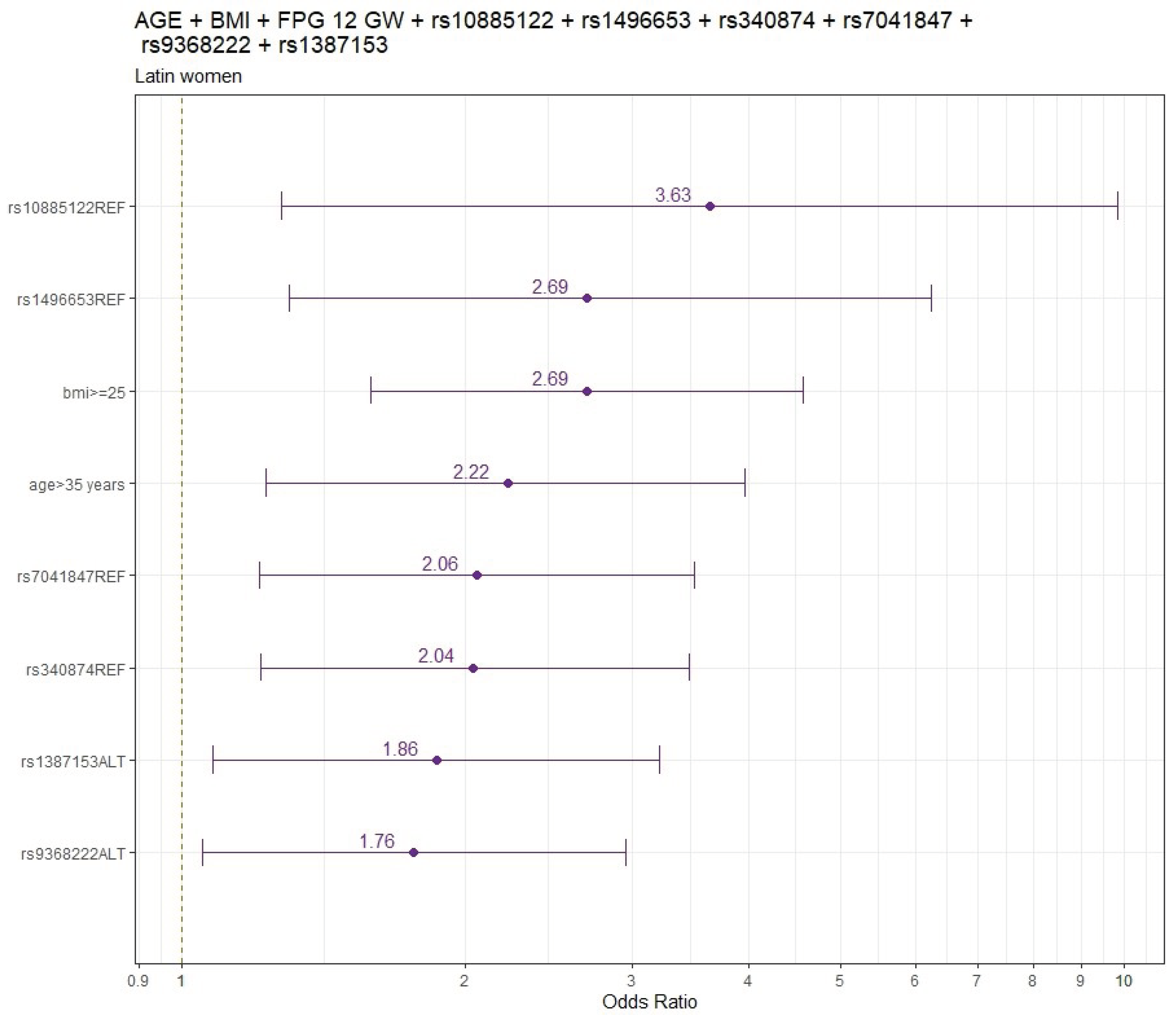
Figure 2 shows the final models’ findings for LAT pregnant women.
Adjusted by age and pre-pregnancy BMI, the genetic variants significantly associated with increased GDM risk (odds ratio: 95%CI) were rs1387153 (1.88:1.09-3.21), rs9368222 (1.77:1.06-2.95) and variants significantly associated with decreased GDM risk were rs10885122 (0.30:0.11-0.84), rs1496653 (0.46;0.21-0.91), rs7041847 (0.49; 0.29-0.82) and rs340874 (0.50;0.30-0.83) (
Table S5).
When FPG values at 12 GW were included in the adjusted model by age and pre-pregnancy BMI the results did not change. The genetic variants significantly associated with increased GDM risk were rs9368222 (1.85:1.10-3.14), rs1387153 (1.85:1.06-3.20) and variants significantly associated with decreased GDM risk were rs10885122 (0.26:0.09-0.73), rs1496653 (0.38:0.16-0.80), rs340874 (0.49:0.29-0.82) and rs7041847 (0.49:0.29-0.84), respectively. (
Table S6).
The logistic regression analysis (ROC) for LAT women is summarized in
Table 3
The power of GDM prediction including only the age was 0.543:0.489-0.597 (
Figure S2 panel a), the age and the pre-pregnancy BMI was 0.651:0.590-0.713 (
Figure S2 panel b) and the age, the pre-pregnancy BMI and the FPG 12 GW was 0.685:0.626-0.745 (
Figure S2 panel c). When we considered the selected SNPs the power of prediction increased to 0.760:0.701-0.819) including the age, pre-pregnancy BMI adding the FPG at 12 GW (
Figure S2 panel d).
3. Discussion
GDM is emerging as a public health concern affecting one in six pregnancies worldwide in 2019 [
13]. It entails maternal, fetal and neonatal complications during pregnancy but also an increased risk of future T2DM and cardiovascular disease for both the mother and the offspring. Therefore, gestation might be a decisive period to an optimal intervention and risk modification. However, some challenges must be overcome to translate this beneficial management to clinical practice. One of these challenges is a correct identification of GDM cases [
14].
Different strategies have been proposed to identify GDM but there is no consensus on the optimal approach [
14]. In our setting, all pregnant women are screened at 24 gestational weeks using a one-step strategy by a 2h 75 g OGTT recommended by the IADPSG endorsed by the American Diabetes Association (ADA), the World Health Organization and the International Federation of Gynecology and Obstetrics [
14,
15,
16,
17]. Nevertheless, an earlier identification of those women more prone to develop GDM might improve management and thus, short- and long-term metabolic outcomes.
Combining well-known environmental GDM risk factors and genetic variants associated with T2DM and GDM might be a more informative strategy than the current one. In fact, genetics of GDM are not fully understood and although some studies evaluated genetic data through genetic risk score, other parameters might be merged to enhance results [
11].
Accordingly, we conducted a case-control study of 1588 CAU and LAT pregnant women to explore the association between GDM risk and some genetic risk score models. We identified different SNPs significantly associated with increased or decreased GDM risk, after performing stratified logistic regression analysis adjusted by age, pre-pregnancy BMI and FPG 12 GW, clinical features significantly different between women with and without GDM in our cohort. Then, six SNPs were selected to build the final genetic risk score models. The power of the prediction enhanced when genetic information was included. Thus, if the risk score is applied at the beginning of the pregnancy, the AUC would increase from 0.573 and 0.644 including only the age and the pre-pregnancy BMI to 0.684 and 0.714 adding the selected SNPs in CAU and LAT, respectively. In line, at the 12 GW, the AUC would increase from 0.651 and 0.685 including only the age, the pre-pregnancy BMI and the FPG at 12 GW to 0.745 in CAU and 0.760 in LAT adding the selected SNPs, showing in the last the highest predictive value of GDM in our study. Therefore, the present risk score could help to identify those women more prone to develop GDM and might allow us to initiate the proper management earlier. In addition, due to the high negative predictive value it might rule out the women with a low risk of GDM.
These results are consistent with previous evidence in which well-recognized risk factors and the combination of genetics increased GDM prediction [
18,
19,
20].
Lamri et. al, found a polygenic risk score of GDM with a power of prediction of AUC=0.62 using a cohort of South Asian women [
18]. Kawai et al., described that the combination of clinical variables and a GRS improved prediction of GDM with AUC=0.70 among Caucasian Women using the GDM Carpentier & Coustan criteria [
20]. Zulueta et al., carried out a case-control study of 139 Mexican women with GDM to build an GDM risk algorithm. They included four phenotypic variables, maternal age, pre-pregestational BMI, family history of T2D and previous pregnancies as well as 11 SNPs showing an AUC of 0.751 [
21].
However, these results are not totally comparable since there are some inconsistencies across studies due to different samples sizes, populations, GDM screening, diagnostic criteria and the statistical analysis.
Regarding the candidate SNPs, we selected those SNPs known to be associated with T2DM and/or GDM and previously SNPs explored in our cohort [
12]. Therefore, the SNPs are located in genetic loci associated to glucose metabolism such as rs1387153 and rs10830963 (melatonin receptor 1b MTNR1B), rs9368222 (cyclin-dependent kinase 5 regulatory subunit associated protein 1-like 1, CDKAL1), rs7651090 (insulin-like growth factor 2mRNA-binding protein 2, IGF2BP2), rs1371614 (dihydropyrimidinase-related protein 5, DPYSL5).
These rs1387153 and rs10830963 polymorphisms of MTN1RB gene are known to be associated with an increased risk of GDM [
22] and also with an increased GDM risk in genetic models of Chinese population, showing similar effect sizes [
19], as well as in genetic models of Caucasian population with modest size effects [
20]. In deep, the rs10830963 variant in MTNR1B has been associated with FPG values and impaired beta-cell function [
23] and might be considered as a pharmacogenetic marker of antenatal insulin therapy in interaction with the pre-pregnancy BMI [
24]. The rs1387153, has been also related in European populations with an increased of FPG and T2DM [
25]. The IGF2BP2 variant 7651090 has been described in Malaysian subjects with glutamic acid decarboxylase antibodies (GADA) negative diabetes [
26]. The rs1371614 DPYSL5 and rs9368222 CDKAL1 variants have been previously reported in GDM women in our cohort [
12].
On the contrary, we identify other SNPs that might confer a protective effect such as the rs7607980 (cordon-bleu protein-like 1, COBLL1), rs10885122 (adrenoceptor alpha 2A, ADRA2A), rs1496653 (ubiquitin-conjugating enzyme E2, UBE2E2), rs340874 (proper homeobox protein-1, PROX1) and rs9368222 (CDK5 regulatory subunit associated protein 1 like 1, CDKAL1) variants with a lower risk of GDM. In line, the rs7607980 has been related to lower fasting insulin in children with overweight and obesity [
27]. However, most of the variants have been reported associated to an increased risk of T2DM development.
Remarkably, we only found genetic variants of the MTNR1B gene in both races. No other SNP were identified in common in CAU and LAT pregnant women.
To the best of our knowledge, this is the first study that examines significant associations between candidate SNPs, environmental factors and GDM risk in genetic models in Spanish population.
Our study exhibits several strengths. We studied our own cohort of subjects representing a broad spectrum of pregnant women. They were carefully phenotyped with clinical, demographic, and anthropometric data confirmed in a face-to-face appointment and with all the patients performing our own OGTT, obtaining a GDM diagnosis based on validated diagnostic criteria.
On the contrary, there are some limitations that need to be acknowledged. Despite the important sample size of our study (1588 pregnant women), studies with larger samples sizes would be needed to validate GRS model. In addition, this is an observational study and we adjusted for multiple confounders in our analysis, however, another confounding factors might still be missed. Finally, our study included only CAU and LAT women. Whether our findings can be generalized and replicated to other ethnic groups needs further investigation.
4. Conclusions
In conclusion, adding genetic variants enhanced the prediction model of GDM risk in CAU and LAT pregnant women in order to facilitate the identification and proper and earlier management of GDM. A Randomized Controlled interventional Trial is underway to demonstrate its usefulness
5. Methods
5.1. Population
The Hospital Clínico San Carlos is a public hospital located in the central area of Madrid and had a healthcare population of 445,000 inhabitants assigned during the performance of this study, between 2015-2017. The prenatal screening consultation is located in the hospital where all pregnant women are admitted between the 9-12 GW to perform the first ultrasound and the screening test to detect the risk of chromosomal alterations.
San Carlos Cohort. The San Carlos Cohort is constituted of women who took part in the studies for the prevention of Gestational Diabetes carried out between 2015-2017 and registered as randomized control trial (ISRCTN84389045;
https://doi.org/10.1186/ISRCTN84389045, accessed on 29 January 2024) [
28] and a prospective real-world study (ISRCTN13389832;
https://doi.org/10.1186/ISRCTN13389832, accessed on 29 January 2024) [
29]. The studies were approved by the Clinical Trials Committee of the Hospital Clínico San Carlos (CI13/296-E, CI16/442-E and CI16/316). All women signed the informed consent.
A total of 3,036 women were enrolled in the studies and 2156 women samples were obtained for genetic study. Normoglycemic < 92 mg/dL, pregnant women at 8-12 GW were invited to participate upon their first ultrasound visit. Gestational age at entry for inclusion was based on the one obtained in this first ultrasound. Inclusion criteria: ≥ 18 years old, single gestation, acceptance of participation in the study and signature of the consent form. Exclusion criteria: gestational age at entry > 14 GW, intolerance to nuts or extra virgin olive oil, medical conditions or pharmacological therapy that could compromise the effect of the intervention and/or the follow-up program.
Genotyping samples were obtained from 1711 (79.4%) women, of which 1645 (96%) were valid. In 1588 (93%), 1069 CAU and 519 LAT, the samples were sufficient to determine > 95 SNPs to be assessed in this study.
5.2. Gestational diabetes mellitus diagnosis
Diagnosis of GDM was performed by a 75-g OGTT at 24-28 GW using the IADPSG criteria [
30].
5.3. Clinical and laboratory parameters
Clinical, demographic and anthropometric data were collected in a face-to-face appointment and examination. FPG levels were determined by the glucose oxidase method in fresh plasma samples and lipid profiles were measured using an analyzer (CobasR 8000 c702, Roche Diagnostics, Basel, Switzerland).
5.4. Lifestyle evaluation
The dietary intake and physical activity were evaluated using the 14-point Mediterranean Diet Adherence Screener (MEDAS) [
31]. This questionnaire considers 14 items and evaluates adherence to Mediterranean Diet. The compliance of each item provides +1 points. A score ≥ 10 is considered as target.
5.5. Genotype analysis
Genomic DNA was extracted from EDTA-stabilized blood samples taken during the OGTT using the Maxwell RSC instrument (Promega, Dubendorf, Switzerland).
Genotyping was performed by IPLEX MassARRAY PCR using the Agena platform (Agena Bioscience, SanDiego, CA). IPLEX MassARRAY PCR and extension primers were designed from sequences containing each target SNP and 150 upstream and downstream bases with AssayDesign Suite (
http://agenabio.com/assay-design-suite-20-software) using the default settings. Single base extension reactions were performed on the PCR reactions with the iPLEX Gold Kit (AgenaBioscience) and 0.8μl of the custom UEP pool. The kit contains mass modified terminator nucleotides that increase the mass difference between extended UEPs, allowing for greater accuracy in genotyping. The mass difference with unmodified terminator nucleotides ranges from 9 to 40 kDa, depending on the two nucleotides compared. With the mass modified terminator nucleotides the mass difference increases to 16–80 kDa. The single base extension reactions were cycled with a nested PCR protocol that used five cycles of annealing and extension nested with a denaturation step in a cycle that was repeated 40 times for a total of 200 annealing and extension steps. The goal was to extend nearly all of the UEPs. Following single base extension, the reactions were diluted with 16μl of water and deionized with 6 ng of resin. After deionizing for 20 min the reactions were dispensed onto SpectroChipArrays with a Nanodispenser (Agena Bioscience). The speed of dispensation was optimized to deliver an average of 20 nl of each reaction to a matrix pad on the SpectroChip. An Agena Bioscience Compact MassArray Spectrometer was used to perform MALDI-TOF mass spectrometry according to the iPLEX Gold Application Guide. The Typer 4 software package (Agena Bioscience) was used to analyze the resulting spectra and the composition of the target bases was determined from the mass of each extended oligo. These panels were designed in collaboration with PATIA and Genotyping was performed at the Agena platform located at the Epigenetics and Genotyping laboratory, Central Unit for Research in Medicine (UCIM), Faculty of Medicine, University of Valencia, Valencia, Spain.
5.6. Statistical analyses
Qualitative variables were summarized by their number and frequencies, normally distributed continuous variables by mean and standard deviation (± SD) and nonnormally distributed continuous variables by median and interquartile range (IQR: P25-P75).
All the analysis was carried out by stratifying the sample by ethnicity, according to the two categories present in the data: Caucasian (CAU) and Latin (LAT).
Differences in clinical, laboratory, anthropometric and genetic variables (age, pre-pregestational BMI, FPG at 12 GW) between women with and without GDM were tested for significance using the student’s t-test.
Independent variables including age (≤ 35 and > 35 years) and pre-pregnancy BMI (<25 and ≥25 kg/m2) were categorized based on known categories of GDM risk. In case of FPB at 12 GW (≤ 83.5 and > 83.5 mg/dL) we define an optimal cut-point value of our cohort performing a ROC curve stratifying by ethnicity. The identification of the cut-point value required the Youden Index which maximizes sum sensitivity and specificity.
A set of 110 SNPs associated with GDM according to bibliographical references [
32,
33,
34,
35,
36,
37,
38] were analyzed [
12]. To evaluate the association between each SNP individually and the GDM risk, univariate binary logistic regression models were used. The genetic analysis was performed using R 4.3.1.
The SNPs were categorized according to the reference allele, the allele indicated in the previous literature was taken as the reference (REF) category. By contrast, the alternate category (ALT) included either heterozygous or homozygous mutations [
12]. For each test, the corresponding p-value was obtained. SNPs previously identified [
12] and/or with a p-value less than 0.05 in the present study were selected for the multivariate analysis.
Finally, three multiple logistic regressions analysis were performed for GDM risk. First, the multivariate completed model; second, the stepwise backward model and third, the stepwise forward model with the 0.05 p-value set for entering and for exclusion. The final model selected an optimal model with the best predictors together. The results of the regression models were presented using the odds ratio (OR), its 95%CI and the p-value.
The ROC analysis was used as a diagnostic tool for the logistic regression. The ROC curve with the AUC for the probabilities predicted by the final models of each analysis and its 95% CIs were calculated and compared.
Supplementary Materials
The following supporting information can be found as: Supplementary Tables (Table S1. The SNP considered for CAU women. Table S2. The SNP considered for LAT women. Table S3. Logistic regression model for CAU women including age and pre-pregnancy BMI. Table S4. Logistic regression model also considering FPG 12 GW for CAU women, Table S5. Logistic regression model for LAT women including age and pre-pregnancy BMI. Table S6. Logistic regression model also considering FPG 12 GW for LAT women) and Supplementary figures (FIGURE S1 ROC for CAU, Panel a for age. Panel b for age and BMI. Panel c for Age, BMI and FPG. Panel d For Relevant Snips and age, BMI and FPG.
Figure 2S ROC for LAT. Panel a for age. Panel b for age and BMI. Panel c for Age, BMI and FPG. Panel d For Relevant Snips and age, BMI and FPG.)
Author Contributions
Conceptualization and design: MA-R, IS, AB, PM-M and ALC-P. Data curation, analysis and interpretation of data: MA-R, IS, MP, AB, PM-M, ALC-P, IM, MC, LdV, VM, JV, RO, VM. Funding acquisition: ALC-P. Investigation: ALC-P, MMN, PdM., AD, AB, AR, MAR, CM, PM-M. Methodology: ALC-P, CF, IM, MPG, MA-R, MJT, IM, PdM, MAR, CM, AB, LdV, VM, JV, RO. Supervision, Validation and Visualization: ALC-P, AR, IS, AB, MP, MA-R. Writing – original draft: MA-R, IS, PM-M, ALC-P. Writing – review & editing: MA-R, IS, ALC-P. All authors have seen and agree with the content of the full last version of manuscript.
Funding
This research was funded by grants from the Instituto de Salud Carlos III/MICINN of Spain under grant number PI20/01758, and European Regional Development Fund (FEDER)‘‘A way to build Europe’’ and Ministerio de Ciencia e Innovación, and Agencia Estatal de Investigación of Spain under grant number PREDIGES RTC2019-007406-1.The design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, and approval of the manuscript; and decision to submit the manuscript for publication are the responsibilities of the authors alone and independent of the funders
Institutional Review Board Statement
The studies were approved by the Clinical Trials Committee of the Hospital Clínico San Carlos (CI13/296-E, CI16/442-E and CI16/316).
Informed Consent Statement
All women signed the informed consent.
Data Availability Statement
The data analysed in this study is subject to the following licenses/restrictions: No restriction. Requests to access these datasets should be directed to acalle.edu@gmail.com
Acknowledgments
We wish to acknowledge our deep appreciation to the administrative personnel and nurses and dieticians from the Laboratory Department (María Dolores Hermoso Martín, María Victoria Saez de Parayuelo), the Pregnancy and Diabetes Unit and to all members of the Endocrinology and Nutrition and Obstetrics and Gynaecology departments of the San Carlos Clinical Hospital, Madrid, Spain
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Diagnostic criteria and classification of hyperglycaemia first detected in pregnancy: a World Health Organization Guideline. Diabetes Res Clin Pract. 2014;103(3):341-63. [CrossRef]
- Wang H, Li N, Chivese T, Werfalli M, Sun H, Yuen L, et al. IDF Diabetes Atlas Committee Hyperglycaemia in Pregnancy Special Interest Group. IDF Diabetes Atlas: Estimation of Global and Regional Gestational Diabetes Mellitus Prevalence for 2021 by International Association of Diabetes in Pregnancy Study Group's Criteria. Diabetes Res Clin Pract. 2022;183:109050. [CrossRef]
- Sweeting A, Wong J, Murphy HR, Ross GP. A Clinical Update on Gestational Diabetes Mellitus. Endocr Rev. 2022;43(5):763-793. [CrossRef]
- HAPO Study Cooperative Research Group; Metzger BE, Lowe LP, Dyer AR, Trimble ER, Chaovarindr U, et al. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med. 2008;358(19):1991-2002. [CrossRef]
- Dabelea D, Hanson RL, Lindsay RS, Pettitt DJ, Imperatore G, Gabir MM, et al. Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes. 2000;49(12):2208-11. [CrossRef]
- Tam WH, Ma RCW, Ozaki R, Li AM, Chan MHM, Yuen LY, et al. In Utero Exposure to Maternal Hyperglycemia Increases Childhood Cardiometabolic Risk in Offspring. Diabetes Care. 2017;40(5):679-686. [CrossRef]
- Vounzoulaki E, Khunti K, Abner SC, Tan BK, Davies MJ, Gillies CL. Progression to type 2 diabetes in women with a known history of gestational diabetes: systematic review and meta-analysis. BMJ. 2020;369:m1361. [CrossRef]
- Sweeting AN, Appelblom H, Ross GP, Wong J, Kouru H, Williams PF, et al. First trimester prediction of gestational diabetes mellitus: A clinical model based on maternal demographic parameters. Diabetes Res Clin Pract. 2017;127:44-50. [CrossRef]
- Hayes MG, Urbanek M, Hivert MF, Armstrong LL, Morrison J, Guo C, et al. Identification of HKDC1 and BACE2 as genes influencing glycemic traits during pregnancy through genome-wide association studies. Diabetes. 2013;62(9):3282-91. Erratum in: Diabetes. 2013;62(10):3641. [CrossRef]
- Kwak SH, Kim SH, Cho YM, Go MJ, Cho YS, Choi SH, et al. A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes. 2012;61(2):531-41. [CrossRef]
- Tian Y, Li P. Genetic risk score to improve prediction and treatment in gestational diabetes mellitus. Front Endocrinol (Lausanne). 2022;13:955821. [CrossRef]
- Ramos-Levi A, Barabash A, Valerio J, García de la Torre N, Mendizabal L, Zulueta M, et al. Genetic variants for prediction of gestational diabetes mellitus and modulation of susceptibility by a nutritional intervention based on a Mediterranean diet. Front Endocrinol (Lausanne). 2022;13:1036088. [CrossRef]
- International Diabetes Federation. IDF Diabetes Atlas. 9th ed. International Diabetes Federation; 2019. https://www.diabetesatlas.org/en. Accessed 4 February 2024.
- Fu J, Retnakaran R. The life course perspective of gestational diabetes: An opportunity for the prevention of diabetes and heart disease in women. EClinicalMedicine. 2022;45:101294. [CrossRef]
- Hod M, Kapur A, Sacks DA, Hadar E, Agarwal M, Di Renzo GC, et al. The International Federation of Gynecology and Obstetrics (FIGO) Initiative on gestational diabetes mellitus: A pragmatic guide for diagnosis, management, and care. Int J Gynaecol Obstet. 2015;131 Suppl 3:S173-211. [CrossRef]
- World Health Organization. Diagnostic Criteria and Classification of Hyperglycemia First Detected in Pregnancy. Worl Helath Organization; 2013. https://apps-who.int/ris/handle/10665/85975. Accessed 18 February 2024.
- American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2020. Diabetes Care. 2020;43(Suppl 1):S14-S31. [CrossRef]
- Lamri A, Mao S, Desai D, Gupta M, Paré G, Anand SS. Fine-tuning of Genome-Wide Polygenic Risk Scores and Prediction of Gestational Diabetes in South Asian Women. Sci Rep. 2020;10(1):8941. [CrossRef]
- Shen Y, Jia Y, Li Y, Gu X, Wan G, Zhang P, et al. Genetic determinants of gestational diabetes mellitus: a case-control study in two independent populations. Acta Diabetol. 2020;57(7):843-852. [CrossRef]
- Kawai VK, Levinson RT, Adefurin A, Kurnik D, Collier SP, Conway D, et al. A genetic risk score that includes common type 2 diabetes risk variants is associated with gestational diabetes. Clin Endocrinol (Oxf). 2017;87(2):149-155. [CrossRef]
- Zulueta M, Gallardo-Rincón H, Martinez-Juarez LA, Lomelin-Gascon J, Ortega-Montiel J, Montoya A, et al. Development and validation of a multivariable genotype-informed gestational diabetes prediction algorithm for clinical use in the Mexican population: insights into susceptibility mechanisms. BMJ Open Diabetes Res Care. 2023;11(2):e003046. [CrossRef]
- Kim JY, Cheong HS, Park BL, Baik SH, Park S, Lee SW, et al. Melatonin receptor 1 B polymorphisms associated with the risk of gestational diabetes mellitus. BMC Med Genet. 2011;12:82. [CrossRef]
- Liu C, Wu Y, Li H, Qi Q, Langenberg C, Loos RJ, et al. MTNR1B rs10830963 is associated with fasting plasma glucose, HbA1C and impaired beta-cell function in Chinese Hans from Shanghai. BMC Med Genet. 2010;11:59. [CrossRef]
- Firneisz G, Rosta K, Al-Aissa Z, Hadarits O, Harreiter J, Nádasdi Á, et al. The MTNR1B rs10830963 Variant in Interaction with Pre-Pregnancy BMI is a Pharmacogenetic Marker for the Initiation of Antenatal Insulin Therapy in Gestational Diabetes Mellitus. Int J Mol Sci. 2018;19(12):3734. [CrossRef]
- Bouatia-Naji N, Bonnefond A, Cavalcanti-Proença C, Sparsø T, Holmkvist J, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41(1):89-94. [CrossRef]
- Salem SD, Saif-Ali R, Ismail IS, Al-Hamodi Z, Poh R, Muniandy S. IGF2BP2 alternative variants associated with glutamic acid decarboxylase antibodies negative diabetes in Malaysian subjects. PLoS One. 2012;7(9):e45573. [CrossRef]
- Mancina RM, Burza MA, Maglio C, Pirazzi C, Sentinelli F, Incani M, et al. The COBLL1 C allele is associated with lower serum insulin levels and lower insulin resistance in overweight and obese children. Diabetes Metab Res Rev. 2013;29(5):413-6. [CrossRef]
- Assaf-Balut C, García de la Torre N, Durán A, Fuentes M, Bordiú E, Del Valle L, et al. A Mediterranean diet with additional extra virgin olive oil and pistachios reduces the incidence of gestational diabetes mellitus (GDM): A randomized controlled trial: The St. Carlos GDM prevention study. PLoS One. 2017;12(10):e0185873. [CrossRef]
- de la Torre NG, Assaf-Balut C, Jiménez Varas I, Del Valle L, Durán A, Fuentes M, et al. Effectiveness of Following Mediterranean Diet Recommendations in the Real World in the Incidence of Gestational Diabetes Mellitus (GDM) and Adverse Maternal-Foetal Outcomes: A Prospective, Universal, Interventional Study with a Single Group. The St Carlos Study. Nutrients. 2019;11(6):1210. [CrossRef]
- Metzger BE, Gabbe SG, Persson B, Buchanan TA, Catalano PA, Damm P, et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33(3):676-82. [CrossRef]
- Schröder H, Fitó M, Estruch R, Martínez-González MA, Corella D, Salas-Salvadó J, et al. A short screener is valid for assessing Mediterranean diet adherence among older Spanish men and women. J Nutr. 2011;141(6):1140-5. [CrossRef]
- Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659-69. [CrossRef]
- Zhang C, Bao W, Rong Y, Yang H, Bowers K, Yeung E, et al. Genetic variants and the risk of gestational diabetes mellitus: a systematic review. Hum Reprod Update. 2013;19(4):376-90. [CrossRef]
- Hayes MG, Urbanek M, Hivert MF, Armstrong LL, Morrison J, Guo C, et al. Identification of HKDC1 and BACE2 as genes influencing glycemic traits during pregnancy through genome-wide association studies. Diabetes. 2013;62(9):3282-91. Erratum in: Diabetes. 2013;62(10):3641. [CrossRef]
- Huerta-Chagoya A, Vázquez-Cárdenas P, Moreno-Macías H, Tapia-Maruri L, Rodríguez-Guillén R, López-Vite E, et al. Genetic determinants for gestational diabetes mellitus and related metabolic traits in Mexican women. PLoS One. 2015;10(5):e0126408. [CrossRef]
- Lowe WL Jr, Scholtens DM, Sandler V, Hayes MG. Genetics of Gestational Diabetes Mellitus and Maternal Metabolism. Curr Diab Rep. 2016;16(2):15. [CrossRef]
- Wu L, Cui L, Tam WH, Ma RC, Wang CC. Genetic variants associated with gestational diabetes mellitus: a meta-analysis and subgroup analysis. Sci Rep. 2016;6:30539. [CrossRef]
- Ding M, Chavarro J, Olsen S, Lin Y, Ley SH, Bao W, et al. Genetic variants of gestational diabetes mellitus: a study of 112 SNPs among 8722 women in two independent populations. Diabetologia. 2018;61(8):1758-1768. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).