You are currently viewing a beta version of our website. If you spot anything unusual, kindly let us know.
Preprint
Article
RNA Extraction from Cumulus Cells for Gene Analysis: The Current Strategy and Review of Literature
Altmetrics
Downloads
152
Views
84
Comments
0
This version is not peer-reviewed
Abstract
RNA extraction from human cumulus cells (CCs) is considered challenging due to low cell’s pop-ulation and small cell. The successful gene expression analysis from cumulus cells ultimately de-pends on an excellent concentration of Ribonucleic acid (RNA) yield by an appropriate technique. Our study aims to consolidate appropriate cumulus cell collection method, preparation and ex-traction techniques to ensure a good yield of RNA for better gene expression analysis. From the results obtained, the optimal strategy found involved mechanical denudation without an enzy-matic process, followed by the use of RNA stabilizer prior to RNA extraction. Subsequently, uti-lizing a smaller homogenizer, preferably manual pressure control with filter (Bio-Masher III®) is paramount for a pure and homogenize sample. Subsequently, during RNA extraction, the use of RNeasy micro kit and RNA carriers is recommended. These sample collection, preparation, and extraction techniques yielded an excellent-quality RNA concentration and successful gene expres-sion analysis. We also conducted a systematic review to consolidate our findings with current evidence of RNA extraction technique. Our results added a value as a current strategy for opti-mizing RNA extraction in CCs for experimental studies. Thus, our findings can contribute to for-mulating a better strategy for optimizing the RNA extraction in CCs for transcriptomic studies.
Keywords:
Subject: Medicine and Pharmacology - Obstetrics and Gynaecology
1. Introduction
In reproductive biology, the intricate interplay of various cell types orchestrates the complex oocyte maturation process, culminating in successful fertilization and embryogenesis [1,2]. Among these crucial players, cumulus cells (CCs) emerge as indispensable companions to oocytes, providing vital support and regulatory functions throughout the maturation process. The CCs, specialized somatic cells, reside near the oocyte within the ovarian follicles. Their intimate association and intricate communication with the oocyte are essential for guiding and facilitating its developmental journey [3]. The cumulus-oocyte complex forms a dynamic microenvironment, fostering bidirectional signaling that influences oocyte growth, meiotic progression, and eventual ovulation. This proximity enables CCs to exchange nutrients, growth factors, and metabolites with the oocyte, thereby ensuring optimal oocyte maturation [1,4].
At the molecular level, the CCs support the oocyte development closely by secreting Hyaluronic acid (HA) while expanding the follicle-stimulating hormone (FSH) stimulation [5]. The mural granulosa cell (MGCs), which supports the growth of follicles, coordinates the endocrine function externally. The ovulation phase occurs due to increased HA secretion by CCs once the rupture of the dominant follicles remarks a luteinizing hormone [6] surge [7]. Morphologically, CCs exhibit diverse shapes and sizes, ranging from spindle-like to cuboidal. These variations in morphology reflect the dynamic nature of their functions and underline their adaptability to the changing needs of oocyte maturation [4,8]. The cumulus cell layer enveloping the oocyte forms a protective shield and a communication bridge between the oocyte and the surrounding ovarian microenvironment [9]. The number of CCs enveloping an oocyte can differ depending on the follicular development stage. During the early stages of folliculogenesis, the number of CCs is relatively sparse. As the follicle progresses and matures, a corona radiata forms around the oocyte, comprising numerous layers of closely packed CCs. This protective cocoon of CCs serves as a conduit for nutrient exchange, signaling molecules, and regulatory factors that profoundly impact oocyte maturation [2,4,9].
The size and morphology of CCs exhibit remarkable diversity, reflecting the complex interplay of various intrinsic and extrinsic factors. Several factors contribute to the variability in CC size [10]. Apart from the follicular stage, the number, size, and morphology of the CCs are influenced by hormone levels, such as FSH and LH, metabolic status, ovarian microenvironment including oxygen tension and local growth factors, genetic variability, and aging [11]. Minimal CCs are observed in cases of low oocyte quality, particularly in women with poor ovarian reserve [12] and in the elderly [13]. Due to its minimal CCs, the challenges in obtaining a good concentration of RNA are even higher.
The cumulative evidence has highlighted the pivotal role of CCs as invaluable indicators of oocyte quality. The molecular and biochemical composition of CCs can provide insightful information about the developmental competence of the oocyte and subsequent embryo quality [14]. CCs are endowed with the potential to harbor critical molecular biomarkers that offer predictive insights into the likelihood of successful fertilization and embryo implantation. This burgeoning field holds promise for revolutionizing assisted reproductive technologies and infertility treatments by allowing more informed decision-making regarding oocyte selection.
Most evidence utilizes the CC’s ribonucleic acid (RNA) to study the critical mechanism of CC’s in oocyte development and maturation. For these matters, the RNA extraction steps are crucial. It required a proper purification of RNA from the CC’s tissue samples. Nevertheless, ribonuclease enzymes in the tissues can complicate it, leading to rapid degradation of RNA material upon extraction. To date, various methods are used in molecular biology to isolate RNA from CCs. The most commonly reported is the guanidinium thiocyanate-phenol-chloroform extraction. It consists of the filter paper-based lysis followed by the elution method, which features high throughput capacity. Despite all the reported RNA extraction methods in CCs, given their minute size and scarcity, they still pose significant challenges to effective RNA extraction. The ranges of RNA concentration is still below 10; ranging from 4 - 7 ng/µl as reported by previous study [15].
Nevertheless, the limited number of CCs obtained from a single follicle necessitates meticulous optimization of RNA extraction protocols to ensure sufficient and high-quality RNA [16,17]. The significance of RNA extraction in CCs must be considered. However, despite its importance, limited published literature suggests the optimal method for achieving a pure final product. Therefore, it is imperative to conduct further research and experimentation to uncover the best approach for extracting RNA in CCs. A notable gap exists in the current shreds of evidence regarding the detailed methodology for RNA extraction from CCs. While numerous original articles delve into the molecular insights gleaned from CCs, the intricacies of RNA extraction should be addressed. Our study addresses a significant gap in gene expression studies in CCs. We aim to provide a comprehensive optimization strategy that covers all aspects of sample collection, preparation, RNA extraction, and purification. By consolidating our findings with current literature, we will offer valuable insights to help researchers prepare high-quality RNA for their studies. Our research will pave the way for more accurate and reliable gene expression studies in the future.
2. Materials and Methods
2.1. Clinical Study
2.1.1. Study Design, Recruitment, and Ethical Approval
The prospective cohort study was conducted in the Advanced Reproductive Center, Hospital Canselor Tuanku Mukhriz (HCTM) UKM Cheras, Kuala Lumpur, from June 2022 to December 2022. This research was approved by the Human Ethical Research Committee (JEP-2022-187), Faculty of Medicine, National University of Malaysia Cheras, Kuala Lumpur, Malaysia. Consent was obtained prior to CC collection during the oocyte retrieval (OR) procedure for in-vitro fertilization (IVF). The CCs were discarded after separation from surrounding oocytes – “denudation” before the IVF procedure. Thus, obtaining these samples does not interfere with the IVF treatment. All women who underwent OR during the study period were included regardless of the causes of the infertility. A total of 80 samples of CCs from 80 follicles to extract RNA and optimize the housekeeping gene (HKG) hypoxanthine–guanine phosphoribosyl transferase (HPRT) amplification was collected and stored in an Eppendorf tube size 1.5 mL filled with 500 µL RNALater® solution (Thermo Fisher USA).
2.1.2. RNA Extraction Method Optimization
For the purpose of our study, the RNA extraction method optimization was divided into three stages:
- A.
- Sample collection optimization
- A.
- B. Sample preparation optimization
- A.
- C. RNA purification optimization
2.1.2.1. Sample Collection Optimization
The oocyte denudation (OD) process was the initial step of CC sample collection. To date, most centers implement the enzymatic protocol for oocyte denudation [18]. In brief, OD was conducted in two phases. Initially, the fresh retrieval oocytes were enzymolyzed in hyaluronidase – hyase-10x TM (Vitrolife® Sweden) to dissolve the bonds between CCs within 40 seconds; then, CCs were further removed from oocytes mechanically by repetitive pipette aspiration and deposition in a series of media without an enzyme. Finally, the “naked” – clean oocytes were examined for their maturity, and only meiosis II stage oocytes (MII) with the presence of polar body were used for IVF procedure either via intra-cytoplasmic sperm injection (ICSI) or conventional IVF. The comparison of Cumulus-Oocytes-Complex (COCs) features with and without hyaluronidase and post-denudation of both techniques were showed in Figure 2. For this study, 40 CCs were collected via conventional method using enzymatic protocol (Group A), whereas 40 other CCs were collected without enzymatic protocol (Group B) – purely mechanical using only manual force created via repetitive pipette aspiration and deposition in a series of media without an enzyme to obtain naked oocytes. The study flow is illustrated in Figure 1.
2.1.2.2 Sample Preparation Optimization
For sample preparation, CCs were lysed to form smaller molecules to achieve excellent RNA extraction [19]. This step required a mixer of RNA carrier solution (QIAamp® Circulating Nucleic Acid Kit) and diluted with buffer RLT, forming the RNA carrier dilution (RCD). The RLT buffer is a lysis buffer used to lyse CCs prior to RNA isolation. All the CC samples were prepared with RCD. In addition, the homogenizers were used as a mechanical disruption agent for CC preparation. This approach helps in forming an even mixture by forcing the CCs through the narrow space via multiple forces and turbulence to distribute the CC molecules as a solution evenly. In our study, we used two types of homogenizers, standard rotor–stator homogenizer [20] or disposable easy grind homogenizer, BioMasher III (Funakoshi®,Japan). In summary, the standard RSH consists of a fast-spinning inner rotor with a stationary outer sheath–stator. It homogenizes CCs through mechanical tearing and shear fluid forces and mixing the CC samples in Eppendorf tubes [21]. The BioMasher III (Funakoshi®, Japan) is formed by a filter tube with an abrasive surface inner wall and combined with a pestle with a textured surface. The pestle in the filtered tube layered on to centrifuge the tube is used for manual grinding [22]. Then, the CC-extracted samples were filtered and placed in the recovery tube. For these steps, every 20 CCs from Groups A and B were homogenized using a rotor–stator homogenizer, and the 20 other CCs from the same groups were manually ground via BioMasher III.
- Rotor–stator homogenizer [20].
Subsequently, the collected CCs were transferred into an empty 1.5 ml centrifuge tube using a sterile pipette tip. Then, 100 µl of RCD was added to the microcentrifuge. Rotor–stator homogenizer was used to disrupt and homogenize the samples. Another 100 µl of RCD was added and centrifuged at 10,000 xg for 30 seconds. Thereafter, the “pass through” homogenized samples were collected for RNA purification.
- 2.
- BioMasher III
The collected CCs, together with their RNA stabilizer, were transferred into the Biomasher III filter column. The CCs were separated from RNA stabilizer through 2000 x g centrifugation for 10 seconds. The filtrate that contained RNA stablizer was subsequently discarded. Then, 100 µl of RCD was added to the filter column. The CCs were ground using the pastel provided. Another 100 µl of RCD was added, and the disrupted samples were homogenized through the filter by centrifugation at 10000 x g for 30 seconds. Subsequently, the “pass through” homogenized samples were collected for RNA purification.
2.1.2.3. RNA Purification Optimization
The RNA from all samples was purified using an RNAeasy® micro kit (Qiagen) with few modifications. In brief, post-homogenized CC samples were collected in a 1.5 mL tube and added with 200 µl of 70% ethanol. Subsequently, the solution was transferred into an RNAeasy column within a 2 mL tube and centrifuged at 8,000 xg for 15 seconds. The flowthrough was then discarded. The 350 mL buffer RW1 was added to the RNAeasy column and centrifuged at 8,000 xg for 15 seconds, and then the flowthrough was discarded. Thereafter, 80 microL of DNAse I dilution was added directly into the spin column RNAeasy. The solution was left to rest on a bench for 15 minutes at room temperature. Then, the 350 µl buffer RW1 was added to the RNAeasy column and centrifuged at 8,000 xg for 15 seconds, and the 2 mL collecting tube (CT) with flowthrough was discarded and replaced with a new 2 mL CT at the RNAeasy column. The 500 µl buffer RPE was added to the RNAeasy column and centrifuged at 8,000 xg for 2 minutes. Subsequently, the CT was discarded. The 500 µl of 80% ethanol was not added in the further modification. Subsequently, the new 2 mL CT was added to the RNAeasy column, with the lid open, centrifuged at full speed for 5 minutes to dry the membrane, and the flowthrough was discarded. Finally, the new 1.5 mL CT was placed at the RNAeasy column, and 14 µL RNAse free water was added at the direct center of the RNAeasy column and set for 5 minutes. The lid was closed gently, followed by centrifugation, which was performed at full speed for one minute to elucidate the final RNA.
2.1.2. RNA Concentration and Purity Measurement
The concentration and purity of the purified RNAs were measured using a Nanodrop. The purity of the samples was evaluated by measuring the ratio of absorbance readings at 260 nm (specific for nucleic acids). Samples with A260/A280 ratios between 1.80 and 2.10 were considered to have no significant RNA contamination, whereas the ratios of A260/A230 were used as a secondary measure of nucleic acid purity and was expected within the range of 2.0–2.2 [23].
2.1.3. cDNA Synthesis and qPCR
Following RNA purification, the cDNA synthesis was conducted accordingly. We considered two types of cDNA synthesis kits, namely, the QuantiTect reverse transcription kit (Qiagen) and the whole genome amplification kit (Qiagen) given that the CC RNA purity was assumed to be smaller than extensive samples/tissues. The cDNA synthesis was performed according to the standard protocol of both kits. Then, the quality of each cDNA synthesized was validated by qPCR amplification. A housekeeping gene (HKG), the hypoxanthine-guanine phosphoribosyl transferase (HPRT), was used to amplify the cDNA. The primer sequence used; Hs_HPRT1_1_SG. QuantiTect Primer Assay (Qiagen®) was used for the qPCR amplification according to the protocol. In brief, genomic DNA elimination was prepared on ice with total volume of 14 µl. Subsequently incubated for 2 minutes in 42°C. The master mix solution also prepared according to protocol. In brief, the master mix final volume is 20 µl including 14 µl from template RNA. Subsequently incubated for 15 minutes in 42°C and then incubated for 3 minutes in 95°C to inactivate the Quantiscript Reverse Transcriptase.
2.2. The Systematic Review
The systematic review was done following the standard guideline protocol based on the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) [24] to identify the best RNA extraction method for cumulus cells.
2.2.1. Information Source and Search Strategy
The PRISMA guideline was used in conducting the systematic review. A thorough search of the literature was conducted with EBSCOhost, PubMed, Science Direct, and Scopus. Relevant research articles that were released up until 1st March 2024, were located. Keywords such as "cumulus cells" and "oocytes" were taken from the Medical Subject Heading (MeSH). MeSH terms from the Cochrane Library were used to generate synonyms for keywords. Through the evaluation of gathered review articles, further text terms were discovered. The following keywords were applied as part of the search strategy: ("cumulus cell" OR "granulosa cell," OR “oocytes” OR “cumulus oocyte complex”) AND (“RNA extraction” OR “RNA purification” OR “RNA isolation”).
2.2.1.1. Inclusion & Exclusion Criteria
Case-control and cross-sectional studies with abstracts investigating the methods used for RNA extraction from cumulus cells, cumulus oocyte complex or oocytes. Only human and animal studies reporting on the RNA concentration and RNA purity of the extracted RNA were included to ensure the homogeneity of the data. In contrast, the editorials, case reports, conference proceedings, and narrative review articles were not included in this review since they had no primary data. Excluded studies included in silico studies, in vitro studies and any intervention studies. Studies that extract DNA from cumulus or oocytes were excluded.
2.2.1.2. Screening of Articles for Eligibility
The articles that were collected from all databases underwent three stages of screening. In the first step, duplicates were removed and all articles with irrelevant titles were excluded. In the second phase, the abstracts of the remaining papers were reviewed and those that did not fit the inclusion criteria were eliminated. Lastly, a thorough review of the full text of remaining articles was conducted. In the stage, every article that did not adhere to the requirements for inclusion was eliminated. All authors involved in the phases of screening, choosing, and extracting data. The PRISMA flow diagram that summarises the article assortment procedure and the grounds for item removal is displayed in Figure 3.
2.2.2. Study Selection, Data Extraction and Risk of Bias Assessment
Based on an initial search, five authors (A.M.F, A.M.A, M.H.I, M.J.N., and A.K.A.K.) screened all titles and abstracts of potential manuscripts. The selection criteria included manuscripts published in English from January 2012 to December 2022, evaluating the RNA extraction method for CCs and oocytes. Following the initial screening - title, and abstract, full-text screening was conducted, excluding manuscripts that used samples other than cumulus cells and oocytes as experimental material, non-English language, case reports, and review articles. The remaining potential manuscripts were then independently reviewed. The final selected manuscripts provided a detailed study design, focusing on RNA extraction methods with the final report of RNA concentration from each method. The conflicts in selection among authors were resolved through detailed discussions and opinions provided by the fifth, sixth, and seventh authors (S.S.E, N.S., and A.A.Z). Additionally, the National Institutes of Health (N.I.H.) tool for observational studies was employed to assess the quality of the selected manuscripts. This evaluation was based on 14 variables, scoring 1 for 'yes,' 0 for 'no,' or 'non-applicable' for N.A. The manuscripts were then categorized as poor (0–5), fair (6–9), or good (10–14) based on their total scores (Supplementary Table S1). Overall, the included studies in our review achieved a minimum fair to good score. Subsequently, the information gathered is as follows: (1) author name (year); (2) article title; (3) country (4) sample size; (5) organism; (6) RNA extraction methods; (7) RNA concentration; and (8) RNA purity. The summary of the collected data is listed in Table 2.
3. Results
3.1. Clinical Study
We collected 80 samples of CCs from 80 follicles to extract RNA and optimize the housekeeping gene (HKG) hypoxanthine–guanine phosphoribosyl transferase (HPRT) amplification. The samples were prepared and analyzed according to the steps outlined in Figure 1. Our women subjects had an average age of 37.19 ± 3.70, which was considered advanced, and their anti-Mullerian hormone (AMH) level was less than 10 picomol/L (7.507 ± 4.33), indicating a low ovarian reserve. After the extraction, we measured RNA quantity and quality for all method combinations. The RNA concentration was generally low, below 10 ng/μl (6.416 ± 5.15), and had an average purity of 1.712 ± 0.244 during RNA purification. However, non-enzymatic handling of CCs resulted in significantly higher RNA concentration at 9.00 ng/μl compared with 3.83 ng/μl (p<0.05). Combining BioMasher III with RNA carrier dilution (RCD) yielded better RNA concentration than the RSH tool at 9.15 ng/μl versus 3.68 ng/μl, although not statistically significant (p=0.18). Our final cDNA and qPCR analysis showed a significant presence of HPRT gene amplification following the use of QuantiTect reverse transcriptase kit (Qiagen) for cDNA synthesis in the BioMasher III and non-enzymatic group compared with another group (p<0.05). The HPRT gene amplification graph and melt peak curve is showed in Figure 4. Unfortunately, the group using whole genome amplification kits for cDNA synthesis with RNA carrier dilution (RCD) and RSH tool with QuantiTect Reverse Transcriptase Kit (Qiagen) for cDNA showed no amplification for their qPCR. All the results are tabulated in Table 1.
3.2. Systematic Review
To date, no available data has been reported that exclusively focuses on the RNA extraction method for cumulus cells (CCs) alone. Most RNA extraction methods have been briefly described in in-vitro studies, intervention, and silico studies, which were excluded from this review. Currently, most evidence for the technique combined CCs with oocytes or Cumulus-Oocytes Complex (COCs). Hence, in our search strategy, initially, no paper yielded only the RNA method for CCs. As a result, we modified the search strategy to include CCs, oocytes, and COCs.
3.2.1. Search Sequence and Quality Assessment
A total of 2603 studies were retrieved during the primary search (Figure 3). After removing 240 duplicates, the remaining 2363 articles were thoroughly screened based on our inclusion criteria. Amongst them, 2,353 articles were excluded, leaving 10 for full-text evaluation. After a detailed evaluation, seven articles were discarded, 3 using samples other than CCs and oocytes, and four papers – no RNA final concentration was reported. Subsequently, three studies that focused on RNA extraction methods with available results of final RNA concentrations were selected for this review. All selected articles were evaluated using the National Institutes of Health (NIH) tool for observational studies to ensure quality and minimize bias. Notably, all four articles obtained a minimum fair to good score, indicating a low risk of bias (Supplementary Table S1).
3.2.2. Studies Characteristics
A total of 1242 samples from three studies were included in this systematic review. All studies collected oocyte for RNA extraction optimization study. Maisarah et al. (2020) include 19 samples of cumulus oocyte complex for RNA extraction [16]. Wiweko et al. (2017) used human samples while Maisarah et al., (2020) and Pavani et al., (2015) were on animal study [16,25,26]. The RNA extraction methods reported included for comparison TRIzol, Guanidium thiocyanate and few types of column based commercial kits. All the relevant information from the included studies has been summarized in Table 2
3.2.3. Main Outcome
Most studies use pool of oocytes and COCs, ranging from 70 to 200 oocytes per pool, to yield the RNA and conclude that the RNA purity is good (1.0 to 2.0) using any preferred methods [16,25,26]. The two TRIzol protocol studies reported a higher RNA concentration with at least 150 ng/ μL than commercial kits [16,26]. Similarly, these two studies compared Modified TRIzol Protocol (MTP) with commercial kits and revealed that MTP is considered the best option for RNA extraction in oocytes[16,26]. In contrast, one study concluded that the yield of RNA using commercial kits is comparable in fresh versus frozen oocytes [25]. Unfortunately, none of these reported for RNA extraction for CCs alone.
4. Discussion
The high RNA concentration and purification are essential in molecular research, specifically in transcriptomic study. However, achieving excellent RNA concentration and purity could be challenging, especially for tiny samples, e.g. CCs [16,22]. Thus, evaluating an appropriate technique as an optimal strategy is crucial to improve the yield of RNA in these sample types. The comparison of multiple methods indicates that the combination of mechanical collection technique, Biomasher III, and RNA carrier improved the RNA concentration and purity obtained from the CCs.
The current practice for oocyte denudation in most in-vitro fertilization (IVF) centers worldwide is via the enzymatic technique, namely, hyaluronidase [27,28]. HYASE-10X™ (Vitrolife® Sweden) is often used to remove CCs from oocytes prior to intracytoplasmic sperm injection (ICSI) in our center. The maturation process in oocytes consists of the accumulation of HA in CCs as a protection mechanism. It is a high-molecular-weight glycosaminoglycan form with an alternate bond of D-glucuronic and N-acetylglucosamine [29]. The HA mainly accumulates within the CC oophorus to support the oocyte developing. These strong attachments of CCs and oocytes form a cumulus–oocytes complex [30], facilitating the supply of nutrients and growth factors for further enhancement and maturation [29,31].
Therefore, separating the CCs from COC for ICSI preparation is technically challenging. To date, the denudation process is conducted in two phases [18,32]. The first step was that the COCs would be enzymolyzed in hyaluronidase, such as HYASE-10X™ (Vitrolife® Sweden), to weaken the bond of HA within the CCs and oocytes. The process is performed rapidly because potential decremental effects can occur if the process takes more than 40 seconds due to enzyme toxicity. Subsequently, the second phase is followed by mechanical denudation [33]. It was conducted using microscope eyepieces and repetitive manipulation of COC in various media without the use of enzymes. This process involved the use of either mouth-controlled or hand-controlled pipettes to expose these oocytes. This process is technically challenging during manipulation because small oocytes within the separate CCs are difficult to identify, resulting in a laborious and demanding task. It is also often reported to vary reproducibility and inconsistency among the operators, and the yield rate and denudation efficiency vary significantly [34].
Thus, in collecting CCs for transcriptomic study, we found that the use of hyaluronidase impairs the end results of RNA yielding compared with mechanical denudation. The effect of these enzymes dissociated the CC composition by breaking the HA bonding in between CCs themselves, leading to over-destruction, thereby decreasing RNA concentration in our study. However, collecting the CCs using mechanical equipment is also laborious. Therefore, we opted to collect the CCs prior to hyaluronidase use via modification of the standard steps of denudation. Our study used the thinnest needle (22G) to enhance mechanical force in separating the CCs from COC to obtain the optimum size of CCs prior to performing the two standard steps of denudation. Hence, the CCs were not exposed to hyaluronidase. Nevertheless, the comparison with and without hyaluronidase significantly concurs with the potential outcome for future recommendations. Therefore, the non-enzymatic group offers higher RNA concentration yield.
The rotor–stator homogenizer [20] and Biomasher III were compared to identify the best method for CC disruption and homogenization. The comparative results show that the Biomasher III significantly improved RNA concentration and purity of the CCs compared with the rotor–stator homogenizer. Incomplete disruption and homogenization can stem from several factors and display cascading effects on downstream analyses and experimental outcomes, including gene expression profiling, given that specific RNA transcripts may be overrepresented or underrepresented due to incomplete disruption [35].
Nevertheless, the partial disruption and homogenization may result from cell and tissue types exhibiting varying degrees of resistance to these processes, influenced by differences in cellular structure and composition [36]. Stiffer tissues or samples with high connective tissue content, such as cartilage or fibrous tissues, are difficult to homogenize effectively [37,38]. Thus, the selection of homogenization methods, such as mechanical disruption, enzymatic digestion, or bead-based methods, can impact the effectiveness of cell or tissue disruption. Complete homogenization may occur if the selected method is optimized for the specific sample type [39].
The use of a homogenizer might raise the temperature of the RNA samples due to the high-frequency vibration, which could eventually lead to sample degradation. Inefficient homogenization may cause the release of endogenous ribonucleases, leading to RNA degradation [40]. Subsequently, these enzymatic activities reduce RNA integrity and compromise downstream applications, such as gene expression analysis [16,41]. Thus, identifying the best homogenizing method for different cell types is crucial. Apart from producing a complete homogenized sample, the use of BioMasher III has been reported to be efficient in processing a high number of samples by shortening the RNA extraction time through one-pot procedure, providing a DNase- and RNase-free condition, preventing cross-contamination, and offering safety [42]. We found that the presence of filter column in BioMasher III improves the purity of the RNA by effectively removing RNA stabilizer and reducing CC loss during cell transfer for the disruption and homogenization process in the RLT buffer. Therefore, despite obtaining RNA yield from the RSH group, potential degradation prevents the amplification of HKG even when using the QuantiTect Reverse Transcription Kit compared with the BioMasher III with QuantiTect Reverse Transcription Kit for cDNA synthesis.
Generally, various methods for RNA extraction from oocytes have been proposed for gene expression profiling studies in this context [43,44]. Most experiments employed the TRIzol method with some modifications and microRNA extraction kits as efficient methods to extract high-quality total RNA from oocytes [45,46]. However, without the column-based system, the TRIzol method demands experience and skills in separating the RNA. Furthermore, the column-based method is more efficient for many samples. Several studies report on the low RNA concentration and quality of the RNeasy Mini Kit [47,48,49]. The RNeasy micro kit was used for the RNA extraction in this study.
Table 2.
The Characteristics Summary of The Included Studies.
| Author, Year (References) | Title | Country | Sample size (n) | Organism | Method for RNA extraction | RNA concentration (ng μL-1) | RNA Purity |
|---|---|---|---|---|---|---|---|
| Maisarah et al. (2020) [16] | The challenge of getting a high quality of RNA from oocyte for gene expression study | Malaysia | COC (19) Oocyte (400) |
Mouse | TRIzol | COC = 151.0 Oocyte = 126.7 |
COC = 1.7 Oocyte = 1.68 |
| RNeasy Mini Kit | COC = 3.8 Oocyte = 1.9 |
COC = 1.68 Oocyte = 10.5 |
|||||
| Wiweko et al. (2017) [25] | The quality of RNA isolation from frozen granulosa cells | Indonesia | Oocyte (28) | Human | QIAamp RNA Blood Mini Kit | 250 | 1.85 |
| Pavani et al. (2015) [26] | Optimisation of Total Rna Extraction from Bovine Oocytes and Embryos for Gene Expression Studies and Effects of Cryoprotectants on Total Rna Extraction | Portugal | Oocyte (795) | Bovine | TRIzol | 152.8 | 1.5 |
| Guanidinium thiocyanate | 47 | 1.18 | |||||
| Commercial kit | 31.2 | 2.06 |
The RNA carrier effectively improved the RNA yield using the RNeasy micro kit protocol. Several studies have shown that adding RNA carriers increases yields and improves PCR amplification performance [50,51]. When working with small sample sizes, limited cell populations, and precious clinical samples, such as CCs, using RNA carriers can reduce RNA loss during extraction. This approach is especially important for obtaining reliable and accurate downstream results. RNA carrier is also commonly used in viral RNA extraction kits to facilitate and aggregate viral RNA [52]. However, we found that the RNA carrier is incompatible with the whole genome amplification kit given that no PCR amplification was detected in the downstream analysis. Nevertheless, specific amplification was observed for samples that utilized QuantiTect reverse transcription kit for the cDNA synthesis.
Concerning the current RNA extraction techniques for micro-size cells, our review revealed that there is no doubt that the Modified TRIzol Protocol (MTP) is considered the best option for RNA yield from oocytes [16,26]. In contrast, commercial kits are considered acceptable for COCs RNA extraction [16]. Although the RNA concentration is low in most commercial kits, the purity is acceptable. To our knowledge, there is no RNA extraction study for CCs alone. Most RNA extraction for CCs was described briefly in most published papers. Thus, our initial review stage found no solid RNA extraction method for CCs papers. Therefore, our clinical study adds value to the current RNA extraction strategy for human CCs for gene expression study, mainly via commercial kits.
4.1. Strengths
Our study delineated an excellent flow of harvesting the CCs to optimize RNA extraction. Based on various studies, we managed to evaluate the optimum strategy for producing a suitable protocol for RNA extraction in CCs for future reference. Utilizing only mechanical denudation for CC collection, Biomasher III as homogenizer, RNA carrier during RNA extraction and QuantiTect reverse transcription kit for the cDNA synthesis results in sound amplification in gene of interest (GOI). Meanwhile, our systematic review also consolidated the current strategy of RNA extraction method for micro cell RNA – mainly COCs and oocytes.
4.2. Limitations
COCs samples were collected mainly from women with Decreased Ovarian Reserve (DOR). As a result, the limited number of CCs from low oocyte count may affect the RNA yield outcome. Thus, for future study regarding RNA yield should consider comparing among normally ovarian-reserved women, who are postulated to have a better composition of COCs. Alternatively, pooling several samples for high throughput analysis, such as NGS, can increase the required RNA concentration. Otherwise, a small number of papers could be added to our review due to our inclusion and exclusion criteria.
5. Conclusions
Our study revealed that the handling of CCs samples should be done without standard enzymatic denudation. The manual pressure control homogenizer, RNeasy micro kit, and RNA carriers provide a better RNA yield. Our findings suggest a better strategy for optimizing the RNA extraction in CCs for experimental studies. Optimizing sample handling in human CCs is also crucial as the baseline strategy for better RNA yielding to be used as molecular markers in assessing outcomes for transcriptomic studies. Otherwise, our review also concluded that the use of MTP is the best choice for RNA extraction in oocyte sample.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Supplementary Table S1.
Author Contributions
Conceptualization, M.H.E and M.F.A; methodology, M.F.A and M.H.E.; software, A.M.A and N.M; validation, N.S., A.A.Z and A.K.A.K.; formal analysis, S.E.S.; investigation, M.F.A.; resources, M.H.E.; data curation, M.F.A; writing—original draft preparation, M.F.A; writing—review and editing, M.H.E and A.K.A.K.; visualization, N.M.; supervision, A.K.A.K.; project administration, A.A.Z.; funding acquisition, N.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundamental Research Grant Scheme (FRGS), awarded by Ministry of Higher Education of Malaysia with a tittle of “Elucidating the GREM1, HAS2 and PTGS2 gene expression as oocytes development competency markers among women with poor ovarian reserve following the in vitro maturation (IVM)”. The grant number FRGS/1/2021/SKK01/UKM/02/1. The APC was funded by Innovation & Research Secretariat (SPI) of Faculty of Medicine National University of Malaysia.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Human Ethics Committee of Faculty of Medicine UNIVERSITI KEBANGSAAN MALAYSIA (JEP-2022-187 and 31.05.2022).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank the Secretariat of Research and Innovation Faculty of Medicine UKM for the esteemed support of the research. We also acknowledged the contribution from all staffs in Advanced Reproductive Centre (ARC) HCTM UKM Cheras, Kuala Lumpur for the esteemed support toward this research. The illustration for Figure 1 was created using Bio Render with approval for publication obtained - Agreement number: LT266DHJHU.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Carvacho, I.; Piesche, M.; Maier, T.J.; Machaca, K. Ion Channel Function During Oocyte Maturation and Fertilization. Front Cell Dev Biol 2018, 6, 63. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhang, T.; Yang, Y.; Wang, C. Mechanisms of Oocyte Maturation and Related Epigenetic Regulation. Front Cell Dev Biol 2021, 9, 654028. [Google Scholar] [CrossRef] [PubMed]
- Turathum, B.; Gao, E.M.; Chian, R.C. The Function of Cumulus Cells in Oocyte Growth and Maturation and in Subsequent Ovulation and Fertilization. Cells 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wells, D. The human oocyte and cumulus cells relationship: new insights from the cumulus cell transcriptome. Mol Hum Reprod 2010, 16, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Assidi, M.; Richard, F.J.; Sirard, M.A. FSH in vitro versus LH in vivo: similar genomic effects on the cumulus. J Ovarian Res 2013, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- van Gijn, J.; Gijselhart, J.P. [Von Willebrand and his factor]. Ned Tijdschr Geneeskd 2011, 155, A2022. [Google Scholar] [PubMed]
- Zhang, C.H.; Liu, X.Y.; Wang, J. Essential Role of Granulosa Cell Glucose and Lipid Metabolism on Oocytes and the Potential Metabolic Imbalance in Polycystic Ovary Syndrome. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Salimov, D.; Lisovskaya, T.; Otsuki, J.; Gzgzyan, A.; Bogolyubova, I.; Bogolyubov, D. Chromatin Morphology in Human Germinal Vesicle Oocytes and Their Competence to Mature in Stimulated Cycles. Cells 2023, 12, 1976. [Google Scholar] [CrossRef]
- Xie, J.; Xu, X.; Liu, S. Intercellular communication in the cumulus-oocyte complex during folliculogenesis: A review. Front Cell Dev Biol 2023, 11, 1087612. [Google Scholar] [CrossRef]
- Sfakianoudis, K.; Maziotis, E.; Karantzali, E.; Kokkini, G.; Grigoriadis, S.; Pantou, A.; Giannelou, P.; Petroutsou, K.; Markomichali, C.; Fakiridou, M.; et al. Molecular Drivers of Developmental Arrest in the Human Preimplantation Embryo: A Systematic Review and Critical Analysis Leading to Mapping Future Research. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Coticchio, G.; Dal Canto, M.; Mignini Renzini, M.; Guglielmo, M.C.; Brambillasca, F.; Turchi, D.; Novara, P.V.; Fadini, R. Oocyte maturation: gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum Reprod Update 2015, 21, 427–454. [Google Scholar] [CrossRef]
- Nana, M.; Hodson, K.; Lucas, N.; Camporota, L.; Knight, M.; Nelson-Piercy, C. Diagnosis and management of covid-19 in pregnancy. BMJ 2022, 377, e069739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Song, S.; Yang, M.; Yan, L.; Qiao, J. Diminished ovarian reserve causes adverse ART outcomes attributed to effects on oxygen metabolism function in cumulus cells. BMC Genomics 2023, 24, 655. [Google Scholar] [CrossRef]
- Gilchrist, R.B.; Smitz, J. Oocyte in vitro maturation: physiological basis and application to clinical practice. Fertility and sterility 2023, 119, 524–539. [Google Scholar] [CrossRef]
- Ferrari, S.; Lattuada, D.; Paffoni, A.; Brevini, T.A.; Scarduelli, C.; Bolis, G.; Ragni, G.; Gandolfi, F. Procedure for rapid oocyte selection based on quantitative analysis of cumulus cell gene expression. J Assist Reprod Genet 2010, 27, 429–434. [Google Scholar] [CrossRef]
- Maisarah, Y.; Hashida, H.N.; Yusmin, M.Y. The challenge of getting a high quality of RNA from oocyte for gene expression study. Vet Res Forum 2020, 11, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Biase, F.H. Isolation of high-quality total RNA and RNA sequencing of single bovine oocytes. STAR Protoc 2021, 2, 100895. [Google Scholar] [CrossRef]
- Tjahyadi, D.; Susiarno, H.; Chandra, B.A.; Permadi, W.; Djuwantono, T.; Wiweko, B. The effect of oocyte denudation time and intracytoplasmic sperm injection time on embryo quality at assisted reproductive technology clinic - A cross-sectional study. Ann Med Surg (Lond) 2022, 80, 104234. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.C.; Yiap, B.C. DNA, RNA, and protein extraction: the past and the present. J Biomed Biotechnol 2009, 2009, 574398. [Google Scholar] [CrossRef]
- Kaivo-oja, N.; Jeffery, L.A.; Ritvos, O.; Mottershead, D.G. Smad signalling in the ovary. Reprod Biol Endocrinol 2006, 4, 21. [Google Scholar] [CrossRef]
- Maa, Y.-F.; Hsu, C. Liquid-liquid emulsification by rotor/stator homogenization. Journal of Controlled Release 1996, 38, 219–228. [Google Scholar] [CrossRef]
- Iwai, H.; Mori, M.; Tomita, M.; Kono, N.; Arakawa, K. Molecular Evidence of Chemical Disguise by the Socially Parasitic Spiny Ant Polyrhachis lamellidens (Hymenoptera: Formicidae) When Invading a Host Colony. Frontiers in Ecology and Evolution 2022, 10. [Google Scholar] [CrossRef]
- Olson, N.D.; Morrow, J.B. DNA extract characterization process for microbial detection methods development and validation. BMC Res Notes 2012, 5, 668. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Wiweko, B.; Utami, R.; Putri, D.; Muna, N.; Aulia, S.N.; Riayati, O.; Bowolaksono, A. The Quality of RNA Isolation from Frozen Granulosa Cells. Advanced Science Letters 2017, 23, 6717–6719. [Google Scholar] [CrossRef]
- Pavani, K.C.; Baron, E.E.; Faheem, M.; Chaveiro, A.; Da Silva, F.M. Optimisation of Total Rna Extraction from Bovine Oocytes and Embryos for Gene Expression Studies and Effects of Cryoprotectants on Total Rna Extraction. Tsitol Genet 2015, 49, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Moura, B.R.; Gurgel, M.C.; Machado, S.P.; Marques, P.A.; Rolim, J.R.; Lima, M.C.; Salgueiro, L.L. Low concentration of hyaluronidase for oocyte denudation can improve fertilization rates and embryo quality. JBRA Assist Reprod 2017, 21, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, Y.; Takeo, T.; Nakao, S.; Yoshimoto, H.; Hirose, Y.; Sakai, Y.; Horikoshi, Y.; Takeuji, S.; Tsuchiyama, S.; Nakagata, N. Prolonged exposure to hyaluronidase decreases the fertilization and development rates of fresh and cryopreserved mouse oocytes. J Reprod Dev 2014, 60, 454–459. [Google Scholar] [CrossRef]
- Nagyova, E. The Biological Role of Hyaluronan-Rich Oocyte-Cumulus Extracellular Matrix in Female Reproduction. Int J Mol Sci 2018, 19. [Google Scholar] [CrossRef]
- Attanasio, M.; Cirillo, M.; Coccia, M.E.; Castaman, G.; Fatini, C. Factors VIII and Von Willebrand Levels in Women Undergoing Assisted Reproduction: Are Their Levels Associated with Clinical Pregnancy Outcome? Mediterr J Hematol Infect Dis 2020, 12, e2020058. [Google Scholar] [CrossRef] [PubMed]
- Salustri, A.; Camaioni, A.; Tirone, E.; D'Alessandris, C. Hyaluronic acid and proteoglycan accumulation in the cumulus oophorus matrix. Ital J Anat Embryol 1995, 100 Suppl 1, 479–484. [Google Scholar]
- Esbert, M.; Florensa, M.; Riqueros, M.; Teruel, J.; Ballesteros, A. Effect of oocyte denudation timing on clinical outcomes in 1212 oocyte recipients. Fertility and sterility 2013, 100, S482. [Google Scholar] [CrossRef]
- Maldonado Rosas, I.; Anagnostopoulou, C.; Singh, N.; Gugnani, N.; Singh, K.; Desai, D.; Darbandi, M.; Manoharan, M.; Darbandi, S.; Chockalingam, A.; et al. Optimizing embryological aspects of oocyte retrieval, oocyte denudation, and embryo loading for transfer. Panminerva Med 2022, 64, 156–170. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, Y.; Fang, Z.; Hu, S.; Li, Z.; Zhu, L.; Jin, L. Performing ICSI within 4 hours after denudation optimizes clinical outcomes in ICSI cycles. Reprod Biol Endocrinol 2020, 18, 27. [Google Scholar] [CrossRef]
- Nouvel, A.; Laget, J.; Duranton, F.; Leroy, J.; Desmetz, C.; Servais, M.D.; de Preville, N.; Galtier, F.; Nocca, D.; Builles, N.; et al. Optimization of RNA extraction methods from human metabolic tissue samples of the COMET biobank. Sci Rep 2021, 11, 20975. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, S. Mechanical/Physical Methods of Cell Disruption and Tissue Homogenization. Methods Mol Biol 2021, 2261, 563–585. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Attanasio, C.; Pedano, M.S.; Cadenas de Llano-Perula, M. Comparison of human dental tissue RNA extraction methods for RNA sequencing. Arch Oral Biol 2023, 148, 105646. [Google Scholar] [CrossRef]
- Choudhary, S.; Choudhary, R.K. Rapid and Efficient Method of Total RNA Isolation from Milk Fat for Transcriptome Analysis of Mammary Gland. Proceedings of the National Academy of Sciences, India Section B: Biological Sciences 2018, 89, 455–460. [Google Scholar] [CrossRef]
- Ali, N.; Rampazzo, R.C.P.; Costa, A.D.T.; Krieger, M.A. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. Biomed Res Int 2017, 2017, 9306564. [Google Scholar] [CrossRef]
- Amiri Samani, S.; Naji, M.H. Effect of homogenizer pressure and temperature on physicochemical, oxidative stability, viscosity, droplet size, and sensory properties of Sesame vegetable cream. Food Sci Nutr 2019, 7, 899–906. [Google Scholar] [CrossRef]
- Pagani, S.; Maglio, M.; Sicuro, L.; Fini, M.; Giavaresi, G.; Brogini, S. RNA Extraction from Cartilage: Issues, Methods, Tips. Int J Mol Sci 2023, 24, 2120. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nakashima, K.; Maruta, Y.; Kiriyama, T.; Sasaki, M.; Sugiyama, S.; Suzuki, K.; Fujisaki, H.; Sasaki, J.; Kaku-Ushiki, Y.; et al. Improved RNA extraction method using the BioMasher and BioMasher power-plus. J Vet Med Sci 2012, 74, 1561–1567. [Google Scholar] [CrossRef]
- Jones, G.M.; Cram, D.S.; Song, B.; Magli, M.C.; Gianaroli, L.; Lacham-Kaplan, O.; Findlay, J.K.; Jenkin, G.; Trounson, A.O. Gene expression profiling of human oocytes following in vivo or in vitro maturation. Human reproduction (Oxford, England) 2008, 23, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.; Patrizio, P. Gene expression profiling of human oocytes at different maturational stages and after in vitro maturation. Am J Obstet Gynecol 2008, 198, 455–e451. [Google Scholar] [CrossRef]
- Trakunram, K.; Champoochana, N.; Chaniad, P.; Thongsuksai, P.; Raungrut, P. MicroRNA Isolation by Trizol-Based Method and Its Stability in Stored Serum and cDNA Derivatives. Asian Pac J Cancer Prev 2019, 20, 1641–1647. [Google Scholar] [CrossRef]
- Duy, J.; Koehler, J.W.; Honko, A.N.; Minogue, T.D. Optimized microRNA purification from TRIzol-treated plasma. BMC Genomics 2015, 16, 95. [Google Scholar] [CrossRef]
- Beltrame, C.O.; Cortes, M.F.; Bandeira, P.T.; Figueiredo, A.M. Optimization of the RNeasy Mini Kit to obtain high-quality total RNA from sessile cells of Staphylococcus aureus. Braz J Med Biol Res 2015, 48, 1071–1076. [Google Scholar] [CrossRef]
- Tavares, L.; Alves, P.M.; Ferreira, R.B.; Santos, C.N. Comparison of different methods for DNA-free RNA isolation from SK-N-MC neuroblastoma. BMC Res Notes 2011, 4, 3. [Google Scholar] [CrossRef]
- Al-Adsani, A.M.; Barhoush, S.A.; Bastaki, N.K.; Al-Bustan, S.A.; Al-Qattan, K.K. Comparing and Optimizing RNA Extraction from the Pancreas of Diabetic and Healthy Rats for Gene Expression Analyses. Genes (Basel) 2022, 13. [Google Scholar] [CrossRef]
- Ramon-Nunez, L.A.; Martos, L.; Fernandez-Pardo, A.; Oto, J.; Medina, P.; Espana, F.; Navarro, S. Comparison of protocols and RNA carriers for plasma miRNA isolation. Unraveling RNA carrier influence on miRNA isolation. PLoS One 2017, 12, e0187005. [Google Scholar] [CrossRef]
- Wright, K.; de Silva, K.; Purdie, A.C.; Plain, K.M. Comparison of methods for miRNA isolation and quantification from ovine plasma. Sci Rep 2020, 10, 825. [Google Scholar] [CrossRef]
- Ogunbayo, A.E.; Sabiu, S.; Nyaga, M.M. Evaluation of extraction and enrichment methods for recovery of respiratory RNA viruses in a metagenomics approach. J Virol Methods 2023, 314, 114677. [Google Scholar] [CrossRef]
Figure 1.
This is a figure of the study flow.
Figure 2.
This is a figure of oocytes denudation process. A: Cumulus Oocytes Complex prepared for mechanical denudation without enzyme. B: Cumulus Oocytes Complex enzymolyzed in hyaluronidase. C: “Naked” oocytes following enzymolyzed in hyaluronidase – clean edge. D: “Naked” oocytes following mechanical denudation – minimal cumulus cell at the edge.
Figure 2.
This is a figure of oocytes denudation process. A: Cumulus Oocytes Complex prepared for mechanical denudation without enzyme. B: Cumulus Oocytes Complex enzymolyzed in hyaluronidase. C: “Naked” oocytes following enzymolyzed in hyaluronidase – clean edge. D: “Naked” oocytes following mechanical denudation – minimal cumulus cell at the edge.
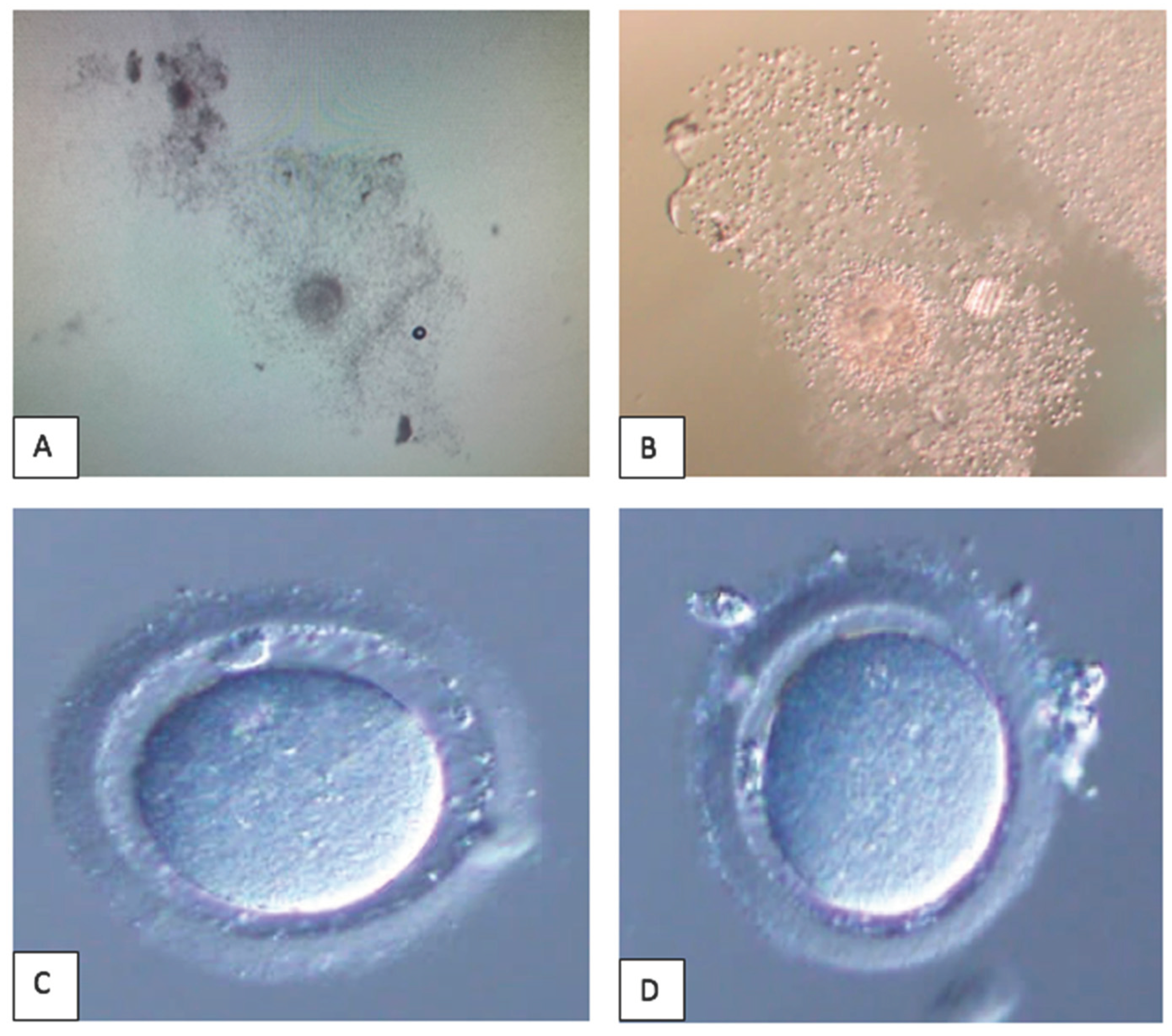
Figure 3.
Prisma Flow Diagram for Systematic Review.
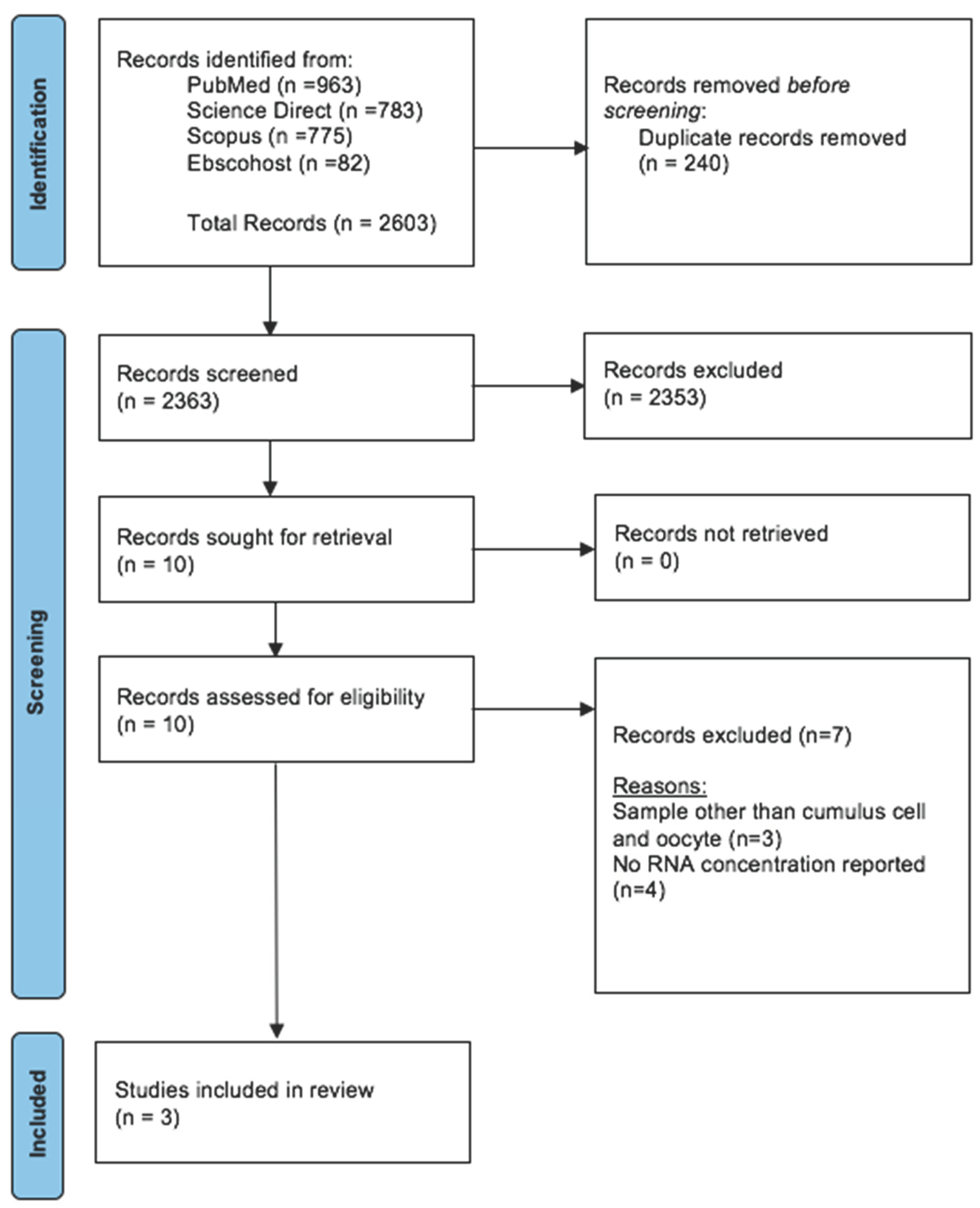
Figure 4.
The HPRT gene amplification with Melt Peak Curve.
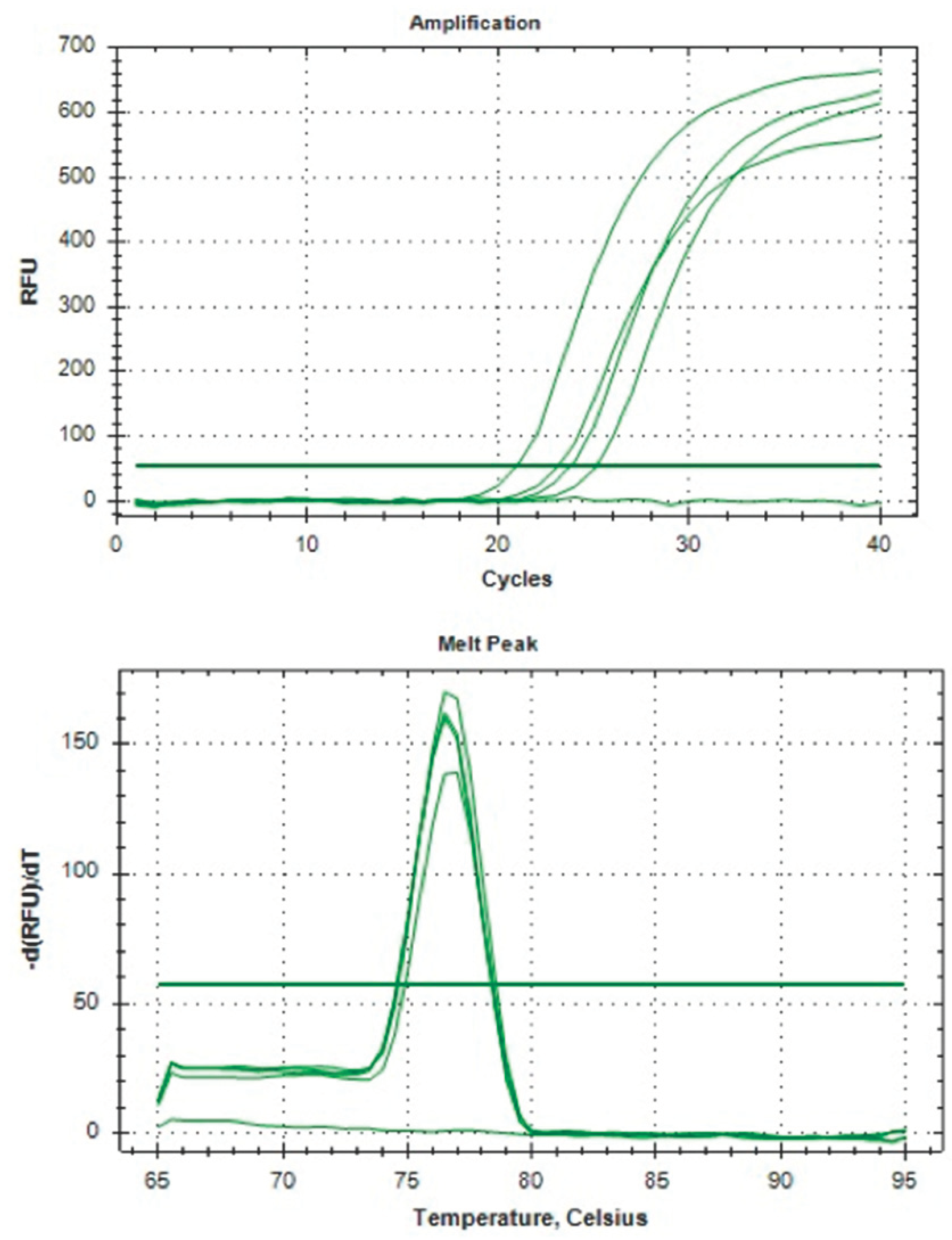
Table 1.
Study Characteristic & Results.
| Parameter | Age (years) |
AMH Level (pmol/L) |
RNA concentration (ng/µl) |
RNA Purification | p- value |
|---|---|---|---|---|---|
| Denudation Technique | |||||
| Recruitment Profile | 37.19 (3.70) | 7.507(4.33) | 6.416(5.15) | 1.712(0.244) | |
| Enzymatic (Hyase) | 36.83 (3.75) | 8.99 (5.35) | 3.83 (1.20) | 1.720 (0.25) | P<0.05* |
| Non-Enzymatic (Non-Hyase) | 37.55 (3.67) | 6.02 (2.17) | 9.00 (6.22) | 1.703 (0.23) | |
| Homogenizer | |||||
| RSH | 37.50 (3.55) | 7.05 (4.05) | 3.68 (1.15) | 1.711(0.23) | P = 0.18* |
| BioMasher III | 36.88 (3.85) | 7.96 (4.59) | 9.15 (6.10) | 1.712 (0.25) | |
| cDNA & qPCR | |||||
| QuantiTect RT | 36.08(3.25) | 7.82(4.12) | 6.82(5.62) | 1.701(0.26) | |
| Whole Genome AK | 38.30 (3.82) | 7.19(4.56) | 6.01(4.68) | 1.722(0.23) | |
| Amplification of HKG | |||||
| Type of cDNA kit | Yes (n, %) |
No (n, %) |
|||
| QuantiTect RT | 20 (50) | 20 (50) | P<0.05# | ||
| Whole Genome AK | 0 | 40 (100) | |||
*Mann–Whitney U test. #Chi-Square test.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Copyright: This open access article is published under a Creative Commons CC BY 4.0 license, which permit the free download, distribution, and reuse, provided that the author and preprint are cited in any reuse.
supplementary.docx (17.65KB )
Submitted:
08 March 2024
Posted:
11 March 2024
You are already at the latest version
Alerts
supplementary.docx (17.65KB )
This version is not peer-reviewed
Submitted:
08 March 2024
Posted:
11 March 2024
You are already at the latest version
Alerts
Abstract
RNA extraction from human cumulus cells (CCs) is considered challenging due to low cell’s pop-ulation and small cell. The successful gene expression analysis from cumulus cells ultimately de-pends on an excellent concentration of Ribonucleic acid (RNA) yield by an appropriate technique. Our study aims to consolidate appropriate cumulus cell collection method, preparation and ex-traction techniques to ensure a good yield of RNA for better gene expression analysis. From the results obtained, the optimal strategy found involved mechanical denudation without an enzy-matic process, followed by the use of RNA stabilizer prior to RNA extraction. Subsequently, uti-lizing a smaller homogenizer, preferably manual pressure control with filter (Bio-Masher III®) is paramount for a pure and homogenize sample. Subsequently, during RNA extraction, the use of RNeasy micro kit and RNA carriers is recommended. These sample collection, preparation, and extraction techniques yielded an excellent-quality RNA concentration and successful gene expres-sion analysis. We also conducted a systematic review to consolidate our findings with current evidence of RNA extraction technique. Our results added a value as a current strategy for opti-mizing RNA extraction in CCs for experimental studies. Thus, our findings can contribute to for-mulating a better strategy for optimizing the RNA extraction in CCs for transcriptomic studies.
Keywords:
Subject: Medicine and Pharmacology - Obstetrics and Gynaecology
1. Introduction
In reproductive biology, the intricate interplay of various cell types orchestrates the complex oocyte maturation process, culminating in successful fertilization and embryogenesis [1,2]. Among these crucial players, cumulus cells (CCs) emerge as indispensable companions to oocytes, providing vital support and regulatory functions throughout the maturation process. The CCs, specialized somatic cells, reside near the oocyte within the ovarian follicles. Their intimate association and intricate communication with the oocyte are essential for guiding and facilitating its developmental journey [3]. The cumulus-oocyte complex forms a dynamic microenvironment, fostering bidirectional signaling that influences oocyte growth, meiotic progression, and eventual ovulation. This proximity enables CCs to exchange nutrients, growth factors, and metabolites with the oocyte, thereby ensuring optimal oocyte maturation [1,4].
At the molecular level, the CCs support the oocyte development closely by secreting Hyaluronic acid (HA) while expanding the follicle-stimulating hormone (FSH) stimulation [5]. The mural granulosa cell (MGCs), which supports the growth of follicles, coordinates the endocrine function externally. The ovulation phase occurs due to increased HA secretion by CCs once the rupture of the dominant follicles remarks a luteinizing hormone [6] surge [7]. Morphologically, CCs exhibit diverse shapes and sizes, ranging from spindle-like to cuboidal. These variations in morphology reflect the dynamic nature of their functions and underline their adaptability to the changing needs of oocyte maturation [4,8]. The cumulus cell layer enveloping the oocyte forms a protective shield and a communication bridge between the oocyte and the surrounding ovarian microenvironment [9]. The number of CCs enveloping an oocyte can differ depending on the follicular development stage. During the early stages of folliculogenesis, the number of CCs is relatively sparse. As the follicle progresses and matures, a corona radiata forms around the oocyte, comprising numerous layers of closely packed CCs. This protective cocoon of CCs serves as a conduit for nutrient exchange, signaling molecules, and regulatory factors that profoundly impact oocyte maturation [2,4,9].
The size and morphology of CCs exhibit remarkable diversity, reflecting the complex interplay of various intrinsic and extrinsic factors. Several factors contribute to the variability in CC size [10]. Apart from the follicular stage, the number, size, and morphology of the CCs are influenced by hormone levels, such as FSH and LH, metabolic status, ovarian microenvironment including oxygen tension and local growth factors, genetic variability, and aging [11]. Minimal CCs are observed in cases of low oocyte quality, particularly in women with poor ovarian reserve [12] and in the elderly [13]. Due to its minimal CCs, the challenges in obtaining a good concentration of RNA are even higher.
The cumulative evidence has highlighted the pivotal role of CCs as invaluable indicators of oocyte quality. The molecular and biochemical composition of CCs can provide insightful information about the developmental competence of the oocyte and subsequent embryo quality [14]. CCs are endowed with the potential to harbor critical molecular biomarkers that offer predictive insights into the likelihood of successful fertilization and embryo implantation. This burgeoning field holds promise for revolutionizing assisted reproductive technologies and infertility treatments by allowing more informed decision-making regarding oocyte selection.
Most evidence utilizes the CC’s ribonucleic acid (RNA) to study the critical mechanism of CC’s in oocyte development and maturation. For these matters, the RNA extraction steps are crucial. It required a proper purification of RNA from the CC’s tissue samples. Nevertheless, ribonuclease enzymes in the tissues can complicate it, leading to rapid degradation of RNA material upon extraction. To date, various methods are used in molecular biology to isolate RNA from CCs. The most commonly reported is the guanidinium thiocyanate-phenol-chloroform extraction. It consists of the filter paper-based lysis followed by the elution method, which features high throughput capacity. Despite all the reported RNA extraction methods in CCs, given their minute size and scarcity, they still pose significant challenges to effective RNA extraction. The ranges of RNA concentration is still below 10; ranging from 4 - 7 ng/µl as reported by previous study [15].
Nevertheless, the limited number of CCs obtained from a single follicle necessitates meticulous optimization of RNA extraction protocols to ensure sufficient and high-quality RNA [16,17]. The significance of RNA extraction in CCs must be considered. However, despite its importance, limited published literature suggests the optimal method for achieving a pure final product. Therefore, it is imperative to conduct further research and experimentation to uncover the best approach for extracting RNA in CCs. A notable gap exists in the current shreds of evidence regarding the detailed methodology for RNA extraction from CCs. While numerous original articles delve into the molecular insights gleaned from CCs, the intricacies of RNA extraction should be addressed. Our study addresses a significant gap in gene expression studies in CCs. We aim to provide a comprehensive optimization strategy that covers all aspects of sample collection, preparation, RNA extraction, and purification. By consolidating our findings with current literature, we will offer valuable insights to help researchers prepare high-quality RNA for their studies. Our research will pave the way for more accurate and reliable gene expression studies in the future.
2. Materials and Methods
2.1. Clinical Study
2.1.1. Study Design, Recruitment, and Ethical Approval
The prospective cohort study was conducted in the Advanced Reproductive Center, Hospital Canselor Tuanku Mukhriz (HCTM) UKM Cheras, Kuala Lumpur, from June 2022 to December 2022. This research was approved by the Human Ethical Research Committee (JEP-2022-187), Faculty of Medicine, National University of Malaysia Cheras, Kuala Lumpur, Malaysia. Consent was obtained prior to CC collection during the oocyte retrieval (OR) procedure for in-vitro fertilization (IVF). The CCs were discarded after separation from surrounding oocytes – “denudation” before the IVF procedure. Thus, obtaining these samples does not interfere with the IVF treatment. All women who underwent OR during the study period were included regardless of the causes of the infertility. A total of 80 samples of CCs from 80 follicles to extract RNA and optimize the housekeeping gene (HKG) hypoxanthine–guanine phosphoribosyl transferase (HPRT) amplification was collected and stored in an Eppendorf tube size 1.5 mL filled with 500 µL RNALater® solution (Thermo Fisher USA).
2.1.2. RNA Extraction Method Optimization
For the purpose of our study, the RNA extraction method optimization was divided into three stages:
- A.
- Sample collection optimization
- A.
- B. Sample preparation optimization
- A.
- C. RNA purification optimization
2.1.2.1. Sample Collection Optimization
The oocyte denudation (OD) process was the initial step of CC sample collection. To date, most centers implement the enzymatic protocol for oocyte denudation [18]. In brief, OD was conducted in two phases. Initially, the fresh retrieval oocytes were enzymolyzed in hyaluronidase – hyase-10x TM (Vitrolife® Sweden) to dissolve the bonds between CCs within 40 seconds; then, CCs were further removed from oocytes mechanically by repetitive pipette aspiration and deposition in a series of media without an enzyme. Finally, the “naked” – clean oocytes were examined for their maturity, and only meiosis II stage oocytes (MII) with the presence of polar body were used for IVF procedure either via intra-cytoplasmic sperm injection (ICSI) or conventional IVF. The comparison of Cumulus-Oocytes-Complex (COCs) features with and without hyaluronidase and post-denudation of both techniques were showed in Figure 2. For this study, 40 CCs were collected via conventional method using enzymatic protocol (Group A), whereas 40 other CCs were collected without enzymatic protocol (Group B) – purely mechanical using only manual force created via repetitive pipette aspiration and deposition in a series of media without an enzyme to obtain naked oocytes. The study flow is illustrated in Figure 1.
2.1.2.2 Sample Preparation Optimization
For sample preparation, CCs were lysed to form smaller molecules to achieve excellent RNA extraction [19]. This step required a mixer of RNA carrier solution (QIAamp® Circulating Nucleic Acid Kit) and diluted with buffer RLT, forming the RNA carrier dilution (RCD). The RLT buffer is a lysis buffer used to lyse CCs prior to RNA isolation. All the CC samples were prepared with RCD. In addition, the homogenizers were used as a mechanical disruption agent for CC preparation. This approach helps in forming an even mixture by forcing the CCs through the narrow space via multiple forces and turbulence to distribute the CC molecules as a solution evenly. In our study, we used two types of homogenizers, standard rotor–stator homogenizer [20] or disposable easy grind homogenizer, BioMasher III (Funakoshi®,Japan). In summary, the standard RSH consists of a fast-spinning inner rotor with a stationary outer sheath–stator. It homogenizes CCs through mechanical tearing and shear fluid forces and mixing the CC samples in Eppendorf tubes [21]. The BioMasher III (Funakoshi®, Japan) is formed by a filter tube with an abrasive surface inner wall and combined with a pestle with a textured surface. The pestle in the filtered tube layered on to centrifuge the tube is used for manual grinding [22]. Then, the CC-extracted samples were filtered and placed in the recovery tube. For these steps, every 20 CCs from Groups A and B were homogenized using a rotor–stator homogenizer, and the 20 other CCs from the same groups were manually ground via BioMasher III.
- Rotor–stator homogenizer [20].
Subsequently, the collected CCs were transferred into an empty 1.5 ml centrifuge tube using a sterile pipette tip. Then, 100 µl of RCD was added to the microcentrifuge. Rotor–stator homogenizer was used to disrupt and homogenize the samples. Another 100 µl of RCD was added and centrifuged at 10,000 xg for 30 seconds. Thereafter, the “pass through” homogenized samples were collected for RNA purification.
- 2.
- BioMasher III
The collected CCs, together with their RNA stabilizer, were transferred into the Biomasher III filter column. The CCs were separated from RNA stabilizer through 2000 x g centrifugation for 10 seconds. The filtrate that contained RNA stablizer was subsequently discarded. Then, 100 µl of RCD was added to the filter column. The CCs were ground using the pastel provided. Another 100 µl of RCD was added, and the disrupted samples were homogenized through the filter by centrifugation at 10000 x g for 30 seconds. Subsequently, the “pass through” homogenized samples were collected for RNA purification.
2.1.2.3. RNA Purification Optimization
The RNA from all samples was purified using an RNAeasy® micro kit (Qiagen) with few modifications. In brief, post-homogenized CC samples were collected in a 1.5 mL tube and added with 200 µl of 70% ethanol. Subsequently, the solution was transferred into an RNAeasy column within a 2 mL tube and centrifuged at 8,000 xg for 15 seconds. The flowthrough was then discarded. The 350 mL buffer RW1 was added to the RNAeasy column and centrifuged at 8,000 xg for 15 seconds, and then the flowthrough was discarded. Thereafter, 80 microL of DNAse I dilution was added directly into the spin column RNAeasy. The solution was left to rest on a bench for 15 minutes at room temperature. Then, the 350 µl buffer RW1 was added to the RNAeasy column and centrifuged at 8,000 xg for 15 seconds, and the 2 mL collecting tube (CT) with flowthrough was discarded and replaced with a new 2 mL CT at the RNAeasy column. The 500 µl buffer RPE was added to the RNAeasy column and centrifuged at 8,000 xg for 2 minutes. Subsequently, the CT was discarded. The 500 µl of 80% ethanol was not added in the further modification. Subsequently, the new 2 mL CT was added to the RNAeasy column, with the lid open, centrifuged at full speed for 5 minutes to dry the membrane, and the flowthrough was discarded. Finally, the new 1.5 mL CT was placed at the RNAeasy column, and 14 µL RNAse free water was added at the direct center of the RNAeasy column and set for 5 minutes. The lid was closed gently, followed by centrifugation, which was performed at full speed for one minute to elucidate the final RNA.
2.1.2. RNA Concentration and Purity Measurement
The concentration and purity of the purified RNAs were measured using a Nanodrop. The purity of the samples was evaluated by measuring the ratio of absorbance readings at 260 nm (specific for nucleic acids). Samples with A260/A280 ratios between 1.80 and 2.10 were considered to have no significant RNA contamination, whereas the ratios of A260/A230 were used as a secondary measure of nucleic acid purity and was expected within the range of 2.0–2.2 [23].
2.1.3. cDNA Synthesis and qPCR
Following RNA purification, the cDNA synthesis was conducted accordingly. We considered two types of cDNA synthesis kits, namely, the QuantiTect reverse transcription kit (Qiagen) and the whole genome amplification kit (Qiagen) given that the CC RNA purity was assumed to be smaller than extensive samples/tissues. The cDNA synthesis was performed according to the standard protocol of both kits. Then, the quality of each cDNA synthesized was validated by qPCR amplification. A housekeeping gene (HKG), the hypoxanthine-guanine phosphoribosyl transferase (HPRT), was used to amplify the cDNA. The primer sequence used; Hs_HPRT1_1_SG. QuantiTect Primer Assay (Qiagen®) was used for the qPCR amplification according to the protocol. In brief, genomic DNA elimination was prepared on ice with total volume of 14 µl. Subsequently incubated for 2 minutes in 42°C. The master mix solution also prepared according to protocol. In brief, the master mix final volume is 20 µl including 14 µl from template RNA. Subsequently incubated for 15 minutes in 42°C and then incubated for 3 minutes in 95°C to inactivate the Quantiscript Reverse Transcriptase.
2.2. The Systematic Review
The systematic review was done following the standard guideline protocol based on the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) [24] to identify the best RNA extraction method for cumulus cells.
2.2.1. Information Source and Search Strategy
The PRISMA guideline was used in conducting the systematic review. A thorough search of the literature was conducted with EBSCOhost, PubMed, Science Direct, and Scopus. Relevant research articles that were released up until 1st March 2024, were located. Keywords such as "cumulus cells" and "oocytes" were taken from the Medical Subject Heading (MeSH). MeSH terms from the Cochrane Library were used to generate synonyms for keywords. Through the evaluation of gathered review articles, further text terms were discovered. The following keywords were applied as part of the search strategy: ("cumulus cell" OR "granulosa cell," OR “oocytes” OR “cumulus oocyte complex”) AND (“RNA extraction” OR “RNA purification” OR “RNA isolation”).
2.2.1.1. Inclusion & Exclusion Criteria
Case-control and cross-sectional studies with abstracts investigating the methods used for RNA extraction from cumulus cells, cumulus oocyte complex or oocytes. Only human and animal studies reporting on the RNA concentration and RNA purity of the extracted RNA were included to ensure the homogeneity of the data. In contrast, the editorials, case reports, conference proceedings, and narrative review articles were not included in this review since they had no primary data. Excluded studies included in silico studies, in vitro studies and any intervention studies. Studies that extract DNA from cumulus or oocytes were excluded.
2.2.1.2. Screening of Articles for Eligibility
The articles that were collected from all databases underwent three stages of screening. In the first step, duplicates were removed and all articles with irrelevant titles were excluded. In the second phase, the abstracts of the remaining papers were reviewed and those that did not fit the inclusion criteria were eliminated. Lastly, a thorough review of the full text of remaining articles was conducted. In the stage, every article that did not adhere to the requirements for inclusion was eliminated. All authors involved in the phases of screening, choosing, and extracting data. The PRISMA flow diagram that summarises the article assortment procedure and the grounds for item removal is displayed in Figure 3.
2.2.2. Study Selection, Data Extraction and Risk of Bias Assessment
Based on an initial search, five authors (A.M.F, A.M.A, M.H.I, M.J.N., and A.K.A.K.) screened all titles and abstracts of potential manuscripts. The selection criteria included manuscripts published in English from January 2012 to December 2022, evaluating the RNA extraction method for CCs and oocytes. Following the initial screening - title, and abstract, full-text screening was conducted, excluding manuscripts that used samples other than cumulus cells and oocytes as experimental material, non-English language, case reports, and review articles. The remaining potential manuscripts were then independently reviewed. The final selected manuscripts provided a detailed study design, focusing on RNA extraction methods with the final report of RNA concentration from each method. The conflicts in selection among authors were resolved through detailed discussions and opinions provided by the fifth, sixth, and seventh authors (S.S.E, N.S., and A.A.Z). Additionally, the National Institutes of Health (N.I.H.) tool for observational studies was employed to assess the quality of the selected manuscripts. This evaluation was based on 14 variables, scoring 1 for 'yes,' 0 for 'no,' or 'non-applicable' for N.A. The manuscripts were then categorized as poor (0–5), fair (6–9), or good (10–14) based on their total scores (Supplementary Table S1). Overall, the included studies in our review achieved a minimum fair to good score. Subsequently, the information gathered is as follows: (1) author name (year); (2) article title; (3) country (4) sample size; (5) organism; (6) RNA extraction methods; (7) RNA concentration; and (8) RNA purity. The summary of the collected data is listed in Table 2.
3. Results
3.1. Clinical Study
We collected 80 samples of CCs from 80 follicles to extract RNA and optimize the housekeeping gene (HKG) hypoxanthine–guanine phosphoribosyl transferase (HPRT) amplification. The samples were prepared and analyzed according to the steps outlined in Figure 1. Our women subjects had an average age of 37.19 ± 3.70, which was considered advanced, and their anti-Mullerian hormone (AMH) level was less than 10 picomol/L (7.507 ± 4.33), indicating a low ovarian reserve. After the extraction, we measured RNA quantity and quality for all method combinations. The RNA concentration was generally low, below 10 ng/μl (6.416 ± 5.15), and had an average purity of 1.712 ± 0.244 during RNA purification. However, non-enzymatic handling of CCs resulted in significantly higher RNA concentration at 9.00 ng/μl compared with 3.83 ng/μl (p<0.05). Combining BioMasher III with RNA carrier dilution (RCD) yielded better RNA concentration than the RSH tool at 9.15 ng/μl versus 3.68 ng/μl, although not statistically significant (p=0.18). Our final cDNA and qPCR analysis showed a significant presence of HPRT gene amplification following the use of QuantiTect reverse transcriptase kit (Qiagen) for cDNA synthesis in the BioMasher III and non-enzymatic group compared with another group (p<0.05). The HPRT gene amplification graph and melt peak curve is showed in Figure 4. Unfortunately, the group using whole genome amplification kits for cDNA synthesis with RNA carrier dilution (RCD) and RSH tool with QuantiTect Reverse Transcriptase Kit (Qiagen) for cDNA showed no amplification for their qPCR. All the results are tabulated in Table 1.
3.2. Systematic Review
To date, no available data has been reported that exclusively focuses on the RNA extraction method for cumulus cells (CCs) alone. Most RNA extraction methods have been briefly described in in-vitro studies, intervention, and silico studies, which were excluded from this review. Currently, most evidence for the technique combined CCs with oocytes or Cumulus-Oocytes Complex (COCs). Hence, in our search strategy, initially, no paper yielded only the RNA method for CCs. As a result, we modified the search strategy to include CCs, oocytes, and COCs.
3.2.1. Search Sequence and Quality Assessment
A total of 2603 studies were retrieved during the primary search (Figure 3). After removing 240 duplicates, the remaining 2363 articles were thoroughly screened based on our inclusion criteria. Amongst them, 2,353 articles were excluded, leaving 10 for full-text evaluation. After a detailed evaluation, seven articles were discarded, 3 using samples other than CCs and oocytes, and four papers – no RNA final concentration was reported. Subsequently, three studies that focused on RNA extraction methods with available results of final RNA concentrations were selected for this review. All selected articles were evaluated using the National Institutes of Health (NIH) tool for observational studies to ensure quality and minimize bias. Notably, all four articles obtained a minimum fair to good score, indicating a low risk of bias (Supplementary Table S1).
3.2.2. Studies Characteristics
A total of 1242 samples from three studies were included in this systematic review. All studies collected oocyte for RNA extraction optimization study. Maisarah et al. (2020) include 19 samples of cumulus oocyte complex for RNA extraction [16]. Wiweko et al. (2017) used human samples while Maisarah et al., (2020) and Pavani et al., (2015) were on animal study [16,25,26]. The RNA extraction methods reported included for comparison TRIzol, Guanidium thiocyanate and few types of column based commercial kits. All the relevant information from the included studies has been summarized in Table 2
3.2.3. Main Outcome
Most studies use pool of oocytes and COCs, ranging from 70 to 200 oocytes per pool, to yield the RNA and conclude that the RNA purity is good (1.0 to 2.0) using any preferred methods [16,25,26]. The two TRIzol protocol studies reported a higher RNA concentration with at least 150 ng/ μL than commercial kits [16,26]. Similarly, these two studies compared Modified TRIzol Protocol (MTP) with commercial kits and revealed that MTP is considered the best option for RNA extraction in oocytes[16,26]. In contrast, one study concluded that the yield of RNA using commercial kits is comparable in fresh versus frozen oocytes [25]. Unfortunately, none of these reported for RNA extraction for CCs alone.
4. Discussion
The high RNA concentration and purification are essential in molecular research, specifically in transcriptomic study. However, achieving excellent RNA concentration and purity could be challenging, especially for tiny samples, e.g. CCs [16,22]. Thus, evaluating an appropriate technique as an optimal strategy is crucial to improve the yield of RNA in these sample types. The comparison of multiple methods indicates that the combination of mechanical collection technique, Biomasher III, and RNA carrier improved the RNA concentration and purity obtained from the CCs.
The current practice for oocyte denudation in most in-vitro fertilization (IVF) centers worldwide is via the enzymatic technique, namely, hyaluronidase [27,28]. HYASE-10X™ (Vitrolife® Sweden) is often used to remove CCs from oocytes prior to intracytoplasmic sperm injection (ICSI) in our center. The maturation process in oocytes consists of the accumulation of HA in CCs as a protection mechanism. It is a high-molecular-weight glycosaminoglycan form with an alternate bond of D-glucuronic and N-acetylglucosamine [29]. The HA mainly accumulates within the CC oophorus to support the oocyte developing. These strong attachments of CCs and oocytes form a cumulus–oocytes complex [30], facilitating the supply of nutrients and growth factors for further enhancement and maturation [29,31].
Therefore, separating the CCs from COC for ICSI preparation is technically challenging. To date, the denudation process is conducted in two phases [18,32]. The first step was that the COCs would be enzymolyzed in hyaluronidase, such as HYASE-10X™ (Vitrolife® Sweden), to weaken the bond of HA within the CCs and oocytes. The process is performed rapidly because potential decremental effects can occur if the process takes more than 40 seconds due to enzyme toxicity. Subsequently, the second phase is followed by mechanical denudation [33]. It was conducted using microscope eyepieces and repetitive manipulation of COC in various media without the use of enzymes. This process involved the use of either mouth-controlled or hand-controlled pipettes to expose these oocytes. This process is technically challenging during manipulation because small oocytes within the separate CCs are difficult to identify, resulting in a laborious and demanding task. It is also often reported to vary reproducibility and inconsistency among the operators, and the yield rate and denudation efficiency vary significantly [34].
Thus, in collecting CCs for transcriptomic study, we found that the use of hyaluronidase impairs the end results of RNA yielding compared with mechanical denudation. The effect of these enzymes dissociated the CC composition by breaking the HA bonding in between CCs themselves, leading to over-destruction, thereby decreasing RNA concentration in our study. However, collecting the CCs using mechanical equipment is also laborious. Therefore, we opted to collect the CCs prior to hyaluronidase use via modification of the standard steps of denudation. Our study used the thinnest needle (22G) to enhance mechanical force in separating the CCs from COC to obtain the optimum size of CCs prior to performing the two standard steps of denudation. Hence, the CCs were not exposed to hyaluronidase. Nevertheless, the comparison with and without hyaluronidase significantly concurs with the potential outcome for future recommendations. Therefore, the non-enzymatic group offers higher RNA concentration yield.
The rotor–stator homogenizer [20] and Biomasher III were compared to identify the best method for CC disruption and homogenization. The comparative results show that the Biomasher III significantly improved RNA concentration and purity of the CCs compared with the rotor–stator homogenizer. Incomplete disruption and homogenization can stem from several factors and display cascading effects on downstream analyses and experimental outcomes, including gene expression profiling, given that specific RNA transcripts may be overrepresented or underrepresented due to incomplete disruption [35].
Nevertheless, the partial disruption and homogenization may result from cell and tissue types exhibiting varying degrees of resistance to these processes, influenced by differences in cellular structure and composition [36]. Stiffer tissues or samples with high connective tissue content, such as cartilage or fibrous tissues, are difficult to homogenize effectively [37,38]. Thus, the selection of homogenization methods, such as mechanical disruption, enzymatic digestion, or bead-based methods, can impact the effectiveness of cell or tissue disruption. Complete homogenization may occur if the selected method is optimized for the specific sample type [39].
The use of a homogenizer might raise the temperature of the RNA samples due to the high-frequency vibration, which could eventually lead to sample degradation. Inefficient homogenization may cause the release of endogenous ribonucleases, leading to RNA degradation [40]. Subsequently, these enzymatic activities reduce RNA integrity and compromise downstream applications, such as gene expression analysis [16,41]. Thus, identifying the best homogenizing method for different cell types is crucial. Apart from producing a complete homogenized sample, the use of BioMasher III has been reported to be efficient in processing a high number of samples by shortening the RNA extraction time through one-pot procedure, providing a DNase- and RNase-free condition, preventing cross-contamination, and offering safety [42]. We found that the presence of filter column in BioMasher III improves the purity of the RNA by effectively removing RNA stabilizer and reducing CC loss during cell transfer for the disruption and homogenization process in the RLT buffer. Therefore, despite obtaining RNA yield from the RSH group, potential degradation prevents the amplification of HKG even when using the QuantiTect Reverse Transcription Kit compared with the BioMasher III with QuantiTect Reverse Transcription Kit for cDNA synthesis.
Generally, various methods for RNA extraction from oocytes have been proposed for gene expression profiling studies in this context [43,44]. Most experiments employed the TRIzol method with some modifications and microRNA extraction kits as efficient methods to extract high-quality total RNA from oocytes [45,46]. However, without the column-based system, the TRIzol method demands experience and skills in separating the RNA. Furthermore, the column-based method is more efficient for many samples. Several studies report on the low RNA concentration and quality of the RNeasy Mini Kit [47,48,49]. The RNeasy micro kit was used for the RNA extraction in this study.
Table 2.
The Characteristics Summary of The Included Studies.
| Author, Year (References) | Title | Country | Sample size (n) | Organism | Method for RNA extraction | RNA concentration (ng μL-1) | RNA Purity |
|---|---|---|---|---|---|---|---|
| Maisarah et al. (2020) [16] | The challenge of getting a high quality of RNA from oocyte for gene expression study | Malaysia | COC (19) Oocyte (400) |
Mouse | TRIzol | COC = 151.0 Oocyte = 126.7 |
COC = 1.7 Oocyte = 1.68 |
| RNeasy Mini Kit | COC = 3.8 Oocyte = 1.9 |
COC = 1.68 Oocyte = 10.5 |
|||||
| Wiweko et al. (2017) [25] | The quality of RNA isolation from frozen granulosa cells | Indonesia | Oocyte (28) | Human | QIAamp RNA Blood Mini Kit | 250 | 1.85 |
| Pavani et al. (2015) [26] | Optimisation of Total Rna Extraction from Bovine Oocytes and Embryos for Gene Expression Studies and Effects of Cryoprotectants on Total Rna Extraction | Portugal | Oocyte (795) | Bovine | TRIzol | 152.8 | 1.5 |
| Guanidinium thiocyanate | 47 | 1.18 | |||||
| Commercial kit | 31.2 | 2.06 |
The RNA carrier effectively improved the RNA yield using the RNeasy micro kit protocol. Several studies have shown that adding RNA carriers increases yields and improves PCR amplification performance [50,51]. When working with small sample sizes, limited cell populations, and precious clinical samples, such as CCs, using RNA carriers can reduce RNA loss during extraction. This approach is especially important for obtaining reliable and accurate downstream results. RNA carrier is also commonly used in viral RNA extraction kits to facilitate and aggregate viral RNA [52]. However, we found that the RNA carrier is incompatible with the whole genome amplification kit given that no PCR amplification was detected in the downstream analysis. Nevertheless, specific amplification was observed for samples that utilized QuantiTect reverse transcription kit for the cDNA synthesis.
Concerning the current RNA extraction techniques for micro-size cells, our review revealed that there is no doubt that the Modified TRIzol Protocol (MTP) is considered the best option for RNA yield from oocytes [16,26]. In contrast, commercial kits are considered acceptable for COCs RNA extraction [16]. Although the RNA concentration is low in most commercial kits, the purity is acceptable. To our knowledge, there is no RNA extraction study for CCs alone. Most RNA extraction for CCs was described briefly in most published papers. Thus, our initial review stage found no solid RNA extraction method for CCs papers. Therefore, our clinical study adds value to the current RNA extraction strategy for human CCs for gene expression study, mainly via commercial kits.
4.1. Strengths
Our study delineated an excellent flow of harvesting the CCs to optimize RNA extraction. Based on various studies, we managed to evaluate the optimum strategy for producing a suitable protocol for RNA extraction in CCs for future reference. Utilizing only mechanical denudation for CC collection, Biomasher III as homogenizer, RNA carrier during RNA extraction and QuantiTect reverse transcription kit for the cDNA synthesis results in sound amplification in gene of interest (GOI). Meanwhile, our systematic review also consolidated the current strategy of RNA extraction method for micro cell RNA – mainly COCs and oocytes.
4.2. Limitations
COCs samples were collected mainly from women with Decreased Ovarian Reserve (DOR). As a result, the limited number of CCs from low oocyte count may affect the RNA yield outcome. Thus, for future study regarding RNA yield should consider comparing among normally ovarian-reserved women, who are postulated to have a better composition of COCs. Alternatively, pooling several samples for high throughput analysis, such as NGS, can increase the required RNA concentration. Otherwise, a small number of papers could be added to our review due to our inclusion and exclusion criteria.
5. Conclusions
Our study revealed that the handling of CCs samples should be done without standard enzymatic denudation. The manual pressure control homogenizer, RNeasy micro kit, and RNA carriers provide a better RNA yield. Our findings suggest a better strategy for optimizing the RNA extraction in CCs for experimental studies. Optimizing sample handling in human CCs is also crucial as the baseline strategy for better RNA yielding to be used as molecular markers in assessing outcomes for transcriptomic studies. Otherwise, our review also concluded that the use of MTP is the best choice for RNA extraction in oocyte sample.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Supplementary Table S1.
Author Contributions
Conceptualization, M.H.E and M.F.A; methodology, M.F.A and M.H.E.; software, A.M.A and N.M; validation, N.S., A.A.Z and A.K.A.K.; formal analysis, S.E.S.; investigation, M.F.A.; resources, M.H.E.; data curation, M.F.A; writing—original draft preparation, M.F.A; writing—review and editing, M.H.E and A.K.A.K.; visualization, N.M.; supervision, A.K.A.K.; project administration, A.A.Z.; funding acquisition, N.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundamental Research Grant Scheme (FRGS), awarded by Ministry of Higher Education of Malaysia with a tittle of “Elucidating the GREM1, HAS2 and PTGS2 gene expression as oocytes development competency markers among women with poor ovarian reserve following the in vitro maturation (IVM)”. The grant number FRGS/1/2021/SKK01/UKM/02/1. The APC was funded by Innovation & Research Secretariat (SPI) of Faculty of Medicine National University of Malaysia.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Human Ethics Committee of Faculty of Medicine UNIVERSITI KEBANGSAAN MALAYSIA (JEP-2022-187 and 31.05.2022).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank the Secretariat of Research and Innovation Faculty of Medicine UKM for the esteemed support of the research. We also acknowledged the contribution from all staffs in Advanced Reproductive Centre (ARC) HCTM UKM Cheras, Kuala Lumpur for the esteemed support toward this research. The illustration for Figure 1 was created using Bio Render with approval for publication obtained - Agreement number: LT266DHJHU.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Carvacho, I.; Piesche, M.; Maier, T.J.; Machaca, K. Ion Channel Function During Oocyte Maturation and Fertilization. Front Cell Dev Biol 2018, 6, 63. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhang, T.; Yang, Y.; Wang, C. Mechanisms of Oocyte Maturation and Related Epigenetic Regulation. Front Cell Dev Biol 2021, 9, 654028. [Google Scholar] [CrossRef] [PubMed]
- Turathum, B.; Gao, E.M.; Chian, R.C. The Function of Cumulus Cells in Oocyte Growth and Maturation and in Subsequent Ovulation and Fertilization. Cells 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wells, D. The human oocyte and cumulus cells relationship: new insights from the cumulus cell transcriptome. Mol Hum Reprod 2010, 16, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Assidi, M.; Richard, F.J.; Sirard, M.A. FSH in vitro versus LH in vivo: similar genomic effects on the cumulus. J Ovarian Res 2013, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- van Gijn, J.; Gijselhart, J.P. [Von Willebrand and his factor]. Ned Tijdschr Geneeskd 2011, 155, A2022. [Google Scholar] [PubMed]
- Zhang, C.H.; Liu, X.Y.; Wang, J. Essential Role of Granulosa Cell Glucose and Lipid Metabolism on Oocytes and the Potential Metabolic Imbalance in Polycystic Ovary Syndrome. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Salimov, D.; Lisovskaya, T.; Otsuki, J.; Gzgzyan, A.; Bogolyubova, I.; Bogolyubov, D. Chromatin Morphology in Human Germinal Vesicle Oocytes and Their Competence to Mature in Stimulated Cycles. Cells 2023, 12, 1976. [Google Scholar] [CrossRef]
- Xie, J.; Xu, X.; Liu, S. Intercellular communication in the cumulus-oocyte complex during folliculogenesis: A review. Front Cell Dev Biol 2023, 11, 1087612. [Google Scholar] [CrossRef]
- Sfakianoudis, K.; Maziotis, E.; Karantzali, E.; Kokkini, G.; Grigoriadis, S.; Pantou, A.; Giannelou, P.; Petroutsou, K.; Markomichali, C.; Fakiridou, M.; et al. Molecular Drivers of Developmental Arrest in the Human Preimplantation Embryo: A Systematic Review and Critical Analysis Leading to Mapping Future Research. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Coticchio, G.; Dal Canto, M.; Mignini Renzini, M.; Guglielmo, M.C.; Brambillasca, F.; Turchi, D.; Novara, P.V.; Fadini, R. Oocyte maturation: gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum Reprod Update 2015, 21, 427–454. [Google Scholar] [CrossRef]
- Nana, M.; Hodson, K.; Lucas, N.; Camporota, L.; Knight, M.; Nelson-Piercy, C. Diagnosis and management of covid-19 in pregnancy. BMJ 2022, 377, e069739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Song, S.; Yang, M.; Yan, L.; Qiao, J. Diminished ovarian reserve causes adverse ART outcomes attributed to effects on oxygen metabolism function in cumulus cells. BMC Genomics 2023, 24, 655. [Google Scholar] [CrossRef]
- Gilchrist, R.B.; Smitz, J. Oocyte in vitro maturation: physiological basis and application to clinical practice. Fertility and sterility 2023, 119, 524–539. [Google Scholar] [CrossRef]
- Ferrari, S.; Lattuada, D.; Paffoni, A.; Brevini, T.A.; Scarduelli, C.; Bolis, G.; Ragni, G.; Gandolfi, F. Procedure for rapid oocyte selection based on quantitative analysis of cumulus cell gene expression. J Assist Reprod Genet 2010, 27, 429–434. [Google Scholar] [CrossRef]
- Maisarah, Y.; Hashida, H.N.; Yusmin, M.Y. The challenge of getting a high quality of RNA from oocyte for gene expression study. Vet Res Forum 2020, 11, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Biase, F.H. Isolation of high-quality total RNA and RNA sequencing of single bovine oocytes. STAR Protoc 2021, 2, 100895. [Google Scholar] [CrossRef]
- Tjahyadi, D.; Susiarno, H.; Chandra, B.A.; Permadi, W.; Djuwantono, T.; Wiweko, B. The effect of oocyte denudation time and intracytoplasmic sperm injection time on embryo quality at assisted reproductive technology clinic - A cross-sectional study. Ann Med Surg (Lond) 2022, 80, 104234. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.C.; Yiap, B.C. DNA, RNA, and protein extraction: the past and the present. J Biomed Biotechnol 2009, 2009, 574398. [Google Scholar] [CrossRef]
- Kaivo-oja, N.; Jeffery, L.A.; Ritvos, O.; Mottershead, D.G. Smad signalling in the ovary. Reprod Biol Endocrinol 2006, 4, 21. [Google Scholar] [CrossRef]
- Maa, Y.-F.; Hsu, C. Liquid-liquid emulsification by rotor/stator homogenization. Journal of Controlled Release 1996, 38, 219–228. [Google Scholar] [CrossRef]
- Iwai, H.; Mori, M.; Tomita, M.; Kono, N.; Arakawa, K. Molecular Evidence of Chemical Disguise by the Socially Parasitic Spiny Ant Polyrhachis lamellidens (Hymenoptera: Formicidae) When Invading a Host Colony. Frontiers in Ecology and Evolution 2022, 10. [Google Scholar] [CrossRef]
- Olson, N.D.; Morrow, J.B. DNA extract characterization process for microbial detection methods development and validation. BMC Res Notes 2012, 5, 668. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Wiweko, B.; Utami, R.; Putri, D.; Muna, N.; Aulia, S.N.; Riayati, O.; Bowolaksono, A. The Quality of RNA Isolation from Frozen Granulosa Cells. Advanced Science Letters 2017, 23, 6717–6719. [Google Scholar] [CrossRef]
- Pavani, K.C.; Baron, E.E.; Faheem, M.; Chaveiro, A.; Da Silva, F.M. Optimisation of Total Rna Extraction from Bovine Oocytes and Embryos for Gene Expression Studies and Effects of Cryoprotectants on Total Rna Extraction. Tsitol Genet 2015, 49, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Moura, B.R.; Gurgel, M.C.; Machado, S.P.; Marques, P.A.; Rolim, J.R.; Lima, M.C.; Salgueiro, L.L. Low concentration of hyaluronidase for oocyte denudation can improve fertilization rates and embryo quality. JBRA Assist Reprod 2017, 21, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, Y.; Takeo, T.; Nakao, S.; Yoshimoto, H.; Hirose, Y.; Sakai, Y.; Horikoshi, Y.; Takeuji, S.; Tsuchiyama, S.; Nakagata, N. Prolonged exposure to hyaluronidase decreases the fertilization and development rates of fresh and cryopreserved mouse oocytes. J Reprod Dev 2014, 60, 454–459. [Google Scholar] [CrossRef]
- Nagyova, E. The Biological Role of Hyaluronan-Rich Oocyte-Cumulus Extracellular Matrix in Female Reproduction. Int J Mol Sci 2018, 19. [Google Scholar] [CrossRef]
- Attanasio, M.; Cirillo, M.; Coccia, M.E.; Castaman, G.; Fatini, C. Factors VIII and Von Willebrand Levels in Women Undergoing Assisted Reproduction: Are Their Levels Associated with Clinical Pregnancy Outcome? Mediterr J Hematol Infect Dis 2020, 12, e2020058. [Google Scholar] [CrossRef] [PubMed]
- Salustri, A.; Camaioni, A.; Tirone, E.; D'Alessandris, C. Hyaluronic acid and proteoglycan accumulation in the cumulus oophorus matrix. Ital J Anat Embryol 1995, 100 Suppl 1, 479–484. [Google Scholar]
- Esbert, M.; Florensa, M.; Riqueros, M.; Teruel, J.; Ballesteros, A. Effect of oocyte denudation timing on clinical outcomes in 1212 oocyte recipients. Fertility and sterility 2013, 100, S482. [Google Scholar] [CrossRef]
- Maldonado Rosas, I.; Anagnostopoulou, C.; Singh, N.; Gugnani, N.; Singh, K.; Desai, D.; Darbandi, M.; Manoharan, M.; Darbandi, S.; Chockalingam, A.; et al. Optimizing embryological aspects of oocyte retrieval, oocyte denudation, and embryo loading for transfer. Panminerva Med 2022, 64, 156–170. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, Y.; Fang, Z.; Hu, S.; Li, Z.; Zhu, L.; Jin, L. Performing ICSI within 4 hours after denudation optimizes clinical outcomes in ICSI cycles. Reprod Biol Endocrinol 2020, 18, 27. [Google Scholar] [CrossRef]
- Nouvel, A.; Laget, J.; Duranton, F.; Leroy, J.; Desmetz, C.; Servais, M.D.; de Preville, N.; Galtier, F.; Nocca, D.; Builles, N.; et al. Optimization of RNA extraction methods from human metabolic tissue samples of the COMET biobank. Sci Rep 2021, 11, 20975. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, S. Mechanical/Physical Methods of Cell Disruption and Tissue Homogenization. Methods Mol Biol 2021, 2261, 563–585. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Attanasio, C.; Pedano, M.S.; Cadenas de Llano-Perula, M. Comparison of human dental tissue RNA extraction methods for RNA sequencing. Arch Oral Biol 2023, 148, 105646. [Google Scholar] [CrossRef]
- Choudhary, S.; Choudhary, R.K. Rapid and Efficient Method of Total RNA Isolation from Milk Fat for Transcriptome Analysis of Mammary Gland. Proceedings of the National Academy of Sciences, India Section B: Biological Sciences 2018, 89, 455–460. [Google Scholar] [CrossRef]
- Ali, N.; Rampazzo, R.C.P.; Costa, A.D.T.; Krieger, M.A. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. Biomed Res Int 2017, 2017, 9306564. [Google Scholar] [CrossRef]
- Amiri Samani, S.; Naji, M.H. Effect of homogenizer pressure and temperature on physicochemical, oxidative stability, viscosity, droplet size, and sensory properties of Sesame vegetable cream. Food Sci Nutr 2019, 7, 899–906. [Google Scholar] [CrossRef]
- Pagani, S.; Maglio, M.; Sicuro, L.; Fini, M.; Giavaresi, G.; Brogini, S. RNA Extraction from Cartilage: Issues, Methods, Tips. Int J Mol Sci 2023, 24, 2120. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nakashima, K.; Maruta, Y.; Kiriyama, T.; Sasaki, M.; Sugiyama, S.; Suzuki, K.; Fujisaki, H.; Sasaki, J.; Kaku-Ushiki, Y.; et al. Improved RNA extraction method using the BioMasher and BioMasher power-plus. J Vet Med Sci 2012, 74, 1561–1567. [Google Scholar] [CrossRef]
- Jones, G.M.; Cram, D.S.; Song, B.; Magli, M.C.; Gianaroli, L.; Lacham-Kaplan, O.; Findlay, J.K.; Jenkin, G.; Trounson, A.O. Gene expression profiling of human oocytes following in vivo or in vitro maturation. Human reproduction (Oxford, England) 2008, 23, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.; Patrizio, P. Gene expression profiling of human oocytes at different maturational stages and after in vitro maturation. Am J Obstet Gynecol 2008, 198, 455–e451. [Google Scholar] [CrossRef]
- Trakunram, K.; Champoochana, N.; Chaniad, P.; Thongsuksai, P.; Raungrut, P. MicroRNA Isolation by Trizol-Based Method and Its Stability in Stored Serum and cDNA Derivatives. Asian Pac J Cancer Prev 2019, 20, 1641–1647. [Google Scholar] [CrossRef]
- Duy, J.; Koehler, J.W.; Honko, A.N.; Minogue, T.D. Optimized microRNA purification from TRIzol-treated plasma. BMC Genomics 2015, 16, 95. [Google Scholar] [CrossRef]
- Beltrame, C.O.; Cortes, M.F.; Bandeira, P.T.; Figueiredo, A.M. Optimization of the RNeasy Mini Kit to obtain high-quality total RNA from sessile cells of Staphylococcus aureus. Braz J Med Biol Res 2015, 48, 1071–1076. [Google Scholar] [CrossRef]
- Tavares, L.; Alves, P.M.; Ferreira, R.B.; Santos, C.N. Comparison of different methods for DNA-free RNA isolation from SK-N-MC neuroblastoma. BMC Res Notes 2011, 4, 3. [Google Scholar] [CrossRef]
- Al-Adsani, A.M.; Barhoush, S.A.; Bastaki, N.K.; Al-Bustan, S.A.; Al-Qattan, K.K. Comparing and Optimizing RNA Extraction from the Pancreas of Diabetic and Healthy Rats for Gene Expression Analyses. Genes (Basel) 2022, 13. [Google Scholar] [CrossRef]
- Ramon-Nunez, L.A.; Martos, L.; Fernandez-Pardo, A.; Oto, J.; Medina, P.; Espana, F.; Navarro, S. Comparison of protocols and RNA carriers for plasma miRNA isolation. Unraveling RNA carrier influence on miRNA isolation. PLoS One 2017, 12, e0187005. [Google Scholar] [CrossRef]
- Wright, K.; de Silva, K.; Purdie, A.C.; Plain, K.M. Comparison of methods for miRNA isolation and quantification from ovine plasma. Sci Rep 2020, 10, 825. [Google Scholar] [CrossRef]
- Ogunbayo, A.E.; Sabiu, S.; Nyaga, M.M. Evaluation of extraction and enrichment methods for recovery of respiratory RNA viruses in a metagenomics approach. J Virol Methods 2023, 314, 114677. [Google Scholar] [CrossRef]
Figure 1.
This is a figure of the study flow.

Figure 2.
This is a figure of oocytes denudation process. A: Cumulus Oocytes Complex prepared for mechanical denudation without enzyme. B: Cumulus Oocytes Complex enzymolyzed in hyaluronidase. C: “Naked” oocytes following enzymolyzed in hyaluronidase – clean edge. D: “Naked” oocytes following mechanical denudation – minimal cumulus cell at the edge.
Figure 2.
This is a figure of oocytes denudation process. A: Cumulus Oocytes Complex prepared for mechanical denudation without enzyme. B: Cumulus Oocytes Complex enzymolyzed in hyaluronidase. C: “Naked” oocytes following enzymolyzed in hyaluronidase – clean edge. D: “Naked” oocytes following mechanical denudation – minimal cumulus cell at the edge.

Figure 3.
Prisma Flow Diagram for Systematic Review.

Figure 4.
The HPRT gene amplification with Melt Peak Curve.

Table 1.
Study Characteristic & Results.
| Parameter | Age (years) |
AMH Level (pmol/L) |
RNA concentration (ng/µl) |
RNA Purification | p- value |
|---|---|---|---|---|---|
| Denudation Technique | |||||
| Recruitment Profile | 37.19 (3.70) | 7.507(4.33) | 6.416(5.15) | 1.712(0.244) | |
| Enzymatic (Hyase) | 36.83 (3.75) | 8.99 (5.35) | 3.83 (1.20) | 1.720 (0.25) | P<0.05* |
| Non-Enzymatic (Non-Hyase) | 37.55 (3.67) | 6.02 (2.17) | 9.00 (6.22) | 1.703 (0.23) | |
| Homogenizer | |||||
| RSH | 37.50 (3.55) | 7.05 (4.05) | 3.68 (1.15) | 1.711(0.23) | P = 0.18* |
| BioMasher III | 36.88 (3.85) | 7.96 (4.59) | 9.15 (6.10) | 1.712 (0.25) | |
| cDNA & qPCR | |||||
| QuantiTect RT | 36.08(3.25) | 7.82(4.12) | 6.82(5.62) | 1.701(0.26) | |
| Whole Genome AK | 38.30 (3.82) | 7.19(4.56) | 6.01(4.68) | 1.722(0.23) | |
| Amplification of HKG | |||||
| Type of cDNA kit | Yes (n, %) |
No (n, %) |
|||
| QuantiTect RT | 20 (50) | 20 (50) | P<0.05# | ||
| Whole Genome AK | 0 | 40 (100) | |||
*Mann–Whitney U test. #Chi-Square test.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Copyright: This open access article is published under a Creative Commons CC BY 4.0 license, which permit the free download, distribution, and reuse, provided that the author and preprint are cited in any reuse.
GDF-9 and BMP-15 mRNA Levels in Canine Cumulus Cells Related to Cumulus Expansion and the Maturation Process
George Ramirez
et al.
Animals,
2020
MDPI Initiatives
Important Links
© 2024 MDPI (Basel, Switzerland) unless otherwise stated







